Abstract
Parkinson's disease (PD) is a major neurodegenerative condition with several rare Mendelian forms. Oxidative stress and mitochondrial function have been implicated in the pathogenesis of PD but the molecular mechanisms involved in the degeneration of neurons remain unclear. DJ-1 mutations are one cause of recessive parkinsonism, but this gene is also reported to be involved in cancer by promoting Ras signaling and suppressing PTEN-induced apoptosis. The specific function of DJ-1 is unknown, although it is responsive to oxidative stress and may play a role in the maintenance of mitochondria. Here, we show, using four independent methods, that DJ-1 associates with RNA targets in cells and the brain, including mitochondrial genes, genes involved in glutathione metabolism, and members of the PTEN/PI3K cascade. Pathogenic recessive mutants are deficient in this activity. We show that DJ-1 is sufficient for RNA binding at nanomolar concentrations. Further, we show that DJ-1 binds RNA but dissociates after oxidative stress. These data implicate a single mechanism for the pleiotropic effects of DJ-1 in different model systems, namely that the protein binds multiple RNA targets in an oxidation-dependent manner.
Keywords: gene expression, oxidative stress, Parkinson's disease, translation
Mutations in any of three genes cause a recessively inherited early-onset movement disorder reminiscent of Parkinson's disease (PD). Parkin is an E3 protein-ubiquitin ligase and PINK1 is a mitochondrial kinase (1). Results from Drosophila models suggest that PINK1 and parkin define a single pathway that, when disrupted, leads to mitochondrial damage (2, 3). The third, and rarest, gene for recessive parkinsonism is DJ-1. The DJ-1 protein responds to oxidative stress evidenced by a pI shift in sporadic PD (4, 5) and in cell (6, 7) and animal (8) models. DJ-1 knockout models also show increased sensitivity to toxins that cause mitochondrial dysfunction or oxidative stress (9–13). Cys-106 of DJ-1, which is oxidized to form a cysteine-sulfinic acid, is critically required for DJ-1 to protect against these types of damage both in vitro (6) and in vivo (12). However, the molecular function of DJ-1 is unclear. DJ-1 is part of the ThiJ/PfPI superfamily but the proteins most similar to DJ-1 form a distinct clade away from other members with known function (14, 15), implying a novel activity. As well as effects on oxidation and mitochondrial function, DJ-1 enhances Ras-mediated oncogenesis (16), modulates the PTEN/Akt survival pathway (17, 18), suppresses Ask1-mediated apoptosis (19), and increases glutathione (GSH) synthesis, Hsp70 (20) and tyrosine hydroxylase (21, 22) expression. DJ-1 is a small, dimeric, single-domain protein (23), so if all of these effects are true then either the protein has multiple functions or there is a single biochemical activity that explains all of them.
One of the original descriptions of the cloning of DJ-1 was as RS, a regulatory component of an RNA binding complex (24). Therefore, we examined whether DJ-1 associates with RNA in vitro and in vivo and looked at the consequences of any interaction. We propose that the apparently pleiotropic effects of DJ-1 relate to the common property of RNA binding.
Results
Purification of mRNA Associated with DJ-1 Protein.
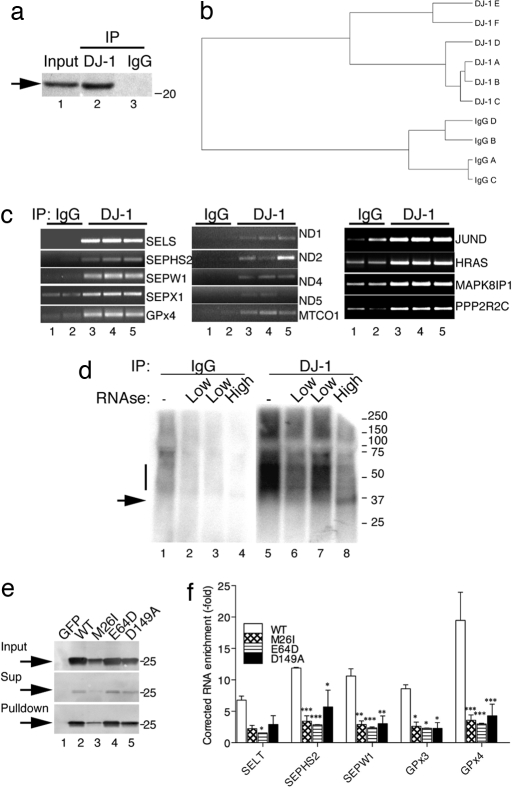
We used immunoprecipitation (IP; Fig. 1a) to isolate endogenous DJ-1 from cultured human dopaminergic neuroblastoma cells (25). The antibody we used was directed to the C-terminal region of DJ-1 that is solvent-accessible and was validated against knockdown cell lines and knockout mouse brains [supporting information (SI) Fig. S1]. We then isolated any associated mRNA and amplified and hybridized this material to microarrays to generate candidate mRNA lists (Table S1; also available at GEO accession no. GSE8632). Using six DJ-1 IP samples and four control IgG IP samples from multiple cell preparations, we identified transcripts significantly enriched in the DJ-1 IP sample that were reliably detected above background (P < 0.001 for detection). Clustering indicated a clear separation of the two groups across all experiments (Fig. 1b). These results suggest that DJ-1 associates with specific mRNA targets.
Fig. 1.
Endogenous DJ-1 binds RNA. (a) DJ-1 was isolated by IP from M17 cells. (b) Associated RNA was isolated, amplified, and hybridized to arrays. Cluster analysis of replicate samples shows a strict separation of DJ-1 and control IPs. (c) Using two control (lanes 1 and 2) and three DJ-1 (lanes 3–5) IP samples, we performed RT-PCR for selenoproteins, mitochondrial transcripts, and components of the PTEN/Akt pathway; gene symbols are listed on the right of each gel. (d) Living M17 cells were subjected to UV cross-linking and immunoprecipitated with a nonspecific IgG (lanes 1–4) or anti-DJ-1 (lanes 5–8). RNA was radiolabeled, showing a smear of label in the DJ-1 IP samples (lane 5, bar on the left of the autoradiogram). A low concentration of RNase (lanes 2, 3, 6, and 7) did not remove label, but a high concentration (lane 8) resolved the DJ-1 samples to a single band of ≈35 kDa (lane 8, arrow). Markers on the right of the autoradiogram are in kDa. (e) M17 cells were transfected with WT (lane 2) or mutant (lanes 3–5) DJ-1 constructs containing a C-terminal V5-His tag. A vector containing GFP was used as a control (lane 1). DJ-1 was purified by using NiNTA beads and subjected to Western blotting with an anti-V5 antibody. (f) We performed qRT-PCR for selenoprotein genes, expressing these results as enrichment relative to GFP. As M26I is partially unstable, the amount of DJ-1 protein in the input and IP is lower, and thus all values were also corrected to amount of DJ-1 in the IP. Values are mean +/− SEM, n = 3–4 replicates per construct. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using one-way ANOVA for each gene with Bonferroni post hoc tests to compare mutants with WT DJ-1.
The transcripts that are associated with DJ-1 are potentially involved in many cellular processes. We chose to examine three groupings of transcripts associated with DJ-1 that might be relevant to the survival of dopaminergic neurons: proteins that are involved in the metabolism of selenocysteine and enzymes that contain selenocysteine, particularly the GSH peroxidases; mitochondrial transcripts, both nuclear and mitochondrially encoded; and components of the PTEN/Akt survival pathway (Tables S2–S4). We validated these interactions by RT-PCR and saw preferential amplification of a series of candidate transcripts (Fig. 1c). We used cross-linked immunoprecipitation (CLIP) (26) as a second independent technique. UV-cross-linked complexes containing DJ-1, and associated RNA were radiolabeled, revealing smear of RNA complexes that were resolved to a single, lower molecular weight band after RNase digestion (Fig. 1d). The apparent mobility of the UV-cross-linked complexes on SDS/PAGE is consistent with a monomer of DJ-1 (22 kDa) plus short RNA tags of 20–50 nt or 7–16 kDa (26). The DJ-1 dimer is dissociated by SDS treatment, and we infer the 35-kDa bands represent individual monomers with cross-linked radiolabeled RNA attached. We cloned a group of short RNA sequences from this material to identify the protected RNA site (see below).
We used a third, nonantibody-based technique and purified 6His-tagged DJ-1 from cells by using Ni-NTA affinity chromatography (Fig. 1e). Pathogenic DJ-1 mutants showed lower binding to mRNA compared with WT protein after correcting all RT-PCR results to the amount of DJ-1 protein in the IP (Fig. 1f). Our estimates of fold enrichment are higher with this approach (6- to 20-fold) than those predicted with arrays (2- to 12-fold over nonspecific IgG), suggesting that the array approach underestimates strength of interaction of DJ-1 with mRNA. The purification using 6His-tagged protein suggests that the observed association does not depend on a specific antibody and is unlikely to represent cross-reactivity.
To address whether such interactions might occur in the brain, we isolated complexes by IP with DJ-1 antibody from WT and DJ-1 knockout (27) mouse brains (Fig. 2a). We validated the interaction of DJ-1 with representative transcripts by using quantitative RT-PCR (Fig. 2b). These observations show that DJ-1 can bind mRNA in vivo.
Fig. 2.
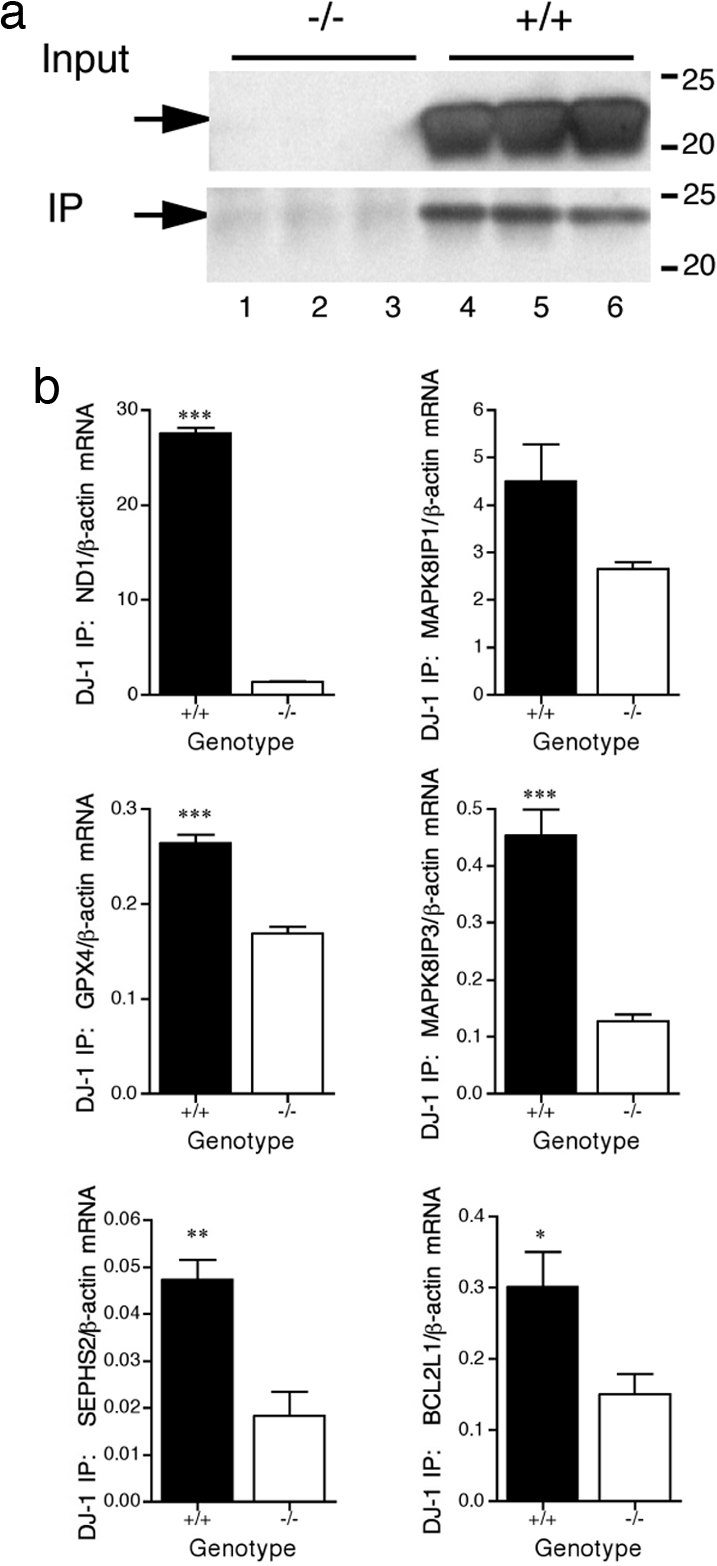
DJ-1 interacts with RNA in vivo. (a) IP for DJ-1 from whole brain lysates from knockout (lanes 1–3) or WT (lanes 4–6) mice. DJ-1 protein is absent from the knockout mice. (b) Validation by qRT-PCR after IP of DJ-1 from WT (closed bars) or knockout (open bars) mice. Error bars indicate the SEM (n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 using t tests to compare genotypes.
Identification of RNA Sequences That Can Bind DJ-1.
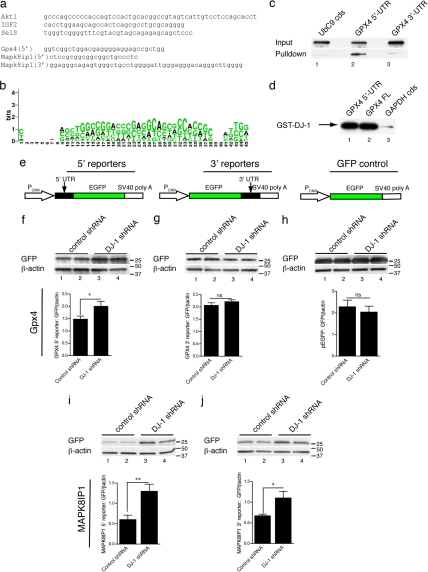
We next used several approaches to determine the region of the mRNAs to which DJ-1 binds. Although the CLIP technique has lower throughput than microarray analysis, it allows for cloning of sequences directly bound to DJ-1 (26). Our CLIP tags included selenoproteins and members of the PTEN/Akt1 pathway (Fig. 3a). The actual sequences all contained multiple copies of GG or CC motifs, reflected in a GG/CC-rich consensus (Fig. 3b). We also recovered many shorter tags (between 10 and 30 nt) that we could not map to specific genes but that were also GG/CC-rich (Table S5). Identified CLIP tags were aligned to multiple regions of RNA, including both UTRs. For example, the 5′ UTR of GPx4 (a candidate confirmed in Fig. 1 c and f) contains a short GG-rich region (Fig. 3a). MAPK8IP1 contains a similar motif in the 5′ UTR but also sequences in the 3′ UTR (Fig. 3a). To test whether DJ-1 was sufficient to bind these sequences, biotinylated single-stranded RNA molecules were prepared by in vitro transcription and incubated with nanomolar amounts of recombinant DJ-1. We found that DJ-1 interacted specifically with the 5′ UTR of GPx4 but not to the 3′ UTR or the coding sequences of two abundant mRNA species (UbC9 and GAPDH), which showed only background binding (Fig. 3c). The binding of DJ-1 to the full-length RNA (including both coding sequence and both UTRs) was no greater than to the 5′UTR of GPx4 (Fig. 3d), suggesting that additional sequences do not contribute to binding. These results indicate that there is an authentic RNA binding sequence in the 5′ UTR of GPx4.
Fig. 3.
DJ-1 interacts directly with GG/CC-rich mRNA sequences. (a) Sequences recovered from CLIP tags for targets also present on the array; each of these was sequenced in duplicate. Similar sequences were seen in other sequences from the array dataset including GPx4 (5′ UTR) and MAPK8IP1 (both 5′ and 3′ UTR). (b) Consensus for all recovered CLIP tags. (c) In vitro binding of recombinant GST-tagged DJ-1 (7.5 nM) was assessed by pull down with biotinylated, single-stranded RNA (50 nM). DJ-1 was coprecipitated with the 5′-UTR of GPx4 (lane 2) but not the 3′-UTR (lane 3) or the coding sequence from UbC9 (lane 1). Data are representative of four independent experiments. (d) The 5′ UTR (lane 1) of GPx4 gives equivalent amounts of pull down compared with the full-length RNA (lane 2) and higher than the coding sequence of GAPDH (lane 3). (e) Diagrams of 5′ and 3′ reporter constructs, which were driven by a CMV promoter (PCMV) and contained an SV40 poly adenylation [poly(A)] sequence. EGFP alone was used as a control for transfection. (f–j) M17 cell lines stably expressing nonspecific shRNA controls (lanes 1 and 2) or shRNA to DJ-1 (lanes 3 and 4) were transfected with reporter constructs where the 5′ UTR (f) or 3′ UTR (g) of GPx4 were cloned adjacent to the 5′ and 3′ end of EGFP. EGFP alone without any additional sequences is shown in h. Similar analyses were performed for the 5′ (i) and 3′ (j) UTR sequences from MAPK8IP1. Cell lysates were blotted for GFP and reprobed for β-actin, and quantitation is shown in the bar grpah below each pair of blots. All reporter construct assays were performed in two shRNA cell lines and each graph shows the average of at least three independent transfections per construct. *, P < 0.05; **, P < 0.01 by t test comparing lines with control shRNA and lines with DJ-1 shRNA.
We created stable dopaminergic neuroblastoma cell lines expressing a nonsense shRNA or two different shRNA sequences to DJ-1 (Fig. S2). Nonsense shRNA did not affect DJ-1 expression levels but the shRNA constructs decreased expression by >85% and >95%, respectively. We made reporter constructs for RNA binding by cloning UTR sequences from GPx4 or MAPK8IP1 into vectors expressing GFP (Fig. 3e). A construct with the 5′ UTR of GPx4 placed 5′ to the GFP coding sequence gave higher protein levels when transfected into DJ-1 knockdown cell lines compared with control shRNA lines (Fig. 3 f and g). In contrast, the 3′ UTR sequence, placed after GFP, was translated as efficiently in both lines (Fig. 3 f and h). Both the 5′ and 3′ UTRs of MAPK8IP1 were able to suppress translation (Fig. 3 i and j). As a control, pEGFP alone without UTR sequences was equally expressed in the presence or absence of DJ-1 (Fig. 3i). These results imply that GG/CC-rich sequences are sufficient for DJ-1 interaction in vitro but are also active in vivo and that DJ-1 partially inhibits translation of its target mRNAs while it is associated with them.
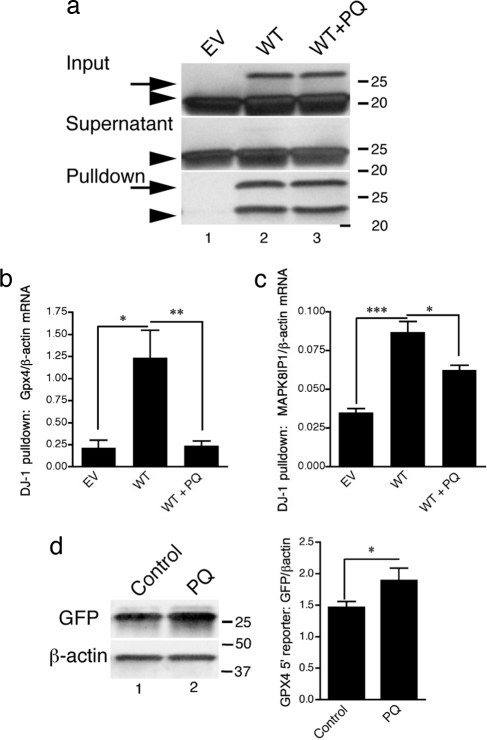
Because DJ-1 is protective under oxidative conditions, we exposed cells to paraquat, which induced a shift in DJ-1 to more acidic isoforms (data not shown) as reported (6). Using NiNTA purification of V5his-tagged WT DJ-1 (Fig. 4a), we found that paraquat treatment significantly decreased the amount of GPx4 (Fig. 4b) and MAPK8IP1 (Fig. 4c) mRNA bound to DJ-1. This decrease in interaction corresponded to an increased expression of the GPx4 5′ UTR-GFP construct in control cell lines (Fig. 4d). This observation suggests DJ-1 releases from target transcripts under oxidative stress.
Fig. 4.
The oxidation state of DJ-1 alters RNA interaction. (a) DJ-1 complexes were isolated from untreated (lane 2) or PQ-treated (lane 3) cells stably transfected with V5–6His-tagged WT DJ-1, or empty vector (lane 1) as a control, using NiNTA beads, then blotted for DJ-1. Arrows show V5–6His-tagged DJ-1, and arrowheads show endogenous DJ-1. (b and c) Associated RNA was analyzed by qRT-PCR (bars show mean signal, error bars indicate the SEM, n = 3 independent PQ treatments and pull downs per construct). PQ treatment decreases the binding of DJ-1 to the RNA for GPx4 and MAPK8IP1. Differences in qRT-PCR were analyzed by one-way ANOVA with Bonferroni's post hoc tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (d) PQ increases the protein levels of the GPx4 5′ UTR GFP construct in cells containing DJ-1. M17 cells were transfected with a reporter construct containing the 5′ UTR of GPx4 (see Fig. 3) placed 5′ to EGFP and were either untreated (Control) or exposed to PQ. Cells were blotted for GFP or β-actin as a loading control. Quantification of n = 4 experiments is shown in the bar graph. *, P < 0.05 by t test.
DJ-1 Effects on RNA and Protein.
Next, we examined the effects of DJ-1 deficiency and oxidative stress on the RNA and protein levels for two candidate targets. We chose GPx4 as a representative selenoprotein and MAPK8IP1 as a representative member of the Akt pathway. In both cases, we were able to obtain high-quality antibodies to the endogenous proteins, although the GPx4 antibody was human-specific (data not shown). In shRNA-mediated knockdown stable cell lines, DJ-1 deficiency was associated with increased GPx4 (Fig. S3 a and b) or MAPK8IP1 (Fig. S3 d and e) protein levels. After oxidative stress, both proteins were induced in control cells but decreased in the knockdown cells. In contrast to the changes in protein levels, mRNA levels for GPx4 or MAPK8IP1 were similar in all cell lines and did not change after paraquat (PQ) treatment (Fig. S3 c and f). To extend this comparison to the in vivo situation, we examined aging DJ-1 knockout mice. We showed that aging is associated with increased oxidation of DJ-1, comparing young (<1 month) and old (24 months old) WT animals (Fig. S3 g and h). MAPK8IP3 protein levels were higher in young knockout animals compared with WT littermates but in aged samples MAPK8IP3 was higher in the WT animals and decreased in the knockouts (Fig. S3 i and j) such that the relative expression levels in knockouts and controls were reversed. Similar patterns were seen with MAPK8IP1 (Fig. S3k). Quantitation of protein levels, correcting for β-actin, revealed a significant effect of age (P = 0.0012 for MAPK8IP3; P = 0.026 for MAPK8IP1) and an interaction between age and genotype (P = 0.0006 for MAPK8IP3; P = 0.0039 for MAPK8IP1) by two-way ANOVA (n = 3–4 animals per group). The steady-state levels of each mRNA did not vary between conditions (data not shown). These data therefore support the hypothesis that DJ-1 binds to RNA for GPx4 and MAPK8IP1/3 in vivo and have a mild suppressive effect on translation.
DJ-1 Knockout Flies Are More Sensitive to Disrupted GSH Peroxidase Function.
Previous studies support the idea that mitochondrial (9) and PTEN/Akt-related (18) DJ-1 targets are important in maintenance of viability in vivo. However, the hypothesis that GSH peroxidases are also targets is novel and led us test the idea in vivo by exposing DJ-1-deficient flies to GSH inhibitors. Flies deficient in DJ-1 showed an increase in sensitivity to the GSH synthesis inhibitor buthionine-S,R-sulfoximine that was similar to that of stressors such as PQ (Fig. S4), supporting a role of GSH in the ability of DJ-1 to protect organisms in vivo.
Discussion
Overall, our results indicate that DJ-1 associates with specific mRNA species in vitro and in vivo. The observation that there are several transcripts that interact may help explain previous observations about the biological roles of DJ-1. By allowing for a simultaneous control of the mRNA for GSH peroxidases, and selenoproteins needed to synthesize functional GPx, DJ-1 may control antioxidant defenses. Targets of DJ-1 within the PTEN/Akt1 pathways may be related to the ability of the protein to suppress cell death, as has been reported in many previous studies, including those demonstrating a specific connection to this pathway (18). The mitochondrial transcripts are also of interest because of the emerging evidence of a mitochondrial basis of recessive parkinsonism (2, 3). DJ-1 can be found on the surface of mitochondria (6) and within the mitochondrial matrix (28), supporting the idea that a small pool of DJ-1 may authentically associate with RNA in the mitochondria. However, this hypothesis requires additional confirmation.
Further work is also required to define the nature of the DJ-1–mRNA interaction. The structural basis of the interaction between the DJ-1 and RNA is unclear as DJ-1 lacks classical RNA binding motifs. However, our data found using in vitro approaches suggests that DJ-1 alone is sufficient to GG/CC-rich sequences at nanomolar concentrations. We have evidence that the RNA targets that DJ-1 binds are distinct from other RNA–protein complexes. In previous large-scale studies of RNA–protein interactions in the brain, FMR-1, associated with mental retardation, binds a distinct set of targets from those discussed here (29), and binding to the splicing factors Nova1/2 are also different (30). Other RNA binding proteins, such as HuR (31), also gave different targets in our studies using the same experimental procedures (data not shown). Therefore, DJ-1 is specific in the sense of binding only a subset of targets.
Our data are consistent with a model whereby DJ-1 interacts with mRNA and dissociates under conditions of oxidative stress. In this respect, when bound to RNA, DJ-1 would slow translation but would allow efficient translation under appropriate local or environmental cues. Rapid translational processes are important in control of stress responses and apoptosis (32), and local translation at synapses is critical for neuronal function. If this interpretation is correct then the increased levels of MAPK8IP1 and MAPKIP3 in the mouse brains with aging would represent successful responses to oxidative stress but the failure of the DJ-1-deficient animals would represent an inadequate ability to do so. The steady-state levels of RNA for these targets are not different in the absence of DJ-1, suggesting that the protein does not modify RNA turnover. However, we cannot exclude that the partial suppression of translation of RNA by DJ-1 is a by-product of another function of the protein. For example, RNA is transported throughout the cell and translation is repressed during transport (33). Future studies will be required to clarify whether DJ-1 has additional activities on its RNA targets.
In summary, we have shown both in vitro and in vivo that DJ-1 is capable of binding a series of target mRNA molecules, identifying a GG/CC-rich sequence that is sufficient for DJ-1 recruitment. The functional classification of these transcripts suggests how DJ-1 may play roles in coordinating responses to oxidative damage and suppression of cell death. Our data support the hypothesis that the apparently pleiotropic roles of DJ-1 may be related to the single function of binding to multiple mRNA transcripts.
Methods
DJ-1 Knockout Mice and Cell Lines.
DJ-1 knockout mice lacking exon 2 have been described in detail (27). DJ-1 expression constructs have been described (6, 34, 35). Stable M17 human neuroblastoma cell lines were cultured as described (6). Clonal M17 cell lines stably expressing different DJ-1 siRNAs were constructed with the Invitrogen BLOCK-iT Lentiviral system, according to the manufacturer's instructions. Two target sequences (GGAAGTAAAGTTACAACACA, GGTCATTACACCTACTCTGAG) and one nonsense control sequence (GCCTAGACGCGATAGTATGGA) were used.
Precipitation of DJ-1 RNA Complexes.
Anti-DJ-1 antibody C-16 (Santa Cruz Biotechnology) had no detectable RNA-degrading activity and was used for IPs. Negative control IPs were performed by using normal goat IgG from the same manufacturer (catalog no. SC-2028). Cell or brain samples were harvested in PLB buffer (0.5% Nonidet P-40, 10 mM Hepes, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, RNase OUT and protease inhibitors) and prepared for RNA-IP as described (31). Briefly, DJ-1 C-16 bound to Protein G Dynabeads (Invitrogen) or Ni-NTA magnetic beads (Qiagen) were incubated with precleared lysate in NT2 buffer (0.05% Nonidet P-40, 50 mM Tris, 150 mM NaCl, 1 mM MgCl2), and then washed four times with NT2 buffer. RNA was extracted by adding DNase for 5 min, discarding the supernatant, then eluting after treatment with Proteinase K. RNA was washed with acid phenol-chloroform and precipitated with 100% ethanol containing sodium acetate and glycoblue (Ambion) overnight. Precipitated RNA was pelleted and washed through 70% ethanol. The concentrations were equalized, and cDNA was generated by using the SuperScript III kit (Invitrogen). CLIP was performed as described (26, 30).
Expression Arrays and Analysis.
Illumina human oligonucleotide arrays were used according to the manufacturer's instructions, starting with 500 ng of total RNA for each sample. Arrays were read on an Illumina Bead array reader confocal scanner. Differential gene expression values were calculated with the Illumina Custom algorithm within the Illumina BeadStudio software suite.
Biotin-Labeled RNA Pull-Down Assays.
Biotin-labeled RNAs corresponding to the UTR sequences for GPx4 were synthesized with T7 polymerase, biotinylated, and used in pull-down assays with recombinant GST-tagged DJ-1 as described (35).
Quantitative RT-PCR (qRT-PCR).
Primers were designed against potential DJ-1 binding targets and validated for use in comparative qRT-PCR with β-actin (25). Primer sequences are available on request. Final quantitation of changes in gene expression was calculated from reactions performed in quadruplicate with four biological replicates per sample.
Protein Analyses.
Analysis of the oxidation state of DJ-1 was performed as described (6) using monoclonal anti-DJ-1 (Stressgen). Antibodies for MAPK8IP1 (JIP1) and MAPK8IP3 (JIP3) were obtained from Santa Cruz Biotechnologies. Quantitation of signal intensities was performed by using a Storm fluorescence scanner, and protein loading was normalized by reprobing the same blots with mAb to β-actin (Sigma; clone AC-15). To make reporter constructs, forward and reverse primers were designed to amplify the 5′ and 3′ UTRs of human GPx4 or MAPK8IP1. Sequences were cloned into the pEGFP-N1 and pEGFP-C1, respectively (Clontech). Constructs were transiently transfected into cell lines by using Lipofectamine 2000 (Invitrogen), and protein was measured by blotting for GFP.
Drosophila melanogaster Viability Experiments.
Drosophila DJ-1b-deleted (DJ-1bΔ93) and double knockout (DKO, DJ-1bΔ93 with DJ-1aΔ72) and exposure to drugs (5% buthionine-S,R-sulfoximine or 1 mM selenium, in 5% sucrose, 1% agar medium) have been described (11).
Supplementary Material
Acknowledgments.
This research was supported in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health. Computational approaches used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov). L.-Y.H. and N.M.B. were supported by National Institute on Aging Grant P01-AG09215. N.M.B. is an Investigator of the Howard Hughes Medical Institute. A.J.M. received grant support from the Verum Foundation and the European Union DIADEM (Delivering Inclusive Access for Disabled or Elderly Members of the community) project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE8632).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708518105/DCSupplemental.
References
- 1.Cookson MR. The biochemistry of Parkinson's disease. Annu Rev Biochem. 2005;74:29–52. doi: 10.1146/annurev.biochem.74.082803.133400. [DOI] [PubMed] [Google Scholar]
- 2.Clark IE, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 3.Park J, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 4.Bandopadhyay R, et al. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson's disease. Brain. 2004;127:420–430. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- 5.Choi J, et al. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J Biol Chem. 2006;281:10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canet-Aviles RM, et al. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kinumi T, et al. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 8.Betarbet R, et al. Intersecting pathways to neurodegeneration in Parkinson's disease: Effects of the pesticide rotenone on DJ-1, α-synuclein, and the ubiquitin-proteasome system. Neurobiol Dis. 2006;22:404–420. doi: 10.1016/j.nbd.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Kim RH, et al. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc Natl Acad Sci USA. 2005;102:5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr Biol. 2005;15:1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 11.Meulener M, et al. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson's disease. Curr Biol. 2005;15:1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 12.Meulener MC, et al. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc Natl Acad Sci USA. 2006;103:12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ved R, et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of α-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J Biol Chem. 2005;280:42655–42668. doi: 10.1074/jbc.M505910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandyopadhyay S, Cookson MR. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol Biol. 2004;4:6. doi: 10.1186/1471-2148-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas JI, Marin I. A new evolutionary paradigm for the Parkinson disease gene DJ-1. Mol Biol Evol. 2007;24:551–561. doi: 10.1093/molbev/msl186. [DOI] [PubMed] [Google Scholar]
- 16.Nagakubo D, et al. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem Biophys Res Commun. 1997;231:509–513. doi: 10.1006/bbrc.1997.6132. [DOI] [PubMed] [Google Scholar]
- 17.Yang Y, et al. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc Natl Acad Sci USA. 2005;102:13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim RH, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7:263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 19.Junn E, et al. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc Natl Acad Sci USA. 2005;102:9691–9696. doi: 10.1073/pnas.0409635102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T α-synuclein toxicity. J Biol Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 21.Xu J, et al. The Parkinson's disease-associated DJ-1 protein is a transcriptional coactivator that protects against neuronal apoptosis. Hum Mol Genet. 2005;14:1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 22.Zhong N, et al. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J Biol Chem. 2006;281:20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 23.Wilson MA, et al. The 1.1-A resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early-onset Parkinson's disease. Proc Natl Acad Sci USA. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hod Y, Pentyala SN, Whyard TC, El-Maghrabi MR. Identification and characterization of a novel protein that regulates RNA–protein interaction. J Cell Biochem. 1999;72:435–444. [PubMed] [Google Scholar]
- 25.Baptista MJ, et al. Coordinate transcriptional regulation of dopamine synthesis genes by α-synuclein in human neuroblastoma cell lines. J Neurochem. 2003;85:957–968. doi: 10.1046/j.1471-4159.2003.01742.x. [DOI] [PubMed] [Google Scholar]
- 26.Ule J, Jensen K, Mele A, Darnell RB. CLIP: A method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 27.Chandran JS, et al. Progressive behavioral deficits in DJ-1-deficient mice are associated with normal nigrostriatal function. Neurobiol Dis. 2008;29:505–514. doi: 10.1016/j.nbd.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L, et al. Mitochondrial localization of the Parkinson's disease-related protein DJ-1: Implications for pathogenesis. Hum Mol Genet. 2005;14:2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 29.Brown V, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 30.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 31.Lopez de Silanes I, et al. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 33.Bramham CR, Wells DG. Dendritic mRNA: Transport, translation, and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 34.Blackinton J, et al. Effects of DJ-1 mutations and polymorphisms on protein stability and subcellular localization. Brain Res Mol Brain Res. 2005;134:76–83. doi: 10.1016/j.molbrainres.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Miller DW, et al. L166P mutant DJ-1, causative for recessive Parkinson's disease, is degraded through the ubiquitin-proteasome system. J Biol Chem. 2003;278:36588–36595. doi: 10.1074/jbc.M304272200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.