Abstract
Methane-oxidizing bacteria (methanotrophs) attenuate methane emission from major sources, such as wetlands, rice paddies, and landfills, and constitute the only biological sink for atmospheric methane in upland soils. Their key enzyme is particulate methane monooxygenase (pMMO), which converts methane to methanol. It has long been believed that methane at the trace atmospheric mixing ratio of 1.75 parts per million by volume (ppmv) is not oxidized by the methanotrophs cultured to date, but rather only by some uncultured methanotrophs, and that type I and type II methanotrophs contain a single type of pMMO. Here, we show that the type II methanotroph Methylocystis sp. strain SC2 possesses two pMMO isozymes with different methane oxidation kinetics. The pmoCAB1 genes encoding the known type of pMMO (pMMO1) are expressed and pMMO1 oxidizes methane only at mixing ratios >600 ppmv. The pmoCAB2 genes encoding pMMO2, in contrast, are constitutively expressed, and pMMO2 oxidizes methane at lower mixing ratios, even at the trace level of atmospheric methane. Wild-type strain SC2 and mutants expressing pmoCAB2 but defective in pmoCAB1 consumed atmospheric methane for >3 months. Growth occurred at 10–100 ppmv methane. Most type II but no type I methanotrophs possess the pmoCAB2 genes. The apparent Km of pMMO2 (0.11 μM) in strain SC2 corresponds well with the Km(app) values for methane oxidation measured in soils that consume atmospheric methane, thereby explaining why these soils are dominated by type II methanotrophs, and some by Methylocystis spp., in particular. These findings change our concept of methanotroph ecology.
Keywords: atmospheric methane, methanotrophs, pmoA
Methane (CH4) is present in the atmosphere at a mixing ratio of ≈1.75 parts per million by volume (ppmv) (1). It is 20–23 times more effective as a greenhouse gas than carbon dioxide, and its atmospheric concentration over the past two centuries has increased at a rate of ≈1% per year (2). Methane-oxidizing bacteria, or methanotrophs, are crucial players in the global cycle of the greenhouse gas methane. They are strict aerobes that use methane as their only source of carbon and energy. The bacteria oxidize methane to formaldehyde, which is then either assimilated into cell biomass or further oxidized to carbon dioxide.
Cultured methanotrophs are classified into types I and II, which differ in the intracellular membrane arrangement, pathways of carbon assimilation, and phospholipid fatty acid (PLFA) composition (3). Type II methanotrophs of the genera Methylocystis and Methylosinus form a distinct clade within the Alphaproteobacteria, whereas the 10 genera currently recognized as type I methanotrophs belong to the Gammaproteobacteria. These well known type I and type II methanotrophs typically inhabit the aerobic interfaces of methanogenic environments, such as natural wetlands and rice paddies, and reduce the potential methane flux to the atmosphere (3–5). Because of the usually high methane supply in these environments, methane is oxidized with low apparent half-saturation constants (Km(app) >1 μM CH4). These Km values are similar to those determined for cultured methanotrophs (Km(app) = 2–12 μM CH4) (6, 7).
In upland soils, methanotrophic bacteria take up methane directly from the atmosphere (3, 6, 8, 9). Methane consumption in these soils is a small but a significant part of the global methane budget, comparable in magnitude to the estimated excess of emissions over sinks in recent years (29 Tg year−1) (9). In contrast to methanotrophs in the aerobic interfaces of methanogenic environments, the methanotrophs active in dry, well aerated upland soils, for example, forest soils, consume methane with high apparent affinity (Km(app) values of 0.01–0.28 μM CH4) (6). Molecular studies have indicated the presence of type II methanotrophs in most upland soils (10, 11) and have provided increasing evidence that, in these soils, the most abundant methanotrophs belong to several uncultivated groups, including upland soil cluster α (USCα) and upland soil cluster γ (USCγ) (10, 12, 13). Members of USCα and USCγ are presumed to be specialized oligotrophs adapted to living solely on atmospheric methane. The methanotrophs cultured to date, however, were thought to be incapable of surviving and remaining active at the trace level of atmospheric methane because of their low affinity for methane.
In the methane-oxidation pathway in all known methanotrophs, except Methylocella spp., the first step is mediated by particulate methane monooxygenase (pMMO) (14, 15). pMMO catalyzes the conversion of methane to methanol. This copper-dependent enzyme is synthesized even when only a minuscule amount of copper is available (16, 17). Under copper-limiting conditions, a subset of the methanotrophs produce a soluble type of MMO (sMMO) (4, 16), an enzyme evolutionarily distinct from pMMO (15). However, to initiate synthesis of pMMO under copper-limiting conditions, some methanotrophs may be able to mobilize and acquire copper from mineral and organic solid phases by releasing the fluorescent chromopeptide methanobactin (17, 18).
The operon encoding pMMO consists of three consecutive ORFs (pmoC1, pmoA1, and pmoB1; pmoCAB1). Two nearly sequence-identical copies of pmoCAB1 are found in both type I and type II methanotrophs (15, 19). Each of the pmoCAB1 operons is transcribed to form a polycistronic mRNA of 3.3 kb (16), which is translated to form the α3β3γ3 main component of functionally active pMMO, particulate methane hydroxylase (pMH) (20). Because pMMO is an integral part of the intracytoplasmic membranes, its crystal structure remained elusive for many years (20). The exact catalytic mechanism of methane oxidation is still hypothetical, involving both activation of oxygen and oxidation of methane (20, 21).
An unusual type of pMMO has recently been detected in the filamentous methanotroph Crenothrix polyspora. This gammaproteobacterium is closely related to type I methanotrophs. Comparative analysis of pmoA gene sequences, however, showed that its deduced PmoA sequence forms a lineage distinct from the monophyletic branch of type I methanotrophs (22).
The current concept of methanotrophy presumes that culturable type I and type II methanotrophs harbor a single type of pMMO. However, in Methylocystis sp. strain SC2, we recently identified, in addition to two copies of pmoCAB1, a pmoCAB-like gene cluster (pmoC2, pmoA2, pmoB2; pmoCAB2) with only a low degree of identity to pmoCAB1 at both the nucleotide (67.4–70.9%) and derived amino acid (59.3–65.6%) sequence levels (23). We have shown by using a PCR-based survey that pmoCAB2 is widely but not universally distributed among type II methanotrophs, and is not present in representative type I methanotrophs of the genera Methylobacter, Methylomicrobium, Methylomonas, Methylococcus, and Methylocaldum (24). The latter two genera are also referred to as type X methanotrophs, a subset of type I methanotrophs that is distinguished by certain physiological, biochemical, and phylogenetic characteristics (3, 25). Among a collection of 27 type II methanotroph strains, 19 strains have pmoCAB2, including most Methylocystis spp. (12 of 16), all strains of Methylosinus sporium (4 of 4), and some strains of Methylosinus trichosporium (3 of 7). pmoA2 sequences obtained from these cultured type II methanotrophs form a coherent phylogenetic cluster distinct from that of pmoA1 (24). The calculation of the relative rate of nonsynonymous (amino acid-changing) to synonymous (non-amino acid-changing) nucleotide substitutions of pmoA1 and pmoA2 sequences indicates that, in recent evolutionary history, a strong purifying selection has been acting on both genes (24). These results provided strong evidence that the gene product of pmoA2 plays an important functional role in type II methanotrophs, but the exact nature of its function remained unknown. We used Methylocystis sp. strain SC2 as a model organism to elucidate this function. Here, we demonstrate that pmoCAB1 and pmoCAB2 encode two pMMO isozymes with different methane oxidation kinetics.
Results and Discussion
Construction of Mutant Strains Defective in pmoCAB Expression.
Sequence-specific fusion PCR enabled the construction of mutant strains defective in a single copy (SC2-P1) or both copies (SC2-P2) of pmoCAB1, in pmoCAB2 (SC2-P3), and in both copies of pmoCAB1 and in pmoCAB2 (SC2-P4) (Table 1). A kanamycin- or tetracycline-resistance marker cassette replaced the target gene(s). The gene deletions and marker insertions were confirmed by diagnostic PCR (data not shown) and Southern hybridization [supporting information (SI) Fig. S1].
Table 1.
pMMO mutants generated from wild-type Methylocystis sp. strain SC2 and used in this study
| Mutant | Relevant trait(s) |
|---|---|
| SC2-P1 | ΔpmoCAB1::kan mutant, deleted in one of the two copies of pmoCAB1 |
| SC2-P2 | ΔpmoCAB1a::kan, ΔpmoCAB1b::kan mutant, deleted in both copies of pmoCAB1 (lacking pMMO1) |
| SC2-P3 | ΔpmoCAB2::kan mutant, deleted in pmoCAB2 (lacking pMMO2) |
| SC2-P4* | ΔpmoCAB1a::kan, ΔpmoCAB1b::kan, ΔpmoCAB2::tet mutant, deleted in both copies of pmoCAB1 and in pmoCAB2 (lacking both pMMO1 and pMMO2) |
*pmo null mutant: growth on methanol but not on methane.
Mutant SC2-P4, defective in both copies of pmoCAB1 and in pmoCAB2, grew on methanol but not on methane. Because Methylocystis sp. strain SC2 does not produce sMMO (4), this inability of mutant SC2-P4 to use methane provided proof that no other, unknown pmo operon was located on its genome.
Growth Experiments under High Methane Mixing Ratios.
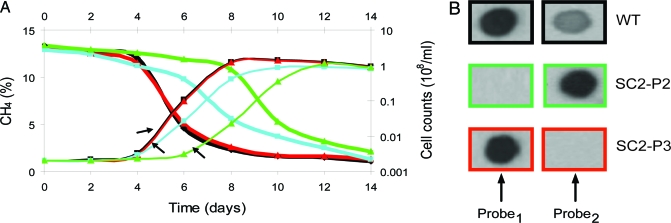
The growth of wild-type and mutants with 10–15% methane in the headspace was compared to determine whether the conventional pMMO1 protein and the novel pMMO2 protein were active (Fig. 1A). Wild-type strain SC2 and the pmoCAB2 mutant SC2-P3 grew exponentially after a lag phase of ≈4 days. Mutant SC2-P1, defective in only one copy of pmoCAB1, grew at a lower rate than wild-type strain SC2 and mutant SC2-P3, which indicates that the expression of both copies of pmoCAB1 is essential for maximum growth. Mutant SC2-P2, defective in both copies of pmoCAB1, grew after a lag phase of ≈6 days and concomitantly oxidized methane; therefore, pmoCAB2 encodes an active pMMO.
Fig. 1.
Methane consumption and growth curves of wild-type Methylocystis sp. strain SC2 and its mutants SC2-P1, SC2-P2, and SC2-P3, shown in relation to the expression of pmoCAB1 and pmoCAB2 mRNA transcripts during exponential growth on methane. (A) Methane consumption (%, vol/vol) over time (solid lines) and cell numbers calculated from OD600 measurements: wild-type (black); mutant SC2-P1, defective in a single copy of pmoCAB1 (blue); mutant SC2-P2, defective in both copies of pmoCAB1 (green); and mutant SC2-P3, defective in pmoCAB2 (red). The pmoCAB null mutant SC2-P4 and uninoculated controls were used as negative growth controls; the methane concentration did not decline in these controls. (B) pmoCAB1 and pmoCAB2 expression detected by Northern blot hybridization of total RNA of wild-type strain SC2 (WT) and mutant strains SC2-P2 and SC2-P3. Total RNA was extracted from aliquots of cultures of wild-type strain SC2 and mutant strains in the early exponential growth phase (arrows, Fig. 1A). Gene probes specifically targeted transcripts of either pmoCAB1 (Probe1) or pmoCAB2 (Probe2). For target specificity of Probe1 and Probe2, see Fig. S1.
We determined the expression of pmoCAB1 and pmoCAB2 in cultures of wild-type strain SC2 and mutants SC2-P2 and SC2-P3 in the early exponential growth phase by Northern blot hybridization of total RNA. Both pmoCAB1 and pmoCAB2 were expressed in wild-type strain SC2 during growth with 10–15% methane in the headspace, and pmoCAB1 was expressed at a higher level than pmoCAB2 (Fig. 1B). As expected, mutant SC2-P2 expressed only pmoCAB2, and mutant SC2-P3 expressed only pmoCAB1. However, the transcript level of pmoCAB2 was much higher in mutant SC2-P2 than in the wild-type; the transcript level of pmoCAB1 in wild-type and mutant SC2-P3 cells appeared to be similar.
The growth rates of mutants SC2-P1 and SC2-P2 during the early exponential phase and correspondingly their doubling times were significantly lower than those of wild-type strain SC2 and mutant SC2-P3 (Table 2). This finding substantiates the conclusion above that, for maximum growth, both copies of pmoCAB1 must be expressed. The methane oxidation rates per cell of mutants SC2-P1 and SC2-P2, in contrast, were similar to those of wild-type strain SC2 and mutant SC2-P3. Apparently, mutant SC2-P2 is in a nonnative, unbalanced metabolic state owing to the deletion of both copies of pmoCAB1. This may have changed the physiological architecture and resulted in an artificial cell response to compensate for the complete deletion of pMMO1 by increased synthesis of pMMO2. However, the increase in pmoCAB2 transcription (Fig. 1B) and subsequently in methane oxidation activity did not provide mutant SC2-P2 with the same growth rate as wild-type strain SC2 or mutant SC2-P3.
Table 2.
Methane consumption and growth rates of Methylocystis sp. strain SC2 and mutant strains SC2-P1, SC2-P2, and SC2-P3 during incubation at high methane concentrations
| Strain | Growth rate, h−1 | Doubling time, h | CH4 oxidation rate, 10−8 μmol·cell−1·h−1 |
|---|---|---|---|
| Wild-type | 0.074 ± 0.001* | 9.4 ± 0.11* | 400.6 ± 29.61* |
| 0.073 ± 0.006† | 9.6 ± 0.82† | 457.9 ± 136.12† | |
| 0.068 ± 0.001‡ | 10.1 ± 0.23‡ | 352.3 ± 29.23‡ | |
| SC2-P1 | 0.045 ± 0.002* | 15.4 ± 0.5* | 403.9 ± 27.7* |
| 0.046 ± 0.003† | 15.2 ± 1.0† | 387.6 ± 63.5† | |
| 0.049 ± 0.002‡ | 14.2 ± 0.6‡ | 468.3 ± 81.6‡ | |
| SC2-P2 | 0.041 ± 0.001* | 16.8 ± 0.2* | 381.6 ± 59.6* |
| 0.041 ± 0.002† | 17.0 ± 0.9† | 360.3 ± 50.2† | |
| 0.042 ± 0.003‡ | 16.6 ± 1.1‡ | 382.4 ± 60.6‡ | |
| SC2-P3 | 0.071 ± 0.002* | 9.7 ± 0.2* | 428.8 ± 11.01* |
| 0.072 ± 0.002† | 9.7 ± 0.3† | 441.8 ± 3.52† | |
| 0.069 ± 0.002‡ | 10.1 ± 0.3‡ | 414.5 ± 16.93‡ |
Parameters were measured during the early exponential growth phase, which occurred between days 4 and 6 after inoculation for wild-type strain SC2 and mutants SC2-P1 and SC2-P3 and between days 6 and 8 after inoculation for mutant SC2-P2 (Fig. 1).
*†‡Growth rate*, doubling time†, and methane oxidation rate‡ per cell shown for three independent growth experiments under a headspace of 10–15% (vol/vol) CH4. Data given for each replicate experiment are means of triplicates ± one standard error of mean.
Growth Experiments under Low Methane Mixing Ratios.
The differences in the growth responses of mutants SC2-P2 and SC2-P3 under high methane mixing ratios led us to assume that (i) pMMO1 and pMMO2 have different functional roles and (ii) the primary role of pMMO1 is to be active at high methane concentrations. Our assumptions were confirmed by growth experiments carried out at successively lower methane concentrations (Table 3) to determine the threshold levels of methane required for growth and survival of wild-type strain SC2 and mutants SC2-P1, SC2-P2, and SC2-P3. These long-term incubation experiments reflected in situ methane concentrations more closely than growth experiments under standard laboratory conditions.
Table 3.
Growth of wild-type Methylocystis sp. strain SC2 and mutants at various low mixing ratios of methane (1,000–1.75 ppmv), based on changes in pseudo-first-order rate constants [10−6 liter·(ml culture)−1·h−1] of methane oxidation over time
| Strain* | Initial rate constant† | Final rate constant‡ | Incubation period, weeks | Growth§ |
|---|---|---|---|---|
| 1,000 ppmv | ||||
| Wild-type | 512 ± 21 | 4,953 ± 213 | 3 | + |
| SC2-P1 | 414 ± 16 | 2,512 ± 34 | 3 | + |
| SC2-P2 | 212 ± 26 | 1,392 ± 24 | 3 | + |
| SC2-P3 | 497 ± 31 | 4,894 ± 10 | 3 | + |
| 700 ppmv | ||||
| Wild-type | 451 ± 13 | 4,248 ± 17 | 3 | + |
| SC2-P1 | 236 ± 11 | 2,135 ± 19 | 3 | + |
| SC2-P2 | 195 ± 24 | 1,358 ± 26 | 3 | + |
| SC2-P3 | 423 ± 20 | 4,193 ± 18 | 3 | + |
| 600 ppmv | ||||
| Wild-type | 384 ± 19 | 2,053 ± 32 | 3 | + |
| SC2-P1 | 351 ± 16 | 1,594 ± 25 | 3 | + |
| SC2-P2 | 289 ± 24 | 1,289 ± 27 | 3 | + |
| SC2-P3 | 296 ± 38 | <1 | 3 | − |
| 100 ppmv | ||||
| Wild-type | 351 ± 56 | 1,549 ± 24 | 5 | + |
| SC2-P1 | 339 ± 37 | 1,640 ± 100 | 5 | + |
| SC2-P2 | 210 ± 23 | 1,295 ± 19 | 5 | + |
| SC2-P3 | 342 ± 37 | <1 | 5 | − |
| 10 ppmv | ||||
| Wild-type | 102 ± 7 | 74 ± 3 | 5 | ± |
| SC2-P1 | 97 ± 5 | 82 ± 12 | 5 | ± |
| SC2-P2 | 112 ± 21 | 80 ± 14 | 5 | ± |
| SC2-P3 | 99 ± 11 | <1 | 5 | − |
| 1.75 ppmv | ||||
| Wild-type | 82 ± 12 | 11 ± 3 | 13 | − |
| SC2-P1 | 79 ± 5 | 9 ± 2 | 13 | − |
| SC2-P2 | 76 ± 6 | 12 ± 4 | 13 | − |
| SC2-P3 | 79 ± 11 | <1 | 13 | − |
Data are means of triplicates ± one standard error of mean.
*For description of strains, see Table 1. Autoclaved inoculum of the wild-type strain and viable cells of mutant SC2-P4 were used as negative growth controls.
†Initial rate constants were measured after overnight incubation at the adjusted mixing ratio.
‡Final rate constants were measured at the end of the incubation periods.
§Growth (+), decline (−), or no change (±) in the cell numbers as determined by direct cell counts (Helber counting chamber).
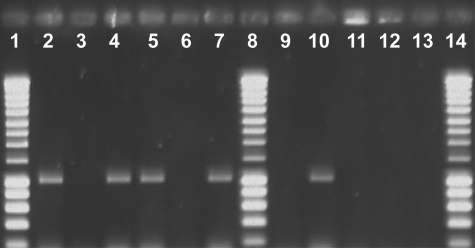
The pmoCAB1 operon was expressed by wild-type strain SC2 and mutants SC2-P1 and SC2-P3 only at mixing ratios >600 ppmv CH4 (Fig. 2). Correspondingly, the pmoCAB2 mutant SC2-P3 grew and concomitantly consumed methane at concentrations ≥700 ppmv, but did not oxidize methane at concentrations ≤600 ppmv (Table 3). Therefore, the methane concentration controls the up- and down-regulation of pMMO1, and growth and concomitant oxidation of methane by wild-type strain SC2 and mutants SC2-P1 and SC2-P2 at methane concentrations <600–700 ppmv is due only to the expression of pmoCAB2 and activity of pMMO2.
Fig. 2.
Detection of pmo mRNA transcripts in wild-type strain SC2 and mutant strains SC2-P1, SC2-P2, and SC2-P3 after a 3-week incubation under ≥700 and ≤600 ppmv CH4. RT-PCR was carried out to specifically detect mRNA transcripts of either pmoCAB1 or pmoCAB2. The expected size of the mRNA RT-PCR products was 1,075 bp for pmoCAB1 and 1,056 bp for pmoCAB2. Lanes 1, 8, and 14, DNA size markers (Smart Ladder, Eurogentec); lanes 2–7, detection of pmoCAB1 (lanes 2, 3, 5–7) and pmoCAB2 (lane 4) mRNA transcripts after a 3-week incubation under ≥700 ppmv CH4; lane 2, wild-type strain SC2; lane 3, negative control (same extract of total RNA as used for the positive control in lane 2 but without reverse transcription); lane 4, wild-type strain SC2 (same extract of total RNA from wild-type strain SC2 as in lane 2 but detection of pmoCAB2 mRNA transcripts); lanes 5–7, mutant strains SC2-P1, SC2-P2, and SC2-P3; lanes 9–13, detection of pmoCAB1 (lanes 9 and 11–13) and pmoCAB2 (lane 10) mRNA transcripts after a 3-week incubation under ≤600 ppmv CH4; lane 9, wild-type strain SC2; lane 10, wild-type strain SC2 (same extract of total RNA from wild-type strain SC2 as in lane 9 but detection of pmoCAB2 mRNA transcripts); lanes 11–13, mutant strains SC2-P1, SC2-P2, and SC2-P3 (as exemplarily shown for wild-type strain SC2, detection of pmoCAB2 was also positive for mutant strains SC2-P1 and SC2-P2 but not for SC2-P3 after a 3-week incubation under ≤600 ppmv CH4).
In good agreement with growth at high methane concentrations (Fig. 1A), cultures of wild-type strain SC2 and mutant SC2-P3 showed similarly high methane oxidation rate constants during incubation at 700 and 1,000 ppmv CH4 (Table 3). The methane oxidation rate constant of mutant SC2-P1 at these CH4 concentrations was ≈50% of that of wild-type strain SC2 and mutant SC2-P3 (Table 3). These findings suggest that growth and concomitant methane consumption of wild-type strain SC2 at methane concentrations ≥700 ppmv are mainly related to the expression of pmoCAB1 and activity of pMMO1 and that each of the two pmoCAB1 operons contribute 50% of the whole-cell methane oxidation activity.
Wild-type strain SC2 and the two mutants having an intact pmoCAB2 (SC2-P1, SC2-P2) oxidized methane at mixing ratios ≤600 ppmv and were even able to consume atmospheric methane (1.75 ppmv) for >3 months (Table 3). Growth occurred at 100 ppmv CH4, and the cell numbers did not decrease during incubation under 10 ppmv CH4. The numbers of wild-type cells and those of mutants SC2-P1 and SC2-P2 increased ≈15- to 17-fold during incubation at 100 ppmv CH4. These results suggested that cells of wild-type strain SC2 may be able to be maintained at methane concentrations as low as 10 ppmv and respond with growth when the methane concentration increases toward 100 ppmv.
After a 3-month incubation with atmospheric methane concentrations, wild-type strain SC2 and mutants SC2-P1 and SC2-P2 grew again when 20% methane was added to the headspace. The pmoCAB2 mutant SC2-P3 did not resume growth under these conditions. Atmospheric concentrations of methane were oxidized when pmoCAB2 was expressed in the wild-type strain SC2 and mutants SC2-P1 and SC2-P2, and pmoCAB1 was not expressed in these strains under these conditions (Fig. S2). These maintenance studies thus provided unequivocal proof that consumption of atmospheric methane was caused by the activity of the pMMO2, encoded by pmoCAB2. pmoCAB2 mRNA transcripts were detected in wild-type strain SC2 after incubation at all methane mixing ratios tested for the expression of pmoCAB2, including 1.75, 600, and 700 ppmv CH4, and 10–15% CH4 (Figs. 1B and 2, Fig. S2), which indicated that pmoCAB2 is constitutively expressed. Constitutive expression could be energetically favorable because regulation would require initiation of expression under limited growth conditions.
Methane Oxidation Kinetics.
The apparent kinetics of the conventional pMMO1 could not be determined by using wild-type strain SC2 because the specific affinity (as0) of pMMO1 in wild-type cells is masked by the constitutive expression of pmoCAB2 and activity of pMMO2. Km(app) of pMMO1 was thus derived from the methane oxidation activity of mutant SC2-P3. Pregrowth of mutant SC2-P3 at either 1,000 ppmv CH4 or 20% CH4 did not significantly affect Vmax(app), which suggested that methane concentrations above the threshold of 600–700 ppmv induce full expression of pmoCAB1. The as0 value was estimated from the exponential decrease of methane over time at 600–700 ppmv, resulting in a Km(app) of 9.2–9.3 μM (Table 4).
Table 4.
Apparent kinetic parameters [Km(app), Vmax(app), as0] determined for pMMO1 (mutant SC2-P3) and pMMO2 (wild-type strain SC2, mutant SC2-P2), and threshold level of methane required for maintenance
| Strain* | Km(app), μM | Vmax(app), ×10−15 mol·cell−1·h−1 | as0†, ×10−12 liter·cell−1·h−1 | CH4 for maintenance, ppmv‡ |
|---|---|---|---|---|
| Wild-type | —§ | —§ | —(pMMO1)§ | 1.75 |
| 0.11¶a | 0.11 ± 0.01¶a | 35.17 ± 2.43 (pMMO2)¶a | ||
| SC2-P2 | 0.12¶a | 0.13 ± 0.01¶a | 34.62 ± 1.65 (pMMO2)¶a | 1.75 |
| 2.20¶b | 2.41 ± 0.14¶b | 37.16 ± 1.66 (pMMO2)¶b | ||
| SC2-P3 | 9.23§a | 2.00 ± 0.11§a | 7.35 ± 0.57 (pMMO1)§a | 600–700 |
| 9.30§b | 1.86 ± 0.06§b | 6.78 ± 1.30 (pMMO1)§b |
*For description of strains, see Table 1.
†as0 describes the slope of the first-order section of a Michaelis–Menten curve and directly indicates how fast a limiting substrate is metabolized (26).
‡The minimum methane mixing ratios enabling cell maintenance for more than 3 months were determined experimentally.
§Km(app), Vmax(app), and as0 of pMMO1. Km(app) could not be determined by using wild-type strain SC2 because of the constitutive expression of pmoCAB2. Mutant strain SC2-P3 was pregrown at either 1,000 ppmv CH4 (§a) or 20% methane (§b) to estimate Vmax(app). Vmax(app) was estimated from the linear decrease of methane over time by using a starting concentration of 0.4–0.5% methane in the headspace of the test tubes. The as0 value of pMMO1 was derived from an exponential decrease of methane during incubation of mutant SC2-P3 at 600–700 ppmv CH4.
¶Km(app), Vmax(app), and as0 of pMMO2. (¶a) Wild-type strain SC2 and mutant SC2-P2 were precultured for >4 weeks at methane concentrations <500 ppmv for estimation of Vmax(app). Different starting concentrations of methane in the headspace of the test tubes had no effect on Vmax(app), regardless whether 500 ppmv CH4 or 0.4–0.5% (vol/vol) methane were initially installed. (¶b) Vmax(app) was estimated by using cells of mutant SC2-P2 growing exponentially with 20% (vol/vol) methane. Incubation of wild-type strain SC2 and mutant SC2-P2 at 50–100 ppmv CH4 resulted in an exponential decrease of methane, from which the as0 value of pMMO2 was calculated.
Km(app) of pMMO2 was derived from the methane oxidation activities of wild-type strain SC2 and mutant SC2-P2. Cells were pregrown for 4 weeks under methane concentrations <500 ppmv, thereby avoiding the expression of pmoCAB1 in the wild-type cells. The Km(app) values determined in both test strains were almost identical (0.11–0.12 μM, corresponding to mixing ratios of 107–120 ppmv) and approximately two orders of magnitude lower than the Km(app) of pMMO1 (Table 4). The specific affinity of pMMO2 was approximately fivefold higher than that of pMMO1. Apparently, pMMO2 operates in wild-type cells close to its maximum in situ activity at mixing ratios between 100 and 600 ppmv and thus below the threshold concentration of methane that induces the expression of pmoCAB1. This finding agrees well with the final rate constants measured for cultures of mutant SC2-P2 and wild-type strain SC2 after incubation at successively lower methane concentrations (Table 3) and highlights the functional role of pMMO2 in allowing growth or maintenance activity of wild-type strain SC2 when cells are exposed to low methane concentrations.
Mutant SC2-P2 exponentially growing under 20% CH4 in the headspace, however, showed a Vmax(app) for methane consumption that was ≈18- to 19-fold higher than that measured for mutant SC2-P2 cells when precultured at methane concentrations <500 ppmv (Table 4), thereby resulting in a Km(app) of 2.2 μM. The strong increase in the Vmax(app) correlates well with both the increase in the expression level of pmoCAB2 (Fig. 1B) and the high methane oxidation rate per cell observed for mutant SC2-P2 during exponential growth at high methane concentrations (Table 2). In fact, Vmax(app) under these growth conditions was similar to that measured for pMMO1 in mutant SC2-P3 (Table 4), thereby confirming that cells of mutant SC2-P2 tried to compensate for the deletion of pmoCAB1 by increasing expression of pmoCAB2.
Ecological Significance.
The as0 values of pMMO2 determined for wild-type strain SC2 and mutant SC2-P2 (Table 4) were higher than those measured for cultured methanotrophs recently tested for their response to low methane mixing ratios (7). Of the strains studied in ref. 7, Methylocystis spp. were the most oligotrophic. Two Methylocystis strains grew at 10–100 ppmv CH4 and one consumed, like strain SC2, methane at atmospheric concentrations for >3 months with little decline in the absolute rates.
Indigenous Methylocystis populations thus should have a sustained capacity for in situ uptake of atmospheric methane. In support of this conclusion, the Km(app) of pMMO2 in wild-type strain SC2 (0.11 μM) overlaps with the apparent Km values measured for methane oxidation in dry upland soils (0.01–0.28 μM, but mostly <0.1 μM CH4) (6, 10, 11). The pMMO2 thus provides a biochemical explanation for why Methylocystis spp. and Methylosinus spp. were detected in most dry upland soils studied by cultivation-independent surveys, in addition to members of USCα and USCγ. Type I methanotrophs were not detected, except for Methylocaldum-like populations (10). Although the pMMO2 presumably allows type II methanotrophs to survive in dry upland soils for extended periods by consuming atmospheric methane, the overall activity response of strain SC2 during incubation at 1.75 ppmv CH4, including the slight decline in cell numbers, implies that these methanotrophic bacteria may not be sufficiently oligotrophic for permanent activity in this type of soil. Rather, the presence of two pMMO isozymes with different thresholds for methane oxidation suggests that these organisms play an important role in consuming atmospheric methane if their growth is periodically supported by methane produced in anoxic microsites, in anoxic deep soil layers, or during temporary flooding (11, 27–29). In support of this view, Methylocystis spp. were recently found to be the most abundant methanotrophs in various hydromorphic soils (11). These soils possess an oxic surface layer and subsurface zones that are usually anoxic and methanogenic because of permanent or periodic water saturation. Hydromorphic soils are usually overall net sinks for atmospheric methane (11). Their apparent Km values (0.1–1.1 μM CH4), however, are higher than those of dry upland soils but lower than those of wetlands. PLFA-labeling experiments with 13C-methane at a low mixing ratio (30 ppmv) have shown that USCα and Methylocystis spp., but not type I methanotrophs, are active at low methane concentrations in the hydromorphic soil tested (11), thereby corroborating the hypothesis that Methylocystis spp. oxidize atmospheric methane in situ.
The existence of two pMMO isozymes with different thresholds for methane oxidation in type II methanotrophs, but not in type I methanotrophs, also has implications for our understanding of niche competition between members of these two major subgroups. Many cultured type I methanotrophs, including Methylococcus capsulatus Bath, Methylocaldum sp. E10a, and Methylobacter luteus, grow only at methane concentrations >1,000 ppmv (7). Copiotrophic growth of type I methanotrophs is corroborated by the PLFA-labeling experiments mentioned above. These organisms became active only at methane mixing ratios as high as 500 ppmv (11), which is similar to the threshold concentration we observed for the activity of the conventional pMMO1 in Methylocystis sp. strain SC2. The pMMO2 thus provides type II methanotrophs with a selective advantage, enabling them to remain in situ at methane concentrations at which most type I methanotrophs do not thrive, presumably <450–600 ppmv. Superiority of type II methanotrophs over type I methanotrophs at low methane concentrations may also be a selective advantage in natural wetlands and rice paddies, where growth is periodically restricted because of fluctuations in the methane supply.
Materials and Methods
Media, Growth Conditions, and General Methods.
NH4Cl mineral salts medium was used throughout this study (4). Methylocystis sp. strain SC2 was grown in 120-ml serum vials containing 60 ml of liquid medium for 3–5 days at 30°C under a CH4:CO2:air headspace (20:5:75, by vol). Escherichia coli strain JM109 (Promega) was maintained on Luria Bertani (LB) agar medium and grown at 37°C in LB broth supplemented with the appropriate antibiotics for plasmid DNA extraction. Genomic DNA from cell cultures of wild-type strain SC2 and mutants was extracted after mechanical lysis as described in ref. 24. Plasmid preparation and agarose gel electrophoresis followed standard protocols (30). Southern hybridization was carried out as described in ref. 31 by using a different set of probes (see Fig. S1).
Generation of pmo Mutants.
Mutants of strain SC2 were generated by knockout mutagenesis. Fusion PCR and antibiotic resistance cassettes were used to generate gene-disruption fragments that enabled specific deletion of either pmoCAB1 or pmoCAB2 by homologous recombination. The gene-disruption fragments were inserted into pUC18, a suicide vector for Methylocystis sp. strain SC2. After electroporation of strain SC2, target mutants were identified based on their resistance to kanamycin but sensitivity to ampicillin. The procedure used to generate pmo mutants is detailed in SI Methods and Tables S1 and S2.
Growth Experiments under High Methane Mixing Ratios.
Growth and concomitant consumption of methane were monitored in cultures of wild-type strain SC2 and mutant strains SC2-P1, SC2-P2, and SC2-P3. Three independent replicate experiments were carried out with three replicate cultures each. All cultures were incubated at 30°C on a rotary shaker (150 rpm) under a headspace of 10–15% CH4 (vol/vol). Starting from an OD600 of <0.001 (<106 cells ml−1) in all three growth experiments, 400-ml cultures were grown in 1,000-ml flasks to an OD600 of ≈1. Uninoculated medium was used as a blank for leakage and sterility control. Additional controls were an autoclaved inoculum of wild-type strain SC2 and viable cells of strain SC2-P4, the pmo null mutant that is able to grow on methanol but not on methane. At daily intervals, the cultures were examined for growth by measuring the OD600 (BioPhotometer, Eppendorf) and by direct cell counts (Helber counting chamber), and the concentration of methane in the headspace was measured on an SRI 8610C gas chromatograph (SRI Instruments Inc.). The cell numbers were deduced from the OD600 values by using a standard calibration curve. An OD600 value of 1 corresponded to ≈1.5 × 108 cells ml−1 in the exponential growth phase.
Northern Blot Detection of pmo mRNA Transcripts.
Total RNA was isolated from ≈1 × 109 cells of wild-type strain SC2 and mutants SC2-P2 and SC2-P3 in the early exponential growth phase as described in ref. 24. Nucleic acid extracts were treated with DNase (Promega), and total RNA was quantified photometrically. Equal amounts of total RNA (5 μg) were used in dot-blot analysis on a Hybond N membrane (Roche Diagnostics) as described in the DIG Application Manual for Filter Hybridization. RNA was UV cross-linked by using a UV Stratalinker 2400 (Stratagene) at 266 nm for 3 min. The pmoCAB1 and pmoCAB2 mRNA transcripts in wild-type strain SC2 und mutants SC2-P2 and SC2-P3 were detected by using Probe1 and Probe2, respectively (see Fig. S1 for probe design). The dot blot was first hybridized with Probe2, stripped, and then hybridized with Probe1 (Fig. 1B).
Growth Experiments under Low Methane Mixing Ratios.
In the early exponential growth phase, wild-type strain SC2 and mutants SC2-P1, SC2-P2, and SC2-P3 were diluted to 5 × 106 cells ml−1 in NH4Cl mineral salts medium. Cell suspensions (10 ml) of wild-type strain SC2 and mutants were incubated at 25°C in 120-ml serum vials on a rotary shaker (150 rpm) under methane mixing ratios ranging from 1,000 to 10 ppmv in the headspace. The methane mixing ratios used for incubation of the three replicate cultures were lowered in 100-ppmv steps to 100 ppmv, and then to 10 ppmv. At each mixing ratio tested, fresh exponentially growing cultures were used for the 3- to 5-week incubations. In addition, maintenance was assessed at 1.75 ppmv CH4 (3-month incubation). In these incubations, the methane mixing ratio did not fall <1.5 ppmv.
The various methane mixing ratios were adjusted and the incubations were carried out by using an experimental device described in ref. 7. In brief, the serum vials were connected to a 54-liter gas reservoir (2 × 27-liter plastigas bags, Linde) via tygon and iso-versinic tubes. The flow (0.2–2.0 liters·min−1) was controlled automatically by gas flow meters coupled to membrane pumps (Fuergut); thereby keeping the installed methane concentrations in the headspace of the cultures constant. Depending on the methane-oxidizing activity of the cultures, the gas reservoir in the plastigas bags was replaced at 6- to 12-h intervals for up to 3 months. Contamination was avoided by passing the in- and out-flowing air through autoclaved cotton wool. Each culture could be disconnected from the system via two-way stopcocks to measure methane oxidation activity in closed vials.
The activity of the cultures was monitored weekly by measuring methane consumption. The methane concentrations in the headspace of the vials were determined by gas chromatography. After the incubation periods, cell viability was verified by detection of pmo mRNA transcripts (see below) and growth recovery under high methane concentrations. For the latter, two 10-ml replicate cultures of wild-type strain SC2 and each mutant strain were transferred into new 120-ml serum vials, mixed with 50 ml of fresh medium, and incubated under standard laboratory growth conditions with 20% CH4 in the headspace.
RT-PCR Detection of pmo mRNA Transcripts.
Cells were harvested by centrifugation and used immediately to detect mRNA transcripts from pmoCAB1 and/or pmoCAB2. Extracts of total RNA were prepared as described in ref. 24. Reverse transcription (RT) of mRNA was carried out in a total volume of 20 μl at 56°C for 60 min by using the Omniscript RT kit (Qiagen). The reaction mixture contained 1 μl of total RNA, 0.5 mM each dNTP, reverse transcriptase buffer, 10 units of RNase inhibitor (Promega), 1.0 μM primer PmoBconv/nov, and 4 units of reverse transcriptase. The primer PmoBconv/nov enabled simultaneous cDNA synthesis from mRNA transcripts of both pmoCAB1 and pmoCAB2. Two primer sets that specifically targeted either pmoCAB1 or pmoCAB2 were used to amplify the synthesized cDNA (Table S3). The reaction mixtures (100 μl total) contained: 1 μl of RT product, 10 μl of 10× reaction buffer, 1.5 mM MgCl2, 200 μM each dNTP, 0.25 μM each primer, and 2.5 units of Taq DNA polymerase (Promega). The thermal profile was as follows: initial denaturation for 3 min at 94°C, followed by 32 cycles of denaturation at 94°C for 40 s, primer annealing at 62°C for 40 s, and elongation at 72°C for 60 s with a final extension step of 7 min. To exclude the possibility of DNA contamination, negative control reactions were carried out for all reactions in which mRNA transcripts were detected by using the same extract of total RNA without the reverse transcriptase step. Aliquots of the amplicons (10 μl) were checked by electrophoresis on a 1% agarose gel. The correct identity of the mRNA transcripts was verified by sequencing.
Methane Oxidation Kinetics [Km(app),Vmax(app), as0].
The apparent kinetic parameters of conventional pMMO1 were determined with the mutant strain SC2-P3, and parameters of the pMMO2 were determined with wild-type strain SC2 and mutant SC2-P2 by using an experimental design described in ref. 7. The Vmax(app) and as0 values of pMMO1 and pMMO2 were determined as detailed in the footnotes to Table 4 and SI Methods. The Km(app) values were calculated as Km(app) = Vmax(app)/as0. Multiplication by the Oswald constant (0.03395 at 25°C) gave the Km(app) as the methane concentration in water.
Supplementary Material
Acknowledgments.
We thank S. Fleissner for technical assistance; P. F. Dunfield, P. Frenzel, and C. Knief for helpful discussions; R. Conrad, S. N. Dedysh, and R. K. Thauer for critical reading of the manuscript and expert advice; and K. A. Brune for editing the manuscript. This work was supported in part by the Deutsche Forschungsgemeinschaft (SFB 395) and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702643105/DCSupplemental.
References
- 1.Dlugokencky E-J, et al. Atmospheric methane levels off: Temporary pause or a new steady-state? Geophys Res Lett. 2003;30:1–4. [Google Scholar]
- 2.Cicerone R-J, Oremland R-S. Biogeochemical aspects of atmospheric methane. Glob Biogeochem Cycles. 1988;2:299–327. [Google Scholar]
- 3.Hanson R-S, Hanson T-E. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heyer J, Galchenko V-F, Dunfield P-F. Molecular phylogeny of type II methane-oxidizing bacteria isolated from various environments. Microbiology. 2002;148:2831–2846. doi: 10.1099/00221287-148-9-2831. [DOI] [PubMed] [Google Scholar]
- 5.Horz H-P, Tchawa Yimga M, Liesack W. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl Environ Microbiol. 2001;67:4177–4185. doi: 10.1128/AEM.67.9.4177-4185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bender M, Conrad R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol Ecol. 1992;101:261–270. [Google Scholar]
- 7.Knief C, Dunfield P-F. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ Microbiol. 2005;7:1307–1317. doi: 10.1111/j.1462-2920.2005.00814.x. [DOI] [PubMed] [Google Scholar]
- 8.Harriss R-C, Sebacher D-I, Day F-P., Jr Methane flux in the Great Dismal Swamp. Nature. 1982;297:673–674. [Google Scholar]
- 9.Smith K-A, et al. Oxidation of atmospheric methane in Northern European soils, comparison with other ecosystems, and uncertainties in the global terrestrial sink. Glob Change Biol. 2000;6:791–803. [Google Scholar]
- 10.Knief C, Lipski A, Dunfield P-F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl Environ Microbiol. 2003;69:6703–6714. doi: 10.1128/AEM.69.11.6703-6714.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knief C, Kolb S, Bodelier P-L-E, Lipski A, Dunfield P-F. The active methanotrophic community in hydromorphic soils changes in response to changing methane concentration. Environ Microbiol. 2006;8:321–333. doi: 10.1111/j.1462-2920.2005.00898.x. [DOI] [PubMed] [Google Scholar]
- 12.Holmes A-J, et al. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–3318. doi: 10.1128/aem.65.8.3312-3318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolb S, Knief C, Dunfield P-F, Conrad R. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ Microbiol. 2005;7:1150–1161. doi: 10.1111/j.1462-2920.2005.00791.x. [DOI] [PubMed] [Google Scholar]
- 14.Dedysh S-N, et al. Methylocella palustris gen. nov., sp. nov., a new methane-oxidizing acidophilic bacterium from peat bogs, representing a novel subtype of serine-pathway methanotrophs. Int J Syst Evol Microbiol. 2000;50:955–969. doi: 10.1099/00207713-50-3-955. [DOI] [PubMed] [Google Scholar]
- 15.Murrell J-C, Gilbert B, McDonald I-R. Molecular biology and regulation of methane monooxygenase. Arch Microbiol. 2000;173:325–332. doi: 10.1007/s002030000158. [DOI] [PubMed] [Google Scholar]
- 16.Nielsen A-K, Gerdes K, Murrell J-C. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol Microbiol. 1997;25:399–409. doi: 10.1046/j.1365-2958.1997.4801846.x. [DOI] [PubMed] [Google Scholar]
- 17.Knapp C-W, Fowle D-A, Kulczycki E, Roberts J-A, Graham D-W. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proc Natl Acad Sci USA. 2007;104:12040–12045. doi: 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim H-J, et al. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 19.Semrau J-D, et al. Particulate methane monooxygenase genes in methanotrophs. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lieberman R-L, Rosenzweig A-C. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–182. doi: 10.1038/nature03311. [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian R, Rosenzweig A-C. Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase. Acc Chem Res. 2007;40:573–580. doi: 10.1021/ar700004s. [DOI] [PubMed] [Google Scholar]
- 22.Stoecker K, et al. Cohn's Crenothrix is a filamentous methane oxidizer with an unusual methane monooxygenase. Proc Natl Acad Sci USA. 2006;103:2363–2367. doi: 10.1073/pnas.0506361103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ricke P, Erkel C, Kube M, Reinhardt R, Liesack W. Comparative analysis of the conventional and novel pmo (particulate methane monooxygenase) operons from Methylocystis strain SC2. Appl Environ Microbiol. 2004;70:3055–3063. doi: 10.1128/AEM.70.5.3055-3063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tchawa Yimga M, Dunfield P-F, Ricke P, Heyer J, Liesack W. Wide distribution of a novel pmoA-like gene copy among type II methanotrophs, and its expression in Methylocystis strain SC2. Appl Environ Microbiol. 2003;69:5593–5602. doi: 10.1128/AEM.69.9.5593-5602.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowman J. The methanotrophs—The families Methylococcaceae and Methylocystaceae. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. Vol 5. New York: Springer; 2006. pp. 266–289. [Google Scholar]
- 26.Button D-K. Nutrient-limited microbial growth kinetics: Overview and recent advances. Antonie van Leeuwenhoek. 1993;63:225–235. doi: 10.1007/BF00871220. [DOI] [PubMed] [Google Scholar]
- 27.Yavitt J-B, Fahey T-J, Simmons J-A. Methane and carbon dioxide dynamics in a northern hardwood ecosystem. Soil Sci Soc Am J. 1995;59:796–804. [Google Scholar]
- 28.Andersen B-L, Bidoglio G, Leip A, Rembges D. A new method to study simultaneous methane oxidation and methane production in soils. Global Biogeochem Cycles. 1998;12:587–594. [Google Scholar]
- 29.Horz H-P, et al. Activity and community structure of methane-oxidising bacteria in a wet meadow soil. FEMS Microbiol Ecol. 2002;41:247–257. doi: 10.1111/j.1574-6941.2002.tb00986.x. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Russell D-W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2003. [Google Scholar]
- 31.Dunfield P-F, et al. Isolation of a Methylocystis strain containing a novel pmoA-like gene. FEMS Microbiol Ecol. 2002;41:17–26. doi: 10.1111/j.1574-6941.2002.tb00962.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.