Abstract
Recently, the extent of copy number variation (CNV) throughout the genome has been shown to be far greater than previously thought. Further, it has been demonstrated that specific copy number variable regions (CNVRs) are associated with particular diseases, suggesting that these genetic variations may have an important biological role. Hence, calling CNVRs and subsequently classifying samples as “losses” or “gains” is of great interest. A number of papers have been published containing classifications of CNVs, and here we show how the presence of pedigree information can be used for assessing the performance of those classification methods. In this article, by examining CNV classifications made in the HapMap samples, we show that estimates of the number of false-positive classifications per individual made by current approaches can be determined. Moreover, commonplace technologies for determining the locations of CNVRs aggregate information across the maternal and paternal chromosomes at the locus of interest. Here, we show that copy number variation on each chromosome can be inferred and, in particular, we discuss the existence of a class of CNVs that are inevitably misclassified and give an estimate of their prevalence. Although our focus is not on the development of calling algorithms per se, we describe and provide an example of how our model might be incorporated into the initial classification procedure to produce more robust results. Finally, we discuss how this methodology might be applied to future studies to obtain better estimates of the extent of CNV across the genome.
Keywords: array CGH, classification, copy number variation, HapMap Project, pedigree information
Copy number variation (CNV) has been found in all mammalian genomes examined thus far (1, 2) and seems to be widespread. CNVs are variations in copy number on a local scale, perhaps just kilobases in length, as distinguished from larger aberrations in copy number, such as those long known to be involved in the onset and development of cancer (3). Within humans, associations have been uncovered between specific copy number variable regions (CNVRs) and the chance of developing adverse phenotypes such as HIV-1 infection (4) and Alzheimer's disease (5). With the advent of high-resolution microarray technology, it has become possible to obtain genome-wide maps of the location of CNVRs. This, in turn, has resulted in an increased appreciation of the scale of human genetic variation, an understanding that has been labeled the scientific breakthrough of 2007 (6).
There are two groups of tools for genome-wide identification of CNVs in mainstream use. We focus here on array Comparative Genomic Hybridization (array CGH) technologies, which directly interrogate the amount of DNA present and complementary to the bacterial artificial chromosomes (BACs) or, increasingly these days, oligonucleotide probes present on the array. The current common alternative is to make inference from SNP genotyping (7), and in the near future, massively parallel resequencing seems set to become popular for such applications.
One of the first comprehensive maps of CNVRs was described by Redon et al. (2) in late 2006. Therein, CNVRs were identified from the array CGH data using a threshold-based approach that called CNV-harboring probes one sample at a time. In a later work (8), a mixture model, fitted across all samples simultaneously, was applied one probe at a time. For each probe, a sample was classified into one of four categories: gained [1], normal [0], lost [−1], or complex [2]. Such calls clearly depend on the choice of reference to be regarded as “normal,” and the paradigm is not ideal but is followed here for consistency and (save for the addition of “complex”) after the example of the database of genomic variants (http://projects.tcag.ca/variation).
To call and classify CNVRs, the technologies described above combine information across copies of chromosomes. Consequently, the data do not lend themselves to making inferences about the extent of CNV on specific chromosomes. In this article, we examine the CNV classifications made previously for a subset of the HapMap population (9) while incorporating the family information contained therein. In doing so, we show (i) that it is possible to model the output from a genome-wide array CGH experiment at a chromosome copy level using information about family structures, (ii) that such a model is a valuable tool for the assessment of CNV classification exercises, (iii) how a previously published set of classifications for the HapMap samples fares under assessment, (iv) the inevitability of the existence and estimated frequency of a class of CNV that is misclassified by such technologies, and (v) the manner in which the model might be incorporated into the initial classification procedure to produce more robust results, with an example of such an application.
Motivating Example.
As has been mentioned, arguably the most comprehensive map of CNVRs was described in late 2006 (2). In this article, nominally diploid lymphoblastoid cell lines from the 270 HapMap samples were analyzed by CGH using high-density BAC arrays. Using these data (in conjunction with complementary information from Affymetrix 500K EA chips), 1,447 CNVRs spanning 360 Mb (≈12% of the genome) were identified.
These data were subsequently reanalyzed by using a mixture model (8), where individuals were classified for each BAC as “lost,” “normal,” “gained,” or “complex” [see supporting information (SI) Fig. S1 for a summary of the analysis leading to these classifications]. The “complex” calls were idiosyncratic to this analysis and broadly represented clones where the mixture model could not identify separate components, yet the variance of the observed log ratios exceeded that which seemed plausible if there was no CNVR. We make no further use nor mention of these complex clones, because (i) the “complex” calls were made “globally,” and thus there is no variation across the pedigree that can be modeled, and (ii) we present these methodologies as a way of assessing the output of a generic array CGH experiment, and most would not offer these “complex” calls.
The HapMap samples include 90 individuals from the Yoruban population, who make up 30 parent–offspring trios. In this article, the classifications from the mixture model are combined with information about the family structure of these Yoruban individuals, information that was not used in the original classifications. We use these data to obtain a (global) measure of the probability that a copy number variant is present on neither, one or both inherited copies of a chromosome in this population.
In Table S1, the classification data are broken down into 27 groups that correspond to the different combinations of classifications that can be observed for each parent–offspring trio. These provide the basis for the modeling approach used in this article.
Use of Familial Information in Classifying CNVs.
We are not the first to advocate the use of family information when analyzing CNV data derived from parent–offspring trios (or other known pedigrees). An approach previously presented, which might represent the gold standard, fully exploited family data at a single locus (10). A Bayesian approach was used to update the actual number of copies present at the locus of interest for each individual in a family, to examine whether the CNV was associated with a particular phenotype.
In this example, however, the copy number had been determined by using PCR. This naturally provides more accurate values than could reasonably be expected from a genome-wide array. Moreover, all measurements were repeated, and the model was applied to the log-ratios rather than classifications. It would not be reasonable to anticipate the application of such a model to our dataset of interest. Not only is the data quality far worse than would arise from focused PCR experiments, with little or no replication, but also the computational burden (and indeed demands on the judgement of the analyst) of a genome-wide application of the model would be prohibitive.
A recent article (11) described a method for calling CNVRs from SNP genotyping data (some of which was generated by using HapMap samples) that made a posteriori use of trio information. By using Bayes' rule, sample-specific CNVR calls were updated to yield the posterior probability of observing a set of classifications within a particular trio. Wang et al. (11) then assumed that the set of classifications with the largest posterior probability represented the true copy number classifications for the trio of interest. This approach makes use of allele frequency information not available from array CGH platforms and (to highlight one difference from the approach presented here) does not distinguish between the case where a mother has two copies of a sequence, one on each of two copies of a chromosome, or two copies of a sequence but both on the same copy of a chromosome. So, for example, under this model if a father has two copies and a mother has two copies, barring a de novo mutation, the offspring has to have two copies.
Our model, by contrast, allows for more flexible inheritance probabilities and naturally distinguishes between the two cases mentioned above. It requires neither allele frequency information nor the actual log ratios, which may be either impossible or difficult to obtain, depending on where and with what technology the data were generated. With additional or better-quality data, previously proposed methods may be preferable, but even in these cases, the computational feasibility of this approach may be appealing.
Results
If a sample is diploid, the CNVR classifications of each probe (gained, normal, or lost), as nominated by array CGH, are a function of the number of copies of that region present on each copy of the relevant chromosome. We assume the region of interest can be normal (N), gained (G), or lost (L) on each chromosome. Six combinations of the two chromosomes are thus possible (GG, GN, LN, LL, NN, and LG), and a CNVR classification determined from the array CGH data can be linked with any one of these. Ideally, the GG and GN pairings would be called as gained, the LL and LN pairings would be called as losses, and any NN pairing would be called as normal.
The estimated probabilities of each chromosome/classification pair are denoted as in Table 1. For example, the probability that two chromosomes are genuinely gains, and that the sample is classified as a gain from the array CGH data, is denoted a1. The other parameters in the third column of Table 1 complete the notation in a similar manner. Further, we note that, because they represent probabilities, their associated values must sum to one.
Table 1.
Parameter estimates
| Chromosomes | CNVR classification | Parameters | Estimate | Conditional estimates | Proportions in each category | Renormalized within a category |
|---|---|---|---|---|---|---|
| GG | 1 | a1 | 0.00607 | 0.1020 | 1 | |
| GG | 0 | a0 | < 10−6 | < 10−6 | 0.1020 | <10−6 |
| GG | −1 | a−1 | < 10−6 | < 10−6 | < 10−6 | |
| GN | 1 | b1 | 0.00012 | 0.0019 | 0.475 | |
| GN | 0 | b0 | 0.00012 | 0.0021 | 0.0040 | 0.525 |
| GN | −1 | b−1 | < 10−6 | < 10−6 | < 10−6 | |
| LN | 1 | c1 | < 10−6 | < 10−6 | <10−6 | |
| LN | 0 | c0 | 0.00008 | 0.0014 | 0.0024 | 0.583 |
| LN | −1 | c−1 | 0.00006 | 0.0010 | 0.417 | |
| LL | 1 | d1 | < 10−6 | < 10−6 | <10−61 | |
| LL | 0 | d0 | < 10−6 | < 10−6 | 0.0660 | < 10−6 |
| LL | −1 | d−1 | 0.00391 | 0.0660 | 1 | |
| LG | 1 | e1 | 0.00021 | 0.0034 | 0.233 | |
| LG | 0 | e0 | 0.00048 | 0.0081 | 0.0146 | 0.555 |
| LG | −1 | e−1 | 0.00019 | 0.0031 | 0.212 | |
| NN | 1 | f1 | < 10−6 | < 10−6 | <10−6 | |
| NN | 0 | f0 | 0.98880 | 0.8110 | 0.8110 | 1 |
| NN | −1 | f−1 | < 10−6 | < 10−6 | < 10−6 |
The first two columns illustrate the different combinations of chromosomes and the corresponding CNVR classifications, as ascertained from the array CGH data. The fourth and fifth columns show the parameters' estimates and estimates conditional on a probe containing a CNVR, respectively. Column 6 shows, for the CNVR probes, the proportion of samples within each of the six chromosome categories. The final column illustrates the proportions in column 5, normalized within each of the chromosome categories.
This model provides an average measure of the parameters (column 3, Table 1) across all probes. Although different effects may be observed for some probes, these measures will yield substantial information about the character of CNV across the genome. Further, the model also assumes that copy number variants do not arise sporadically (i.e., they must be inherited). There are some reasons to believe this will be the case (12), and additional evidence for the rarity of such events is provided by the ease with which the populations within the HapMap project cluster together (8). This assumption could be relaxed, because the arguments for it are not conclusive; however, in the absence of a sensible prior for the mutation rate, distinguishing between misclassifications and de novo mutations would prove difficult.
The vast majority of probes were not called as CNVRs. Thus, the most common occurrence in the population is that individuals have two normal copies of a chromosome and are classified as harboring no copy number aberration. The probabilities of observing each chromosome/classification pair, conditional on the overall probe being called as a CNVR, are given in column 5 of Table 1. As was observed by Redon et al. (2), there are more copy number gains than losses.
These results provide much information regarding the performance of the original classification algorithm. In particular, by examination of the values associated with parameters b0 and c0, we see indications that, half of the time, GN and LN pairings are misclassified as “normal.” Many of the probes called as CNVRs across the entire HapMap population show little variation within the Yoruban population (albeit they might, for example, all be classified as “gains” within the Yoruban population), so the proportions of GN pairings and LN pairings seen here are lower than might be anticipated. That the combination of the technology and classification algorithm seems unable to correctly identify these individuals as being variant may be an even greater concern if considering a more heterogeneous population.
Similarly, it appears those individuals who are NN, GG, or LL are called correctly on nearly all occasions. It should be noted that this is probably because if it were not easy to classify GG and LL individuals, then the probe would have been unlikely to have been called in the first place. We can therefore say little about the true false negative rate, but the false negative classification rate conditional on being in a probe called as a CNVR can be estimated, and this rate and perhaps those in previous studies seem to be driven by an inability to classify correctly individuals who harbor a CNV only on one copy of the chromosome.
We can deduce that (for a particular locus) this leads to the situation that an offspring with (only) one parent classified as having a copy number gain/loss will inherit from that parent a chromosome with a gain/loss at that locus 97.5%/97.0% of the time but will be classified as gained/lost from the array CGH data only 44.8% and 38.5% of the time, respectively (see Methods for details of these calculations). Although an apparent inheritance rate of a little under 50% would not cause alarm, the disparity from the true inheritance rate is a concern.
In summary, it appears from these data that approximately 4% of “loss” classifications made within CNV harboring probes are incorrect, and approximately 3% of “gain” classifications and 1% of “normal” classifications are similarly erroneous. Note that these are not comparable to the validation values in the previous studies, because those values also considered errors in the initial calling of a probe as being CNV harboring.
If we extrapolate to the entire HapMap population classification (8), trusting the initial calling of probes as being CNV harboring as accurate, then we anticipate that approximately 1,300 (≈ 30,031 × 0.0442) classifications of “loss” are in error. We anticipate that ≈1,400 (≈ 43,449 × 0.0317) classifications of “gain” are in error, and that ≈4,000 (≈ 303,324 × 0.0141) classifications of “normal” are in error. This is equivalent to five individuals having been misclassified at each CNV harboring probe.
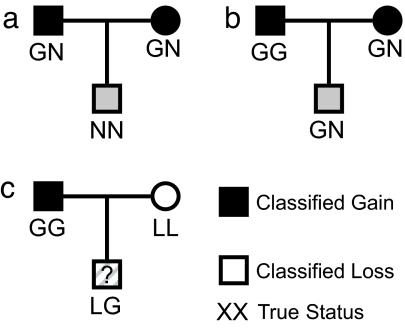
When estimating the error associated with a platform/classification method, the LG combination of chromosomes is of special interest (see Fig. 1 for an example of how such a situation could arise). Previous studies have shown there are sites for which both gains and losses are common within the same population (albeit conditional on the choice of reference sample, as must be all of these classifications), and so it seems inevitable there will be children who inherit both. Although the other discordant examples, GN and LN, ought to be classified as “gains” and “losses,” respectively, it is not clear a priori whether there is any net gain or loss in an LG combination, or how current methods will classify such samples, or indeed how they ought to be classified. Thus, any instance of an LG must be considered misclassified, because none of the available classifications (“gain,” “loss,” “normal”) are adequate.
Fig. 1.
Examples of trios that may cause confusion. Two examples illustrating how the trio of classifications (mother and father “gain,” offspring “normal”) can arise either from the parents having discordant copies (a) or the child being misclassified (b). The third trio (c) shows the ease with which an offspring with LG status can arise.
LG combinations account for one and a half percent of all states (conditional on being a CNVR) in this sample, and we have already noted that other samples will be more heterogeneous than this, and thus may have higher percentages as a whole. In Table 2 we see the potential for LG calls among the HapMap population, with 54 probes having substantial numbers of both “loss” and “gain” calls.
Table 2.
The potential for the LG status in the HapMap population
| Number of classifications (C) | 10 | 20 | 30 | 40 | 50 | 60 | 70 |
| Number of probes | 54 | 24 | 19 | 11 | 7 | 4 | 2 |
Presented are the numbers of probes for which, among the HapMap population, at least C ″loss″ classifications and C ″gain″ classifications were originally made (8) for varying values of C. These are the probes where one might naturally anticipate LG pairings being inherited.
Considering the posterior reclassification of samples (as detailed in Methods) using the results generated from this model, the vast majority of trios/probes are extremely unlikely to display a change in the updated classification. In general, for a particular CNVR, most samples are classified as “normal” to begin with, and will remain classified as “normal” after the exercise. Because of this, and the considerable computational burden of fitting the model to one trio/probe at a time, we do not advise attempting such a comprehensive reclassification. Rather we recommend applying the updating scheme to selected probes.
When the probes were originally classified for each individual, this was done by use of a probabilistic mixture model and, whereas individuals were ultimately placed in one of three bins, a measure of the certainty of that classification is available. It is natural then to apply such reclassification as a priority to those trios containing individuals for which the original classification was doubtful.
An example of its effectiveness is shown in Table 3. For this trio/probe, when family information is not used, only the offspring is classified as having a gain at this locus. However, once the trio information is incorporated, both offspring and father are called as having a gain. Given that copy number variants have been shown to be highly heritable, this change in classification makes biological sense.
Table 3.
Example of a change in classification
| M | F | O | |||||||
|---|---|---|---|---|---|---|---|---|---|
| log2 ratio | 0.0126 | 0.0745 | 0.1745 | ||||||
| CNVR classification | − | 0 | + | − | 0 | + | − | 0 | + |
| Probability ignoring family | < 10−4 | 0.999 | < 10−4 | < 10−4 | 0.863 | 0.137 | < 10−4 | < 10−4 | 0.999 |
| Probability using family | < 10−4 | 0.999 | < 10−4 | < 10−4 | 0.380 | 0.620 | < 10−4 | < 10−4 | 0.999 |
The probabilities of a family member's CNVR classification at a particular probe are shown, both before and after family information is accounted for. M refers to a mother, F to a father, and O to their offspring. Values in bold font correspond to the highest probability (and hence the CNVR classification) for each individual before and after family information is accounted for.
Discussion
This article illustrates the value of pedigree information for assessing CNV classifications. A question arises as to whether a study might include such structures for the sole purpose of evaluating the resultant CNV classifications. Many studies will naturally contain such family structures as a mechanism for conducting their investigations of interest, and these in particular should benefit from this methodology. For other studies, the forced inclusion of families will use arrays that could have been used to run replicates of existing samples (providing some scope for technical validation) or additional samples (improving the quality of estimates produced). It is not appropriate to suggest that the quality of a study should routinely be compromised so its performance might be quantifiable. Nevertheless, such quantification may be necessary in many situations, and there will likely be enough studies containing pedigrees as a matter of course for this methodology to be useful.
Using such methodology, we have demonstrated (and quantified) a significant limitation in current CNV analysis methods, namely, the inability to deal with discordant variants. That is, those where the CNV status of the region of interest is different on each inherited copy of a chromosome; in the language of this article, the NGs, NLs, and LGs. Previous observations that gains are more frequent than losses may have simply reflected a greater power to detect gains in the classification algorithms used. Our model suggests that, on the contrary, more gains are also being missed than are losses, and so we find no evidence from the pedigree information to suggest this is a technical rather than biological result, although the choice of reference is obviously influential.
Although we illustrated our methodology on data generated using array CGH technology, these problems will also affect other technologies. This is particularly important at present, because new studies that aim to examine CNVs in more depth, using both higher-resolution array platforms and larger cohorts, are being planned. When SNP genotyping arrays are used to investigate CNV, a measure of copy number is typically obtained by combining data from each allele-specific probe. Where a different base is present on each inherited copy of a chromosome, the genotype data can be used to partially infer the allele-specific nature of the CNV, and family information can be used to further strengthen the inference. However, the full inference will require the incorporation of family information in a manner such as we have described and will rely on SNPs being located conveniently, which, although often the case, is not guaranteed. Irrespective of the technology used, in the situation where no family information is available, it is not clear whether computational approaches will be able to shed light on the character of the data and, instead, novel experimental approaches may have to be developed.
As well as providing a methodology for assessing CNV classifications, our approach allows for investigation of discordant CNVs in the presence of family data, in particular the LG cases. This is especially important, because our analysis suggests these variants are likely to account for the vast majority of misclassifications and so need to be considered for methodological improvements to be made. Our method also has the advantage that it can be used to generate improved classifications of CNVRs. It would be wrong, however, to attempt to iterate the process. If the CNVR classifications are updated via this approach, then the model cannot be used to assess those new classifications in an unbiased manner. However, alternative estimates of parameters of interest such as the false discovery rate may still be obtained by considering the numbers of classifications that change in the process.
We have used a motivational example where the probes used in the array CGH technology were relatively large and few in number. Therefore, neighboring probes could be treated as independent without causing great distress, in both the original classification and our model. By contrast, SNP technologies and array CGH technologies using oligonucleotides have (increasingly) larger numbers of much shorter probes and may be classified by analyzing several neighboring probes at a time, both to deal with the quantities of data and to address issues of dependence among serial probes.
Adjustment of our approach for such data will have to be tailored in each case depending both on how the classifications were made, and how they are being interpreted. There are, though, three immediately apparent general strategies that might be considered for any circumstance. First, it is common for such data that classifications are made for regions of successive probes and not for individual probes. For many purposes, interpretation of a single probe called as a CNV would be difficult at best. Thus, by applying the methodology to regional classifications, one would approximate the application to the large BACs used in our example. The second is simply to apply the model to subsamples of the data, as we did here for validation. Because one starts with a larger number of probes, fitting the model to a subset of the data need cause no difficulty, and because the retained probes will be sparser, the dependence among them will be weakened.
Finally, one could make the model more complicated to incorporate such dependence. This would need to be done at two levels, once for the dependence of the “truth” and once for the classification. The likelihood is easily, if crudely, adjusted for the believed underlying truth by subtracting a penalty function that increases in magnitude the more often neighboring classifications would be predicted to differ. Adjusting for dependence in the classification would have to be specific to the algorithm used; however, it is not difficult to imagine increasing the number of states modeled from three (G, L, N) to representing “gain with a neighboring gain,” “gain with a neighboring normal” and so on, or to even more complicated representations. This step would increase the dimension of the parameter space considerably, but the large increase in observations in these datasets would make such a model practicable.
In summary, the work described in this article can be used better to model data generated to examine CNVRs across the genome when family information is present. This method can be used to evaluate and/or update calls generated by any calling/classification scheme where the probability of a region being classed as a CNVR for an individual can be calculated. By taking account of chromosome-specific copy number changes, this model and other similar approaches are vital if we are to obtain better insights into the extent and role of CNV in the genome.
Methods
Data Processing.
For each individual in one of the 30 Yoruban parent–offspring trios, we downloaded the CNVR classifications (described in ref. 8) for probes mapped to an autosomal chromosome.
Constructing the Likelihood.
Using the parameters described in the third column of Table 1, we can write the probability that an individual is classified as a gain by summing over all chromosome/classification pairs where the classification is a gain:
P(classification = 1) = a1 + b1 + c1 + d1 + e1 + f1.
Further, we can write the joint probability of a child inheriting a chromosome-type from a particular parent and of observing the CNVR classification assigned to their parent. For example,
P(child inherits a gain and parent classified as 1) = a1 +½(b1 + e1).
Subsequently, we can find the probability that a child has, for example, a gained copy of a chromosomal segment conditional on their parent's CNVR classification. Defining this probability as G1, we obtain
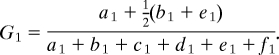 |
Using similar arguments, we can obtain formulae for Gi, Ni and Li, where i denotes the parent's CNVR classification, and G, N and L represent the situations where a child inherits, from a specified parent, a chromosome with a gained, normal or lost copy in the region of interest.
We can now construct the likelihood. The first step is to write down the probability that a child has a particular classification given their parents'. For each trio, we need first to define the probability that parents have an offspring with a specific CNVR classification, given their own classifications. Define this probability as Pj,i, where j corresponds to the joint classification of the parents (e.g., {gain, gain} represented as 11), and i is the classification of the child. Using the probabilities defined above, we have (for example)
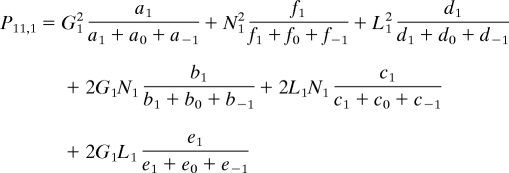 |
The data in Table S1 can be modeled as a multinomial distribution with these probabilities.
However, to construct the likelihood we need also use the probabilities that two parents with particular CNVR classifications meet. Define these probabilities as Quw where u,w ∈ {1,0,−1}. Thus, assuming that two parents meet at random within the population, we have (for example) Q11 = (a1 + b1 + c1 + d1 + e1 + f1)2. Consequently, supposing the parameters (a0, a1,…, f−1) are contained in a vector, θ, we can write the log-likelihood as:
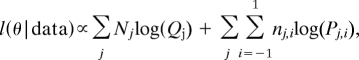 |
where nj,i is the number of trios where the parents have the joint classification j and the offspring has classification i, and Nj is the number of parents with joint classification j (Table S1).
Optimizing the Likelihood.
To obtain the estimates of the parameters, we maximized the likelihood using a constrained numerical optimizer (column 4, Table 1). To assess whether the optimizer converged to the global optimum, we started from a variety of initial values. Further, to check that correlations between genomically adjacent probes were not affecting the parameter estimates, we reestimated them using every tenth probe; this yielded the same estimates, thus providing confidence in the efficacy of the model. Finally, we used a bootstrap-based approach to estimate 95% confidence intervals for the parameters; these are provided in Table S2.
Renormalizing the Parameters.
To ease their interpretation, we renormalized the parameter estimates by removing the effect of probes not classed as CNV harboring. To do this, we excluded probes classed as complex and calculated the proportion of the remaining probes not called as CNVRs. We then subtracted this value from f0 and, to obtain the modified parameter estimates, we scaled this value and the other parameter estimates so that they summed to one.
Inheritance Rates of Classifications.
Where we comment on the inheritance rates of classifications, e.g., the probability of a child being classified as a “gain” given that exactly one of their parents is classified as a “gain,” we calculate the probabilities as follows. For this particular example, we first calculate the probabilities of inheriting a gained copy, normal copy or loss copy from the parent classified as a gain. These have already been denoted G1, N1, and L1. In a similar manner, we then calculate the three probabilities of inheritance from the parent not classified as a “gain.”
From these two sets of probabilities, it is possible to calculate the probabilities of the six possible states that the child may possess (GG, GN, etc.), and from the results in Table 1, we can calculate the probabilities of the three possible classifications that the child may be given.
Updating the Classifications.
To obtain the prior probabilities of observing different combinations of CNVR classifications within a trio, we used the optimized parameter values obtained after conditioning on a probe containing a (noncomplex) CNVR (column 5, Table 1). For example, suppose the mother is normal, the father is normal, and the offspring has a gain. We can then define the prior probability of observing this trio as pr0,0,1, where pr0,0,1 = Q00 P00,1. We can define the prior probabilities of observing the remaining 26 combinations similarly.
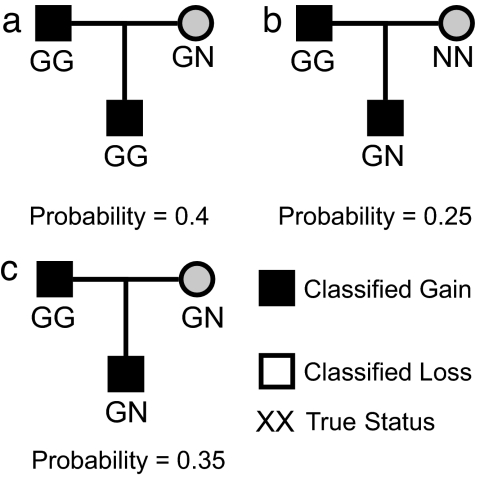
In ref. 8, for each probe, the EM algorithm was used to fit a Normal mixture model to the log2 ratios observed across all of the HapMap samples. The fitted model was then used to classify a sample as a copy number loss or gain, or as having a normal copy number status for that probe. For a specific probe/trio, we can refit this model when the classifications of the trio of interest are fixed a priori. Estimation of the parameters in the mixture model and the allocation of the other samples to components of the fitted model were performed by using the EM algorithm. For the probe/trio of interest, we can fit this model 27 times, corresponding to the different combinations of CNVR classifications that are possible (see the previous paragraph and Table S2) and thus obtain 27 different likelihoods. Subsequently, using Bayes' rule, we can multiply the prior probabilities and the likelihoods together to obtain a value proportional to the posterior probability of observing a particular set of CNVR classifications for the trio of interest. For example, define the posterior probability of observing the trio where the mother and father are both classified as normal, and the offspring is classified a gain as π0,0,1. Then, π0,0,1 ∝ pr0,0,1 × likelihood0,0,1, and naturally we can normalize these values to estimate π0,0,1.
We can calculate the probability, M1, that the mother should be classified as a copy number gain by summing the probabilities over all families consistent with this classification. Probabilities for loss and gain and for the father and offspring can be obtained by using the same approach. Subsequently, the updated classification for each individual is found by considering the classification for which the probability is largest. Note that this differs from the approach of Wang et al. (11), who take the classification associated with the most probable family (Fig. 2). Finally, we can compare these classifications with those found in the original mixture model, where no family information was incorporated, and examine whether there are any changes (e.g., Table 3).
Fig. 2.
A hypothetical example of the final step of the Bayesian reclassification. Illustrated are three plausible true statuses for the situation where the classifications made to father and offspring were “gain” and that to mother was “normal.” If the posterior probabilities associated with each were calculated to be as shown, then methods choosing the most probable family would choose a. We, however, are looking to reclassify the individual and so, for the individuals, sum the probabilities for families that have common states. In this case, the posterior probability that the child is GN is 0.6, and so the posterior statuses assigned would correspond to family (c).
Web Resources.
The code used to perform the calculations described throughout this article and some additional information on the likelihood calculation are available at www.compbio.group.cam.ac.uk/resources.html.
Supplementary Material
Acknowledgments.
We thank two anonymous referees for helpful comments. J.C.M., S.T., and A.G.L. were supported by grant C14303/A8646 from Cancer Research UK. S.T. is a Royal Society/Wolfson Research Merit Award Holder. M.W. was supported by the University of Cambridge.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711252105/DCSupplemental.
References
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kallioniemi A, et al. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzalez E, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 5.Rovelet-Lecrux A, et al. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet. 2006;38:24–26. doi: 10.1038/ng1718. [DOI] [PubMed] [Google Scholar]
- 6.Pennisi E. Breakthrough of the year: Human genetic variation. Science. 2007;318:1842–1843. doi: 10.1126/science.318.5858.1842. [DOI] [PubMed] [Google Scholar]
- 7.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 8.Marioni JC, et al. Breaking the waves: improved detection of copy number variation from microarray-based comparative genomic hybridization. Genome Biol. 2007;8:R228. doi: 10.1186/gb-2007-8-10-r228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosta K, et al. A Bayesian approach to copy-number-polymorphism analysis in nuclear pedigrees. Am J Hum Genet. 2007;81:808–812. doi: 10.1086/520096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, et al. PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Locke DP, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.