Abstract
Respiratory syncytial virus (RSV) infects polarized epithelia, which have tightly regulated trafficking because of the separation and maintenance of the apical and basolateral membranes. Previously we established a link between the apical recycling endosome (ARE) and the assembly of RSV. The current studies tested the role of a major ARE-associated protein, Rab11 family interacting protein 2 (FIP2) in the virus life cycle. A dominant-negative form of FIP2 lacking its N-terminal C2 domain reduced the supernatant-associated RSV titer 1,000-fold and also caused the cell-associated virus titer to increase. These data suggested that the FIP2 C2 mutant caused a failure at the final budding step in the virus life cycle. Additionally, truncation of the Rab-binding domain from FIP2 caused its accumulation into mature filamentous virions. RSV budding was independent of the ESCRT machinery, the only well-defined budding mechanism for enveloped RNA viruses. Therefore, RSV uses a virus budding mechanism that is controlled by FIP2.
Keywords: Pneumovirinae, Rab11 protein, virus replication, virus shedding
Respiratory syncytial virus (RSV) is the most common viral cause of serious lower respiratory tract illness in infants and children worldwide. RSV is a negative-sense, single-stranded RNA virus of the Paramyxoviridae family, which encodes 11 proteins. Infection is limited to the most superficial layer of polarized ciliated cells in the respiratory tract epithelium, entering through the apical surface (1, 2). Late steps of the RSV life cycle include assembly and budding of the virus, which also occur at the apical membrane in polarized cells (3).
RSV virions are pleomorphic, with a spherical form varying in size from 150 to 250 nm in diameter. The filamentous form of the virus has a smaller diameter of 50 nm and can have a length from 1 to 10 microns, depending on the cell line in which virus is grown (4). Filamentous RSV has been observed budding and fusing with cells during the spread of RSV infection (5). These long filaments destabilize into spherical forms that can be similar to the size of spherical particles collected during infection (4). Other related viruses also make filamentous virions such as influenza, Ebola, and parainfluenza viruses (6, 7, 8). In polarized cell culture monolayers, influenza virus is predominantly filamentous, and this filamentous form has a higher specific infectivity (9). A previous analysis of the RSV cell-surface filaments in live infected cells showed that these filaments are dynamic, demonstrating rotation, directed motion, and diffusion (10).
The minimal viral protein requirements for budding of RSV virus-like particles (VLPs) are fusion (F), matrix (M), nucleoprotein (N), and phosphoprotein (P) (11). The F protein follows the secretory pathway through the endoplasmic reticulum and Golgi and is directed to the apical membrane through a lipid raft-associated mechanism (12). The other viral structural proteins and the RNA genome must also traffic to the apical membrane from the cytoplasm and viral inclusion bodies through an undetermined mechanism. Lipid rafts have been associated with the assembly of RSV virions, possibly as sites of coordinated assembly, yet the mechanism by which membrane microdomains function during budding remains obscure (12, 13).
Polarized epithelial cells exhibit a complex distribution of proteins that is maintained by a polarized cytoskeleton and a series of discrete endosomal compartments. The apical recycling endosome (ARE) is located just below the apical membrane and is marked by the presence of Rab11a (14). The ARE serves as a slow recycling endosome as well as the final destination for basolateral to apical membrane transcytosing proteins (15). A number of Rab11-interacting proteins have been identified to date including myosin Vb (MVb) and the Rab11 family interacting proteins (Rab11-FIPs) (16, 17). Previously, we have shown that if a dominant-negative form of MVb is present, then RSV does not assemble properly at the apical membrane (18).
In the current study, we analyzed the role of another Rab11 interacting protein, Rab11-FIP2 (FIP2), in the RSV life cycle (Fig. 1A). FIP2 domains include an N-terminal C2 domain that binds preferentially to membrane lipids (19), a central MVb-binding domain (20), three NPF motifs (21), and a C-terminal Rab-binding domain (RBD) that is conserved among all FIPs (16, 22). We now demonstrate that expression of a dominant-negative form of the FIP2 protein lacking the C2 domain (FIP2-ΔC2) in polarized Madin–Darby canine kidney (MDCK) cells blocks RSV budding, causing retention of assembled virus on infected cells. Additionally, inhibition of the previously identified major mechanism of RNA virus budding, the ESCRT machinery, did not affect virus titers. These results suggest that RSV uses an RNA-enveloped virus budding mechanism controlled by FIP2.
Fig. 1.
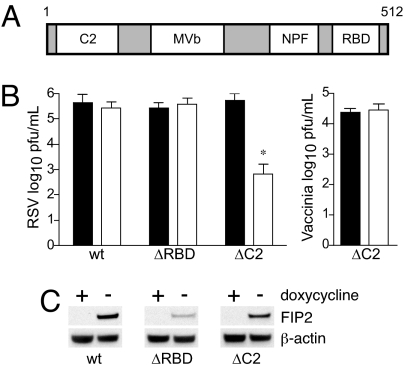
FIP2 facilitates production of supernatant-associated virus. (A) FIP2 contains an N-terminal C2 domain, a MVb-binding domain, and three NPF motifs, followed by a C-terminal Rab11-binding domain. MDCK cells stably transfected with DNA encoding FIP2-GFP variants (FIP2-WT; C2 domain deletion, FIP2-ΔC2; or RBD deletion, FIP2-ΔRBD) under doxycycline repression, expressing (open square) or not expressing (filled square), were infected with RSV or Vaccinia virus. (B) Five days after infection, supernatants were collected, and the virus titer was measured by plaque assay. Representative data from three independent experiments are shown. (C) Relative expression levels for each FIP2 variant in either the presence or absence of doxycycline along with β-actin loading control. *, P = 0.002; Wilcoxon rank sum test. Data shown are mean ±SD.
Results
FIP2 Function Is Necessary for RSV Production.
MDCK cell lines expressing FIP2 variants under doxycycline repression were tested to determine whether dominant-negative forms of FIP2 would alter RSV replication. Expression of the FIP2 variants had no observable effect on cell viability. Tight control of FIP2 variant expression and relative expression levels are shown in Fig. 1C. As expected, expression of FIP2-WT did not affect RSV supernatant titer measured at 5 days after infection (Fig. 1B). Expression of a mutant lacking the RBD (FIP2-ΔRBD) also did not inhibit the RSV supernatant titer. In contrast, a mutant that lacks the C2 domain (FIP2-ΔC2) reduced the RSV titer ≈1,000-fold. None of the three FIP2 variants affected vaccinia virus replication (Fig. 1B and data not shown). There was no reduction in entry or viral RNA production in the FIP2-ΔC2 cells as measured by fluorescent focus or real-time RT-PCR assays (data not shown).
Production of RSV Was Delayed and Reduced.
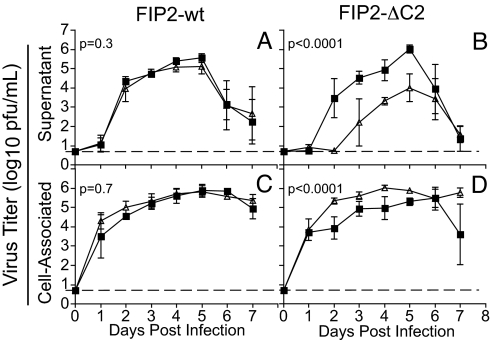
To determine whether the reduction of virus shedding observed at day 5 was an effect of delayed kinetics or reduced magnitude, we performed a 7-day growth curve. Supernatant and cell-associated fractions were collected daily and RSV titer measured by plaque assay. There was no alteration in RSV kinetics in the presence of FIP2-WT (Fig. 2A). In contrast, when FIP2-ΔC2 was expressed, the time to achieve 50% maximal peak titer of budded virus was delayed by 2 days, and the titer was reduced by 10- to >100-fold over the first 5 days (Fig. 2B). Although production of RSV was delayed and reduced, the peak titer still occurred on day 5, followed by a steep decline of RSV titer.
Fig. 2.
FIP2-ΔC2 delays and reduces RSV budding. The MDCK FIP2-WT cell line (A and C) or FIP2-ΔC2 cell line (B and D) was infected with RSV. Viral titer was measured in both expressing (open triangle) and not expressing (filled square) cells over a 7-day period for both supernatant-associated virus (A and B) and cell-associated virus (C and D). Dotted lines indicate the limit of detection. Least-squares regression was used to estimate the best-fitting quadratic curves for viral titer over time, and P values were computed by using likelihood ratio tests comparing these curves between expressing and not expressing cells. Triplicate values were determined; error bars indicate ±SD.
RSV Accumulates on the Cell in the Presence of FIP2-ΔC2.
The titer of RSV in the cell-associated fraction was measured simultaneously with that in the supernatant fraction. There was no alteration in viral kinetics when FIP2-WT was expressed (Fig. 2C). Interestingly, when FIP2-ΔC2 was expressed, there was an increase in cell-associated, viral titer through day 6 (Fig. 2D). This statistically significant accumulation of RSV suggested that the FIP2-ΔC2 block occurs at a late budding step. This phenotype differs dramatically from that observed with the previous MVb-tail experiments in which the cell-associated fractions were decreased in titer ≈10,000-fold (18). Therefore, MVb acts earlier than FIP2 in the assembly and budding process of RSV.
Apical Filaments Are Fully Assembled Virus Structures.
We determined the location of the structural proteins required for budding. The four necessary major structural proteins (F, M, N, and P) were localized to cell-surface filaments in both nonexpressing (data not shown) and FIP2-ΔC2-expressing, RSV-infected cells [supporting information (SI) Fig. S1]. Molecular beacon RNA probes specific for the RSV gene-start consensus sequence were used to identify the viral genomic RNA within the viral filaments. These data suggested that cell-surface filaments in RSV-infected FIP2-ΔC2-expressing cells are assembled, infectious, late-stage viral particles. Additionally, to determine whether these viral structures contained cytoskeletal elements, we stained infected cells for actin, microtubules, and ezrin (Fig. S2). None of these proteins were present in the viral filaments for either nonexpressing or expressing FIP2-ΔC2 cell lines. We repeated the actin localization studies by using visualization of a YFP-actin fusion construct or by using actin-specific antibodies, both of which confirmed the absence of actin from viral filaments (data not shown).
FIP2-ΔC2-Mediated Viral Inhibition Induced a Tethered Late-Domain-Like Phenotype.
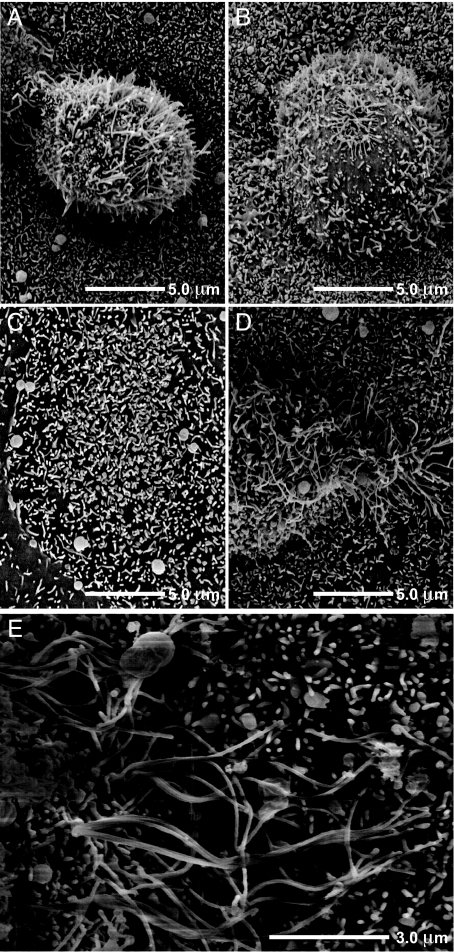
To determine how FIP2-ΔC2 affected the morphology of RSV budding from the apical membrane of polarized cells, we used scanning EM to visualize the viral filaments. Typically the filaments were 1–3 μm in length on the apical surface of infected MDCK cells (Fig. 3A). Microvilli, usually <0.5 μm in length, were observed in surrounding uninfected cells. When the MVb-tail mutant was expressed, we observed a loss of viral filaments from the apical plasma membrane, which was expected because of the known preassembly block (Fig. 3B). Finally, when FIP2-ΔC2 was expressed, we observed a significant elongation of viral filament length to ≈3–10 μm, suggesting that the filamentous virions were unable to undergo the fission process and continued to extend in length (Fig. 3 D and E). This cell morphology is markedly different from that of an uninfected monolayer expressing FIP2-ΔC2 (Fig. 3C).
Fig. 3.
FIP2 facilitates termination of RSV filament growth and virus budding. (A, B, D, and E) Scanning EM was performed to view the length of RSV filaments at the apical surface of MDCK cells expressing FIP2-WT, a dominant negative (A), an MVb-tail construct (B), or FIP2-ΔC2 (D or E) (images from separate cells at two resolutions). (C) Uninfected cells expressing FIP2-ΔC2.
The RSV Budding Mechanism Is Vps4-Independent.
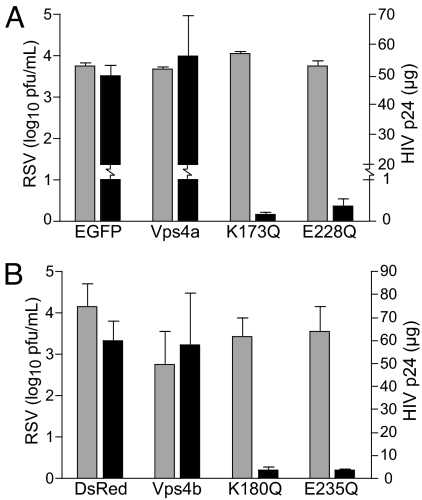
The primary mechanism for budding used by many enveloped RNA viruses is through hijacking the host ESCRT machinery, which is responsible for multivesicular body formation (23). Using WT Vps4a or each of the two dominant-negative Vps4a mutants K173Q or E228Q transfected into 293T cells, we tested whether RSV used this machinery for assembly and budding. No reduction of RSV titer was observed in the presence of either WT or dominant-negative Vps4a mutants (Fig. 4A). In contrast, a HIV pseudotyped with vesicular stomatitis virus (VSV) G protein (HIVNL4-3), exhibited >90% reduced supernatant p24 levels in the presence of the dominant-negative Vps4a constructs, as expected (Fig. 4A). Experiments with similar Vps4b constructs also yielded the same results (Fig. 4B). The ubiquitination cycle is important for many viruses that use late domains (L-domains). However, three separate proteasome inhibitors did not affect budded RSV titers (Fig. S3D), again suggesting that RSV assembly and budding uses a mechanism independent of the ESCRT machinery.
Fig. 4.
RSV uses a Vps4-independent budding mechanism. Cells (293T) transfected with Vps4a constructs: pEGFP vector (EGFP) or Vps4a-EGFP (Vps4a), or dominant-negative mutants Vps4a-K173Q-EGFP (K173Q) or Vps4a-E228Q-EGFP (E228Q) (A), or Vps4b constructs: pDsRed vector (DsRed) or Vps4b-DsRed (Vps4b), or dominant-negative mutants Vps4b-K180Q-DsRed (K180Q) or Vps4a-E235Q-DsRed (E235Q) (B). Cells were infected 24 h after transfection at a moi of 1.0 with either WT RSV or HIVNL4−3. Supernatant virus was collected 48 h after infection and assayed for either RSV by viral titer (gray bars) or HIV p24 by ELISA (black bars).
Addition of Strong L-Domains to RSV M Did Not Render RSV Sensitive to Vps4a.
To determine whether RSV possesses a cryptic or weak L-domain, we introduced the L-domain from the paramyxovirus PIV5 into the RSV M protein. A second mutant construct was made by adding the first 13 aa of the Ebola VP40 protein (containing a functional L-domain) to the N terminus of RSV M protein (Fig. S3A). These proteins were expressed in cells during RSV infection. Neither of the variant M proteins affected the magnitude of budded or cell-associated virus (Fig. S3B). Additionally, coexpression of the M protein variants in infected cells with WT Vps4a or either of the two dominant-negative Vps4a mutants did not affect RSV titer of supernatant or cell-associated virus (Fig. S3C and data not shown, respectively). These data further indicated that the FIP2-ΔC2 block of RSV budding is independent of the ESCRT machinery.
FIP2 Variants Localize with Viral Structures.
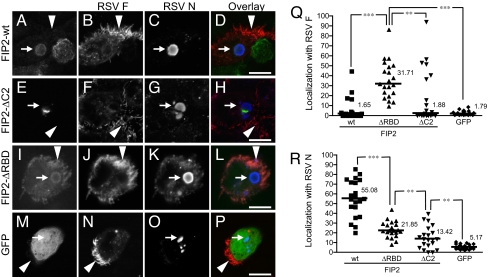
Next we determined whether FIP2 localized with viral structures. Infected cell monolayers expressing FIP2-WT, FIP2-ΔC2, or FIP2-ΔRBD were stained for viral filaments (marked by RSV F protein) and viral inclusions (marked by RSV N protein). FIP2-WT showed 55% colocalization with viral inclusion bodies and little localization with viral filaments (Fig. 5). FIP2-ΔC2-containing tubulovesicular membranes appeared to be docked with viral inclusions, and colocalization was reduced to 13%. FIP2-ΔRBD also demonstrated colocalization with viral inclusion bodies (21% greater than FIP2-ΔC2, P = 0.009), but removal of the RBD caused a relocalization to the viral filaments (31%, P < 0.0001). Finally, when MDCK cells were infected with recombinant RSV, which expresses GFP in the cytoplasm, GFP was excluded from viral filaments (Fig. 5). These data suggest that truncation of the RBD blocks the cycling of FIP2 away from the apical membrane, causing it to assemble into RSV filaments.
Fig. 5.
FIP2 associates with both viral inclusions and viral filaments. (A–P) MDCK cells expressing FIP2-GFP variants [FIP2-WT (A–D), FIP2-ΔC2 (E–H), or FIP2-ΔRBD (I–L)] infected with RSV or MDCK cells infected with recombinant RSV expressing GFP (M–P) were stained by indirect immunofluorescence for both RSV F (B, F, J, and N) and RSV N (C, G, K, and O). In the overlays (D, H, L, and P), FIP2-GFP variants and GFP are pseudocolored green, RSV F is pseudocolored red, and RSV N is pseudocolored blue. The arrows with tails indicate viral inclusion body location, whereas the arrows without tails indicate an area of dense viral filament formation. (Q and R) Percentage of the total integrated pixel intensity indicating colocalization of FIP2-GFP variants or GFP with RSV F (Q) or RSV N (R) were calculated. The median value is shown. ***, P < 0.0001; **, P ≤ 0.009; Wilcoxon rank sum test. No significant difference was measured among WT, ΔC2, and GFP groups in graph Q. Additional comparisons on graph R demonstrate that the WT differs significantly from ΔC2 and GFP (both P < 0.0001); ΔRBD differed significantly from GFP (P < 0.0001).
Discussion
In this study we sought to define the role of the ARE in late steps of the RSV life cycle. Using a panel of FIP2 mutant-expressing MDCK cells, we determined that FIP2 is required at a late stage of RSV budding, most likely fission. Our studies show RSV does not use a Vps4-dependent virus budding mechanism, indicating that FIP2 acts in a previously uncharacterized pathway for enveloped virus budding.
It has been shown that RSV is released from the apical surface in polarized cells, and the minimal requirements for virus-like particle formation are the RSV F, M, N, and P proteins (11). We observed all four proteins and the RSV genome in the viral filaments. In addition, when these viral proteins were cotransfected, we observed predominantly filamentous structures at the surface of cells (data not shown). Studies testing the effect of inhibitors of RSV have shown that loss of supernatant virus titer coincides with loss of surface filaments (24, 25). Not only does this study show that FIP2 controls fission of surface filaments, but the data also suggest that these filaments are virion particles.
It has been speculated that the filamentous structures at the surface of RSV-infected cells are actin-based structures (26, 27), yet our studies show that neither actin nor apical microvilli-associated proteins are present in the filaments. These findings do not eliminate the possibility that actin coordinates the assembly of virus at the base of filaments. In fact, some viral filaments appeared to be extensions of polymerized actin structures. High-resolution EM studies have recently suggested that the cortical actin network may be linked to the base of these viral filaments (28). Additionally, the lack of free diffusion of cytoplasmic GFP into filaments indicated that these viral filaments are specifically and tightly packaged, not passively filled tubular structures.
The only previously well delineated mechanism responsible for negative-sense RNA virus budding is the hijacking of the ESCRT machinery. The ESCRT mechanism of viral trafficking and virion formation is governed by viral proteins containing L-domains. RSV proteins do not contain any identifiable L-domains. Budding through the use of such L-domains depends on the AAA-ATPase Vps4. When dominant-negative forms of Vps4 are expressed in cells infected with viruses containing L-domains, virions appear to be tethered to the membrane through a thin stalk because of a loss of fission (23, 29). We studied RSV infection in cells expressing the Vps4a and Vps4b dominant-negative proteins and found there was no reduction in virus budding.
Unlike many viruses, infectious RSV virions are predominantly cell-associated in culture. This observation suggests that either RSV contains an inefficient L-domain or uses an undescribed budding mechanism that is less efficient. Therefore, we engineered recombinant forms of the RSV M protein that contained either the Ebola virus VP40 L-domain or the L-domain of the related paramyxovirus PIV5 (30, 31). Although L-domains are able to act in a location-independent manner, we inserted the sequences at positions similar to other viruses (32). Overexpression of these altered M proteins in cells during WT RSV infection did not increase budding or render RSV sensitive to the dominant-negative Vps4a proteins, suggesting that an inefficient or occult RSV L-domain is not a determining factor in the RSV budding pathway. A caveat of this interpretation is that WT M protein expressed by the virus was also present in cells. Although not entirely comparable, it has been shown that insertion of the Ebola virus L-domain into VSV M protein altered VLP budding (33).
Many viruses require ubiquitination for entry into the ESCRT pathway, although this correlation does not exist for all viruses. VSV is sensitive to proteasome inhibitors but does not appear to be sensitive to the dominant-negative Vps4a protein (33). Equine infectious anemia virus contains a Vps4a-dependent L-domain but is not sensitive to proteasome inhibitors (34). In our studies, RSV budding was not inhibited by proteasome inhibitors, suggesting that the FIP2-associated budding machinery belongs to a third category of budding mechanisms that is both Vps4-independent and ubiquitination-independent. Recent studies have demonstrated that both human cytomegalovirus and Semliki Forest virus budding also are independent of Vps4 and ubiquitination (35, 36). It will be interesting to determine whether the FIP2 mechanism, like ESCRT, is a common mechanism used by other enveloped viruses.
Analysis to determine whether FIP2 associated with viral structures demonstrated a significant localization with viral inclusion bodies yet little apparent localization in viral filaments. This finding may be due to a low concentration required for function at the base of the filaments or a fast recycling rate. The dynamics of FIP2 recycling are demonstrated by the wide spread of values for localization of FIP2-WT with viral inclusion bodies. Further analysis of the FIP2 truncation mutants demonstrated that removal of the C2 domain blocked colocalization, yet viral inclusions appear to be docked to FIP2-ΔC2-containing vesicles. FIP2-ΔC2 expression blocks transcytosis from the basal to apical membrane at a transitional step between the common endosome and ARE (37). Together these findings indicate that the association of FIP2 with the viral inclusion body depends on endosomal transcytotic trafficking events through the ARE that occur after passage through the common endosome.
The other FIP2 truncation mutant, FIP2-ΔRBD, localized with both cytoplasmic viral inclusion bodies and apical viral filaments. These data suggest that the RBD is required for the recycling of FIP2 from the apical membrane back to the ARE and the viral inclusion body. Thus the dissociation of FIP2-ΔRBD from Rab11-dependent recycling allowed FIP2-ΔRBD to assemble into viral filaments. The RBD of FIP2 is required not only for binding Rab11 but also for homodimerization (21). The lack of FIP2-ΔRBD inhibition of RSV budding indicates that FIP2 may have functions independent of dimerization and Rab11 binding (38).
EM analysis of viral inclusion bodies suggest that they are not membrane-bound structures (39). Removal of the RBD also blocks the Rab-dependent association with membranes at the ARE through Rab myristoylation, which causes a cytoplasmic diffuse localization of FIP2-ΔRBD in the absence of infection (21). The localization of FIP2-ΔRBD and FIP2-WT to viral inclusion bodies suggests that FIP2 may also function in trafficking nonmembrane-bound cargo.
If this budding mechanism proves to be as complex as the ESCRT mechanism, FIP2 may be only one host protein from a large group of host proteins functioning together to facilitate virus budding. Current proteomic analysis of ARE vesicles is targeted at discovering this remaining host machinery involved in late steps of the RSV assembly and budding pathway. It is tempting to speculate that the mechanism hijacked by RSV may be an ARE-regulated mechanism for cellular deflagellation or shedding of microvilli because the N complex in filamentous virions mimics the periodicity of actin in microvilli or tubulin in cilia (4, 40).
Materials and Methods
Mammalian Cell Lines and Virus.
MDCK T23 cell lines (Clontech) expressing tetracycline-repressible WT or mutant human EGFP-FIP2 were constructed as described (37). Suppression or induction of FIP2-GFP F protein expression was regulated by the presence or absence of 20 ng/ml doxycycline. MDCK and HEp-2 cell lines were maintained as described (18). The 293T cells were grown in DMEM and Ham's F-12 medium supplemented with 10% FBS, 320 μg/ml l-glutamine, 1% (vol/vol) nonessential amino acids, 2.7 μg/ml amphotericin B, and 45 μg/ml gentamicin. All cell lines were maintained at 37°C in 5% CO2. The RSV WT strain A2 (41), recombinant RSV-GFP (42), VSV-G-pseudotyped HIVNL4-3 (43), and vaccinia virus strain Western Reserve have been described (18).
Viral Infection Assay.
Cells were seeded in triplicate onto 48-well tissue culture plates (Costar) at 20% confluence and allowed to form fully-confluent cell monolayer cultures in the presence or absence of doxycycline to control expression of GFP-FIP2 variant proteins. Once confluent, the cell culture monolayers were inoculated with RSV at a multiplicity of infection (moi) of 0.25 plaque-forming units per cell. Virus was allowed to adsorb for 1 h at 37°C. After adsorption, virus infection medium was removed, tissue culture wells were washed three times with PBS, and 300 μl of growth medium (with or without doxycycline) was added. Infected cells were incubated at 37°C in 5% CO2.
Virus Recovery and Titration.
Tissue culture supernatant containing virus was collected and brought to a volume of 300 μl. After the collection of the supernatant fraction, tissue culture wells were washed three times with 250 μl of PBS. Cell-associated virus was harvested by scraping MDCK cell culture monolayers into 150 μl of medium, followed by an additional washing of the well with 150 μl of medium. Virus was released from harvested cells by using rapid freeze–thaw with movement between an ethanol dry-ice bath and a 37°C water bath, which was repeated three times. Recovered RSV was quantified by plaque assay as described (44). HIV p24 assay was performed as described (43). Vaccinia virus titer was quantified by plaque assay on BSC-40 cell monolayers, followed by staining with crystal violet (18).
Transfection.
Forty micrograms of DNA was added to 2.5 μM calcium chloride, followed by dropwise addition of 2× BBS. This solution was incubated for 25 min and then added dropwise to 293T cells plated at 40% confluency in 10-cm dishes, followed by 24 h of incubation at 35°C in 3% CO2. Transfected cells were washed with PBS and fresh DMEM and Hanks' F-12 medium was added. Transfection efficiency was assessed by visual inspection using microscopy with epifluorescence. Cells were inoculated with RSV at a moi of 5 on day 3 after transfection. RSV M protein variants were transfected into HEp-2 cells by using Effectene (Qiagen) with or without Vps4a constructs and then infected with RSV at an moi of 0.25 at 24 h after transfection.
Scanning EM.
MDCK cells (with or without doxycycline) were seeded on 12-mm cover slips in 24-well plates. Cells were infected with RSV at a moi of 0.25 as described above. Samples were fixed 24 h after infection in 4% glutaraldehyde (Sigma) in 0.1 M cacodylate buffer (Sigma), treated with 1% osmium (Sigma), and dehydrated through a series of 70–100% ethanol washes. Dehydrated cells were critical-point dried, sputter-coated with gold, and visualized by using a Hitachi S4200 scanning electron microscope.
Fluorescence Assays.
A molecular beacon molecule specific for the RSV genome at 1 mM was delivered by using transient permeablization with streptolysin O of live infected cells as described (10). Cells were fixed by using 3.7% formaldehyde (Sigma) in PBS, followed by 3.7% formaldehyde with 0.3% Triton X-100 (Sigma) in PBS. Cells were blocked by using 3% BSA (Sigma) and then stained with a primary antibody against F (palivizumab; MedImmune), M (clone B135), N (clone B130), P (clone 4.14.4), ezrin (Sigma), or α-tubulin (AbCam) in block. Cells were washed, and then antibodies were conjugated with Alexa Fluor 568 or 647 and/or phalloidin 647 (Invitrogen) in block were added. Slides were mounted by using ProLong Antifade (Invitrogen). Images were acquired on a Zeiss LSM 510 Meta confocal microscope. Analysis of colocalization was performed by using MetaMorph 7.1 software (Molecular Devices) on 21 fields from three separate assays. Western blot analyses were performed by using NuPAGE reagents (Invitrogen) following the manufacturer's instructions. Primary antibodies were against GFP (Clontech) or β-actin (AbCam). Secondary antibody was goat anti-mouse conjugated to HRP (KPL), followed by visualization with Chemiluminescent Substrate (Pierce).
Proteasome Inhibitor Assays.
MG-132, lactacystin, and epoxomicin (EMD) were dissolved in DMSO (Sigma) and used within 2 weeks at a final concentration of 1.0 μM. HEp-2 cells were infected with either HIVNL4-3 or RSV at a moi of 2.0. Fresh media with inhibitor or DMSO vehicle were added 24 h after infection. Virus was collected 48 h after infection.
PCR Mutagenesis of RSV M Protein.
A cDNA encoding the RSV strain A2 M protein was sequence-optimized for mammalian expression and synthesized by GENEART (designated Mopt). L-domain sequences from Ebola virus VP40 protein (amino acids PTAPPEY) or PIV5 M protein (amino acids FPIV) were designed for integration into, or as an addition to, the M sequence. PCR extension was performed to generate the chimeric RSV M proteins, designated RSV Mopt-EbV or Mopt-PIV5; primers are available upon request. All constructs were cloned into the Invitrogen plasmid vector pcDNA3.1(+) with restriction sites 5′-BamHI-EcoRI-3′.
Supplementary Material
Acknowledgments.
We thank Sarah Sewell for assistance with the SEM and Natalie Thornburg for assistance with Vaccinia experiments. Confocal microscopy and SEM experiments were performed in part through the use of the Vanderbilt University Medical Center Cell Imaging Shared Resource. We thank Wes Sundquist (Department of Biochemistry, University of Utah, Salt Lake City) for the Vps4 DNA constructs and Peter Collins (National Institute of Allergy and Infectious Disease, National Institutes of Health, Bethesda, MD) and Mark Peeples (Center for Vaccines and Immunity, The Research Institute at Nationwide Children's Hospital, Columbus, OH) for RSV-GFP. J.E.C. holds a Clinical Scientist Award in Translation Research from the Burroughs Wellcome Fund. This work was supported by National Institutes of Health (NIH) Training Grant T32 GM08554 (to T.J.U.) and NIH Grant DK48370 (to J.R.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0712144105/DCSupplemental.
References
- 1.Wright PF, et al. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J Virol. 2005;79:8651–8654. doi: 10.1128/JVI.79.13.8651-8654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L, Peeples ME, Boucher RC, Collins PL, Pickles RJ. Respiratory syncytial virus infection of human airway epithelial cells is polarized, specific to ciliated cells, and without obvious cytopathology. J Virol. 2002;76:5654–5666. doi: 10.1128/JVI.76.11.5654-5666.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts SR, Compans RW, Wertz GW. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J Virol. 1995;69:2667–2673. doi: 10.1128/jvi.69.4.2667-2673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachi T, Howe C. Morphogenesis and ultrastructure of respiratory syncytial virus. J Virol. 1973;12:1173–1180. doi: 10.1128/jvi.12.5.1173-1180.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bachi T. Direct observation of the budding and fusion of an enveloped virus by video microscopy of viable cells. J Cell Biol. 1988;107:1689–1695. doi: 10.1083/jcb.107.5.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellis DS, et al. Ultrastructure of Ebola virus particles in human liver. J Clin Pathol. 1978;31:201–208. doi: 10.1136/jcp.31.3.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosley VM, Wyckoff RWG. Electron micrography of the virus of influenza. Nature. 1946;157:263. doi: 10.1038/157263a0. [DOI] [PubMed] [Google Scholar]
- 8.Yao Q, Compans RW. Filamentous particle formation by human parainfluenza virus type 2. J Gen Virol. 2000;81:1305–1312. doi: 10.1099/0022-1317-81-5-1305. [DOI] [PubMed] [Google Scholar]
- 9.Ada GL, Perry BT, Edney M. Infectivity of influenza virus filaments. Nature. 1957;180:1134. doi: 10.1038/1801134a0. [DOI] [PubMed] [Google Scholar]
- 10.Santangelo PJ, Bao G. Dynamics of filamentous viral RNPs prior to egress. Nucleic Acids Res. 2007;35:3602–3611. doi: 10.1093/nar/gkm246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teng MN, Collins PL. Identification of the respiratory syncytial virus proteins required for formation and passage of helper-dependent infectious particles. J Virol. 1998;72:5707–5716. doi: 10.1128/jvi.72.7.5707-5716.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown G, et al. Analysis of the interaction between respiratory syncytial virus and lipid-rafts in HEp2 cells during infection. Virology. 2004;327:175–185. doi: 10.1016/j.virol.2004.06.038. [DOI] [PubMed] [Google Scholar]
- 13.McCurdy LH, Graham BS. Role of plasma membrane lipid microdomains in respiratory syncytial virus filament formation. J Virol. 2003;77:1747–1756. doi: 10.1128/JVI.77.3.1747-1756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casanova JE, et al. Association of Rab25 and Rab11a with the apical recycling system of polarized Madin–Darby canine kidney cells. Mol Biol Cell. 1999;10:47–61. doi: 10.1091/mbc.10.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Apodaca G, Katz LA, Mostov KE. Receptor-mediated transcytosis of IgA in MDCK cells is via apical recycling endosomes. J Cell Biol. 1994;125:67–86. doi: 10.1083/jcb.125.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hales CM, et al. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- 17.Lapierre LA, et al. Myosin vb is associated with plasma membrane recycling systems. Mol Biol Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brock SC, Goldenring JR, Crowe JE., Jr Apical recycling systems regulate directional budding of respiratory syncytial virus from polarized epithelial cells. Proc Natl Acad Sci USA. 2003;100:15143–15148. doi: 10.1073/pnas.2434327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay AJ, McCaffrey MW. The C2 domains of the class I Rab11 family of interacting proteins target recycling vesicles to the plasma membrane. J Cell Sci. 2004;117:4365–4375. doi: 10.1242/jcs.01280. [DOI] [PubMed] [Google Scholar]
- 20.Hales CM, Vaerman JP, Goldenring JR. Rab11 family interacting protein 2 associates with Myosin Vb and regulates plasma membrane recycling. J Biol Chem. 2002;277:50415–50421. doi: 10.1074/jbc.M209270200. [DOI] [PubMed] [Google Scholar]
- 21.Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 22.Meyers JM, Prekeris R. Formation of mutually exclusive Rab11 complexes with members of the family of Rab11-interacting proteins regulates Rab11 endocytic targeting and function. J Biol Chem. 2002;277:49003–49010. doi: 10.1074/jbc.M205728200. [DOI] [PubMed] [Google Scholar]
- 23.Garrus JE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 24.Gower TL, et al. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J Virol. 2005;79:5326–5336. doi: 10.1128/JVI.79.9.5326-5336.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oomens AG, Bevis KP, Wertz GW. The cytoplasmic tail of the human respiratory syncytial virus F protein plays critical roles in cellular localization of the F protein and infectious progeny production. J Virol. 2006;80:10465–10477. doi: 10.1128/JVI.01439-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernie BF, Gerin JL. Immunochemical identification of viral and nonviral proteins of the respiratory syncytial virus virion. Infect Immun. 1982;37:243–249. doi: 10.1128/iai.37.1.243-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulloa L, Serra R, Asenjo A, Villanueva N. Interactions between cellular actin and human respiratory syncytial virus (HRSV) Virus Res. 1998;53:13–25. doi: 10.1016/s0168-1702(97)00121-4. [DOI] [PubMed] [Google Scholar]
- 28.Jeffree CE, et al. Ultrastructural analysis of the interaction between F-actin and respiratory syncytial virus during virus assembly. Virology. 2007;369:309–323. doi: 10.1016/j.virol.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Mebatsion T, Weiland F, Conzelmann KK. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol. 1999;73:242–250. doi: 10.1128/jvi.73.1.242-250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Licata JM, et al. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: Involvement of host proteins TSG101 and VPS-4. J Virol. 2003;77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79:2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74:7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irie T, Licata JM, McGettigan JP, Schnell MJ, Harty RN. Budding of PPxY-containing rhabdoviruses is not dependent on host proteins TGS101 and VPS4A. J Virol. 2004;78:2657–2665. doi: 10.1128/JVI.78.6.2657-2665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patnaik A, Chau V, Li F, Montelaro RC, Wills JW. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J Virol. 2002;76:2641–2647. doi: 10.1128/JVI.76.6.2641-2647.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraile-Ramos A, et al. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol. 2007;9:2955–2967. doi: 10.1111/j.1462-5822.2007.01024.x. [DOI] [PubMed] [Google Scholar]
- 36.Taylor GM, Hanson PI, Kielian M. Ubiquitin depletion and dominant-negative VPS4 inhibit rhabdovirus budding without affecting alphavirus budding. J Virol. 2007;81:13631–13639. doi: 10.1128/JVI.01688-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducharme NA, et al. Rab11-FIP2 regulates differentiable steps in transcytosis. Am J Physiol Cell Physiol. 2007;293:C1059–C1072. doi: 10.1152/ajpcell.00078.2007. [DOI] [PubMed] [Google Scholar]
- 38.Ducharme NA, et al. MARK2/EMK1/Par-1B alpha phosphorylation of Rab11-family interacting protein 2 is necessary for the timely establishment of polarity in Madin–Darby canine kidney cells. Mol Biol Cell. 2006;17:3625–3637. doi: 10.1091/mbc.E05-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: Formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology. 1993;195:243–247. doi: 10.1006/viro.1993.1366. [DOI] [PubMed] [Google Scholar]
- 40.Norrby E, Marusyk H, Orvell C. Morphogenesis of respiratory syncytial virus in a green monkey kidney cell line (Vero) J Virol. 1970;6:237–242. doi: 10.1128/jvi.6.2.237-242.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coates HV, Alling DW, Chanock RM. An antigenic analysis of respiratory syncytial virus isolates by a plaque reduction neutralization test. Am J Epidemiol. 1966;83:299–313. doi: 10.1093/oxfordjournals.aje.a120586. [DOI] [PubMed] [Google Scholar]
- 42.Hallak LK, Spillmann D, Collins PL, Peeples ME. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J Virol. 2000;74:10508–10513. doi: 10.1128/jvi.74.22.10508-10513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Varthakavi V, et al. The pericentriolar recycling endosome plays a key role in Vpu-mediated enhancement of HIV-1 particle release. Traffic. 2006;7:298–307. doi: 10.1111/j.1600-0854.2005.00380.x. [DOI] [PubMed] [Google Scholar]
- 44.Murphy BR, Sotnikov AV, Lawrence LA, Banks SM, Prince GA. Enhanced pulmonary histopathology is observed in cotton rats immunized with formalin-inactivated respiratory syncytial virus (RSV) or purified F glycoprotein and challenged with RSV 3–6 months after immunization. Vaccine. 1990;8:497–502. doi: 10.1016/0264-410x(90)90253-i. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.