Abstract
The results of genetic studies in Arabidopsis indicate that three proteins, the RNase III DICER-Like1 (DCL1), the dsRNA-binding protein HYPONASTIC LEAVES1 (HYL1), and the C2H2 Zn-finger protein SERRATE (SE), are required for the accurate processing of microRNA (miRNA) precursors in the plant cell nucleus. To elucidate the biochemical mechanism of miRNA processing, we developed an in vitro miRNA processing assay using purified recombinant proteins. We find that DCL1 alone releases 21-nt short RNAs from dsRNA as well as synthetic miR167b precursor RNAs. However, correctly processed miRNAs constitute a minority of the cleavage products. We show that recombinant HYL1 and SE proteins accelerate the rate of DCL1-mediated cleavage of pre- and pri-miR167b substrates and promote accurate processing.
Keywords: microRNA, biogenesis, Dicer, Arabidopsis
Micro RNAs (miRNAs) are ≈21-nt regulatory RNAs found in viruses, plants, and animals. miRNAs inhibit gene expression by translational repression and by pairing with their target mRNA to promote their cleavage (1, 2). miRNA regulation is known to play an important role in development, stress responses, and carcinogenesis (3, 4). miRNAs are transcribed by RNA polymerase II as long primary transcripts, termed pri-miRNA, which are capped and polyadenylated (5, 6). In animals, the pri-miRNA, which contains the miRNA sequence embedded within a hairpin, is processed in the nucleus and the cytoplasm sequentially by two RNase III-family enzymes (7). The class II RNase III Drosha, together with the dsRNA-binding protein (dsRBP) DGCR8/Pasha, cleaves the stem loop of pri-miRNA in the nucleus to a hairpin RNA (pre-miRNA) of ≈70 nt (8–11). The pre-miRNAs are transported out of the nucleus by the Ran-binding protein exportin 5 (12–14). Dicer, another class III RNase III, cleaves the pre-miRNA in the cytoplasm to release the ≈22-nt miRNA/miRNA* duplex (15–17). Like Drosha, the animal Dicer has dsRBP partners, including Loquacious/R3D1-L in Drosophila (18–20) and TRBP and PACT in mammals (21–23). There is also a Drosha-independent pathway for generating certain intronic miRNAs (24, 25).
The plant miRNA biogenesis mechanism is somewhat different from that of animals. The pri-miRNAs transcripts appear to be RNA polII transcripts in plants, as they are in animals, but the hairpins are substantially more variable in length (26, 27). Plants contain multiple Dicer homologs, termed the Dicer-like (DCL) enzymes. Of the four Dicer homologs in Arabidopsis, DCL1 carries out both cleavage steps in the nucleus to generate miRNAs (28–31). DCL2 produces 24-nt natural antisense siRNAs from complementary overlapping mRNA transcripts involved in the salt stress response (32). DCL3 generates the 24-nt DNA repeat sequence-associated siRNAs that direct heterochromatin formation (33), whereas DCL4 produces 21-nt trans-acting siRNAs that control leaf development (34–36). In addition, DCL4 and DCL2 generate 21- and 22-nt virus-derived small RNAs (vsRNA) redundantly functioning in antiviral defense (37).
Animal Dicers and plant DCL1 enzymes are large proteins of ≈220 kDa that contain an RNA helicase domain, a PAZ domain, two RNase III domains, and either one (animal) or two (plant) dsRNA-binding domains (dsRBD) (Fig. 1A). Based on the observation that pri-miRNA levels increase and pre-miRNA levels decrease in the weak dcl1-9 insertion mutant, DCL1 is believed to be required to cleave pri-miRNA to pre-miRNA (31, 38). Several lines of evidence suggest that the dsRNA-binding protein (dsRBP) HYL1 and a C2H2 zinc-finger protein SE (Fig. 1A) are DCL1 cofactors. The mature miRNA levels are low in both hyl1 and se mutants (39–42), and pri-miRNAs accumulate (38, 43). DCL1 and HYL1 recombinant proteins form a complex in vitro (43, 44), and HYL1 has been reported to interact with SE (45). DCL1 and HYL1 colocalize with DCL1 in small nuclear bodies containing pri-miRNAs, as does a fraction of the nuclear SE (38, 46). A protein complex immunoprecipitated by using anti-HYL1 antiserum has been reported to process miR169 pri-mRNAs into mature miRNAs (47). A nuclear methyltransferase, HUA ENHENCER1 (HEN1), methylates the 2′ hydroxyl group of 3′end of mature miRNAs (28, 48, 49). In addition, HASTY, the Arabidopsis homolog of exportin 5, is required for miRNA accumulation and may transport miRNA into the cytoplasm (50).
Fig. 1.
Recombinant DCL1 cleavage of dsRNA. (A) Domains of DCL1, HYL1, and SE. The 6× His, HA, and FLAG epitope tags are indicated. (B) Purified recombinant His-HA-DCL1-FLAG, His-HYL1, and His-SE proteins were fractionated on 4–12% SDS/PAGE and stained with Coomassie blue G-250. (C–F) DCL1 cleavage of dsRNA substrates. Recombinant DCL1 protein was incubated with 200 ng of a 94-bp dsRNA substrate with a 2-nt 3′ overhang (see Methods). The reaction products were phenol/chloroform purified and analyzed on 15% 8 M urea PAGE. (C) Time course of cleavage using 200 ng of DCL1. (D) dsRNA cleavage as a function of DCL1 protein concentration for 60 min. (E) The effect of NaCl concentration on dsRNA cleavage for 60 min with 200 ng of DCL1. (F) The effect of ATP and Mg2+ on DCL1 cleavage of dsRNA. EDTA (2 mM) was added to the reaction lacking the Mg2+ (last lane). The arrowheads indicates the dsRNA substrate, and the arrows indicate the 21-nt RNA products.
Although DCL1, HYL1, SE, HEN1, and HASTY have all been identified as playing a role in miRNA biogenesis in Arabidopsis, the detailed mechanism remains unknown. In the present study, we analyzed the ability of recombinant DCL1, HYL1, and SE proteins to process miRNA precursors in vitro. We show that recombinant DCL1 cleaves dsRNA, pre-miRNA, and pri-miRNA to release short RNAs of predominantly 21 nt, requiring only divalent cations and ATP. Both HYL1 and SE stimulate DCL1 activity on both pri- and pre-miRNA substrates and markedly increase the fidelity of cleavage. These results suggest that DCL1, HYL1, and SE form a heteropolymeric complex that catalyzes efficient and accurate processing of pri-miRNA to miRNA.
Results
Preparation of Recombinant DCL1, HYL1, and SE Proteins.
Recombinant His-HA-DCL1-FLAG, His-HYL1, and His-SE were expressed in insect cells. To overcome the protease cleavage routinely observed in insect cell extracts expressing recombinant DCL1, we epitope-tagged the DCL1 with a 6× His tag at the N terminus and a FLAG tag at the C terminus (Fig. 1A). We obtained intact DCL protein by a two-step affinity purification procedure. Recombinant His-HYL1 and His-SE proteins were purified by using Talon (cobalt) resin, followed by NTA/Ni resin. As judged by SDS/PAGE analysis, >90% of each purified protein preparation had the expected mobility of the intact protein (Fig. 1B). In addition, we confirmed the identity and integrity of the purified recombinant His-HA-DCL1-FLAG protein by Western blot analysis with both anti-FLAG and anti-His sera (data not shown).
Recombinant DCL1 Processes dsRNA to 21-nt RNA.
We first tested the activity of recombinant DCL1 protein using a dsRNA substrate with 2-nt 3′ overhangs. Consistent with a previous report that used immunoprecipitated DCL1 (51), we observed the release of predominantly 21-nt small RNAs in a reaction that was time- and DCL1 protein concentration-dependent (Fig. 1 C and D). The optimal salt concentration for the DCL1-mediated cleavage reaction was found to be 25–50 mM NaCl (Fig. 1E), similar to that observed for human Dicer (52). At 50 mM NaCl, DCL1 requires both a divalent cation and ATP to cleave dsRNA; addition of EDTA inhibited the reaction completely (Fig. 1F). No activity was observed in the absence of ATP, and the activity was substantially reduced when ATP was replaced by GTP (Fig. 1F). Moreover, DCL1 recombinant protein is able to hydrolyze ATP in vitro (data not shown). We conclude that DCL1 cleavage is ATP-dependent (52, 53).
Recombinant DCL1 Processes Pre-miR167b into Mature miRNA.
Because pre-miRNAs have an imperfect dsRNA hairpin structure with a 2-nt 3′ overhang at one end, we reasoned that DCL1 might itself be able to cleave the pre-miRNA to the mature 21-nt miRNA. Because miR167 level were substantially reduced in both hyl1 and se mutant plants (40, 42, 45), we chose miR167b precursor RNA as substrate. We prepared a synthetic substrate corresponding to the pre-miR167b sequence (Fig. 2A), labeled it at the 5′ end, and incubated it with DCL1. We found that recombinant DCL1 alone generates 21-nt cleavage products (Fig. 2B). However, there is genetic evidence that two additional proteins, HYL1 and SE, are required in vivo for miRNA biogenesis (38–43, 45). There is also cytological evidence that HYL1 and DCL1 consistently colocalize, whereas a fraction of SE also colocalizes with DCL1 as well as with miRNA precursors (38, 46). We therefore investigated the effect of recombinant HYL1 and SE proteins on the cleavage of pre-miR167b by recombinant DCL1 in vitro. Addition of either recombinant HYL1 or recombinant SE significantly increased the initial rate of DCL1-mediated cleavage of pre-miR167b to 21-nt small RNAs (Fig. 2 B and C). HYL1 and SE have additive effects on the initial cleavage rate, as shown in Fig. 2 B and C, which depend on the concentration of each protein (Fig. 2 D and E). Hence, HYL1 and SE stimulate the cleavage of the pre-miRNA substrate by DCL1.
Fig. 2.
HYL1 and SE facilitate DCL1 cleavage of pre-miRNA. (A) Schematic representation of the miR167b pre-miRNA substrate. (B) DCL1 cleavage of the miR167b pre-miRNA substrate. Ten nanograms of the 5′ end-labeled 93-nt pre-miRNA were incubated with 15 ng of recombinant DCL1 for the indicated time. Recombinant HYL1 (150 ng) and recombinant SE (150 ng) were added to the reactions as indicated. Reactions without recombinant proteins (first lane) and with HYL1 (150 ng) and SE (150 ng), but without DCL1, were used as controls (last lane). (C) Quantification of the miR167b small cleavage products of the reactions shown in B. The activity was defined as the relative intensity of the band corresponding to the small RNA cleavage products (quantified by using ImageJ software). The ordinate shows the ratio of the intensity of the small RNA band observed with each combination of proteins to that observed with DCL1 alone at 1 min (lane 2). (D) The effect on DCL1 activity of varying amounts of HYL1 and SE singly and together. HYL1 and SE were each added singly or together at 40, 75, and 150 ng to a reaction containing 10 ng of the pre-miR167b substrate and 15 ng of DCL1. Control reactions (lanes 1–3) contained reaction buffer only or HYL1 (150 ng) or SE (150 ng) only. All of the reactions were incubated for 2 min at 37°C. (E) The 21-nt miRNA products were quantified as described in D and expressed as a ratio of the intensity of the band obtained with each combination of proteins to that observed with DCL1 alone (lane 4). The arrowheads indicates the pre-miR167b substrate, and the arrows indicates the cleavage products. D, DCL1; H, HYL1; S, SE.
Recombinant DCL1 Processes pri-miR167b into Mature miRNA.
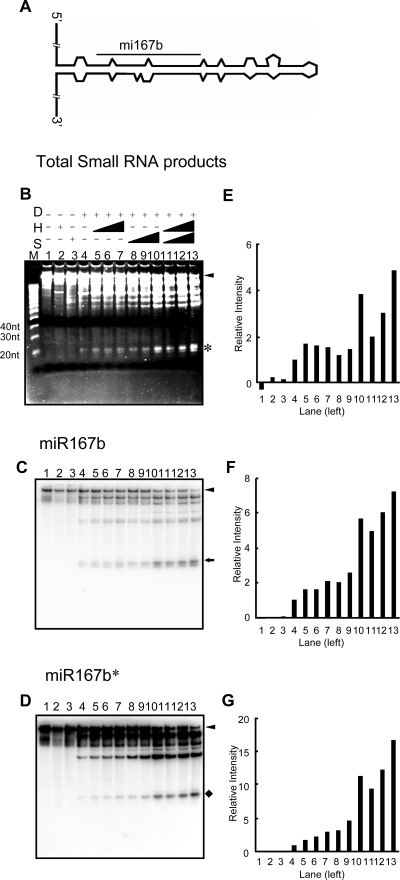
To ask whether DCL1 can process pri-miRNA, we incubated DCL1 protein with a synthetic pri-miR167b substrate containing 93-nt pre-miRNA flanked by an additional 115 and 90 nt on the 5′ and 3′ sides of the pre-miRNAs, respectively (Fig. 3A). We observed 21-nt cleavage products as well as a weaker 19-nt band (Fig. 3B, lane 4). We confirmed that the small RNA products contain both miR167b and miR167b* by Northern blot analysis with oligonucleotides (Fig. 3 C and D). To determine whether all of the cleavage products correspond to mature miR167b and miR167b*, the short digestion products were cloned and sequenced (see Methods). After removal of the sequences that do not match the pri-miR167b substrate sequence or that are shorter than 19 nt, we obtained 83 sequences. Among them, nine were identical to mature miR167b, one was a close variant of miR167b, and one was derived from the vicinity of the miR167b* sequence (Fig. 4A). We note that the small RNA databases of Arabidopsis contain, in addition to the 21-nt miR167b sequence, 20-, 22-, 23-, 24-, and 1-nt offset reads (54, 55), suggesting that the shorter and offset products also result from DCL1 cleavage. The remaining 72 sequences were derived from the other part of the pri-miR167b substrate, comprising ≈86% of the total. These sequences mostly matched the 3′ end of the pri-miR167b substrate, indicating that DCL1 alone predominantly processes the substrate incorrectly.
Fig. 3.
In vitro processing of pri-miR167b. (A) Schematic representation of the pri-miR167b substrate. (B) RNA digestion products fractionated on an 8 M urea denaturing gel and stained with Sybr gold. The pri-miR167b substrate (125 ng) was incubated with 40 ng of recombinant DCL1 protein alone (lane 4) or with increasing amounts of either HYL1 (lanes 5–7, 30 ng, 60 ng, and 120 ng) or SE (lanes 8–10, 20 ng, 40 ng, and 80 ng) alone or both together (lanes 11–13, HYL1 at 30 ng, 60 ng, and 120 ng and SE at 20 ng, 40 ng, and 80 ng). Controls (lanes 1–3) contained reaction buffer, HYL1 only (120 ng), and SE only (80 ng), respectively. (C–D) Northern blots of RNA digestion products from the gel in B probed with the P32 end-labeled anti-miR167b (C) and anti-miR167b* (D) oligonucleotides, respectively. (E) Quantification of ≈21-nt RNA products shown in B by using ImageJ software. The ordinate shows the ratio of the intensity of the small RNA band observed with various combinations of proteins to that observed with DCL1 alone (lane 4). (F and G) Quantification of 21-nt bands detected by anti-miR167b (F) or anti-miR167b* (G) probes as described in E. Arrowheads, pri-miR167b substrates; asterisk, ≈21-nt RNA products; arrows, ≈21-nt small RNAs detected by miR167b antisense probe; diamond, ≈21-nt miR167b* small RNAs detected by the miR167b* antisense probe; D, DCL1; H, HYL1; S, SE.
Fig. 4.
Effects of HYL1 and SE on the processing accuracy of DCL1. (A–D) Distribution of sequenced small RNAs from in vitro processing reactions within that of the pri-miR167b substrate. Each small bar represents a single small RNA sequence. The recombinant proteins added to each reaction is indicated; n is the number of small RNAs sequenced from each reaction shown in Fig. 3B lanes 4, 7, 10, and 13. The positions of miR167b and miR167b* are indicated by black lines. (E) Processing accuracy. Small RNA products of DCL1 cleavage were cloned and sequenced. The accuracy of processing is defined as the percentage of sequences identical to miR167b or its complement miR167b* (precise) or identical sequences together with the fraction of sequences that are either offset by 1–2 nt or are 1- to 2-nt shorter than the miR167b sequence or its complement (almost precise). Incorrect cleavage is defined as the fraction of ≈21 -nt sequences derived from other parts of the pri-miR167b sequence, predominantly corresponding to its 3′end. I and II, in vivo processing accuracy or inaccuracy at MIR167b locus from Rajagopalan deep sequencing data (55) and ASRP database (54). The accuracy of in vivo processing is defined as representation of sequences identical to miR167b or its complement miR167b* as a fraction of all small RNA sequences derived from the MIR167b locus. Incorrect cleavage is defined as the fraction of ≈21-nt sequences derived from other parts of the MIR167b locus. D, DCL1; H, HYL1; S, SE.
HYL1 and SE Increase the Accuracy of pri-miR167b Processing.
In view of the reports that the hyl1 and se mutations cause substantial reductions in the accumulation of mature miRNAs (39–41) and that miscleavage of miR163 pri-miRNA is observed in hyl1 mutant plants (43), we next asked whether recombinant HYL1 and SE proteins affect either the rate or the accuracy of DCL1 processing of pri-miR167b. We performed reactions using a fixed DCL1 concentration and increasing amounts of HYL1, SE, or both proteins. When judged by the amount of ≈21-nt RNA produced, we found that at the highest protein concentration used, HYL1 increased the amount of the cleavage product by ≈1.5-fold (Fig. 3 B and E, lanes 5–7), SE by 3.8-fold (Fig. 3 B and E, lanes 8–10), and both together by almost 5-fold (Fig. 3 B and E, lanes 11–13). When the miR167b present in the digest was estimated by using an antisense oligonucleotide probe on a Northern blot, the added HYL1 and SE proteins appeared to have an even more pronounced stimulatory effect. Quantification of the results indicates that HYL stimulated DCL1-mediated release of miR167b by 2-fold (Fig. 3 C and F, lanes 5–7), SE almost 6-fold (Fig. 3 C and F, lanes 8–10), and both together by >7-fold (Fig. 3 C and F, lanes 11–13). Quantification of miR167b* by image analysis of Northern blots shows an even more pronounced stimulatory effect of HYL1 and SE proteins (HYL1, 3-fold; SE, 11-fold; and HYL1+SE, 17-fold; Fig. 3 D and G). We also tested the influence of HYL1 and SE on the cleavage of a pri-miR171a substrate and observed a similar stimulatory effect of both proteins [supporting information (SI) Fig. S1]. These results suggested that both proteins improve the accuracy with which DCL1 processes pri-miRNA to mature miRNA.
To determine directly whether HYL1 and SE improve the accuracy of cleavage, we cloned and sequenced the small RNAs from reactions containing DCL1 and each protein alone as well as both proteins together. We determined the accuracy of cleavage by measuring the frequency with which the short RNAs correspond precisely or almost precisely to the miRNA sequence or its complement. The location of the sequenced small RNAs cleavage products in the sequence of the pri-miR167b substrate is shown in Fig. 4 A–D. Precise processing was defined as the percentage of the small RNA sequences identical to miR167b or its complement. HYL1 increased the accuracy from 11% to 22%, whereas SE increased the accuracy to 41%. When both HYL1 and SE were added to the reaction, 68% products corresponded to the miR167b sequence (Fig. 4E). If shorter 19-nt sequences and sequences offset from the miR167b sequence (or its complement) by <2 nt were included (termed “almost precise” in Fig. 4E), the accuracy increased from 13% for DCL1 alone to 26% for DCL1 + HYL1, 58% for DCL1 + SE, and 81% for DCL1 + HYL1 + SE (Fig. 4E). Conversely, the fraction of short RNA sequences derived from the end of the pri-miRNA substrate (termed “incorrect” in Fig. 4E) decreased when either HYL1 or SE or both were included in the reaction (Fig. 4E).
The in vivo processing accuracy is ≈100%, as judged by the representation of small RNAs corresponding to the mature miRNA or its complement as a fraction of all small RNAs in the small RNA deep-sequencing databases derived from the MIR167b locus (Fig. 4E) (54, 55). The miR167b* sequence is substantially underrepresented among the cloned short RNA sequences, which may be a cloning artifact, because the complementary sequence can be detected by Northern blot analysis. Correction of the error introduced by this underrepresentation would further increase the fidelity of in vitro processing of the pri-miRNA to mature miRNA by DCL1 in the presence of HYL1 and SE. Combining the increasing bias toward correct cleavage with the extent of stimulation of the cleavage rate, it appears likely that the HYL1 and SE proteins function by stimulating correct cleavage rather than by suppressing incorrect cleavage of the precursor by DCL1. We conclude that both HYL1 and SE are important in defining the cleavage sites of DCL1 on miR167b pri-miRNA, and the two proteins appear to function synergistically.
Discussion
The results of the present studies on DCL1, one of the four DCL enzymes in Arabidopsis, reveal that the plant enzyme shows significant mechanistic differences from its animal counterparts DCL and Drosha in miRNA biogenesis. It was reported that DCL1 immunoprecipitated from plant tissue extracts was able to cleave dsRNA to 21-nt small RNAs (51). We characterized the nuclease activity of purified recombinant DCL1 and found that it is able to cleave dsRNA with 2-nt 3′ overhangs to 21-nt products, requiring a divalent cation and ATP for activity.
DCL1 alone is capable of cleaving both the hairpin RNA structure of the pre-miR167b and a longer pri-miR167b containing flanking single-stranded regions to release 21-nt small RNAs. However, when we analyzed the sequence content of the small RNA digestion products by cloning and sequencing those released upon digestion of the pri-miR167b substrate with DCL1 alone, we found those corresponding to the miR167b miRNA to be a minority (13%). Based on both genetic and cytological observations that implicate the involvement of the HYL1 and SE proteins in miRNA biogenesis (38–42, 45, 46), we studied the effects on the rate and accuracy of the DCL1-mediated processing of pri-miRNA, using pri-miR167b substrates. We found that each protein increased the rate of cleavage, as well as its accuracy, as judged by the fraction of short RNA sequences that correspond nearly or exactly to miR167b and its complement miR167b*. Addition of both proteins further enhanced the rate and accuracy of processing, suggesting that the proteins act synergistically. Judging from the increase in both the rate and the accuracy of cleavage, it appears that the HYL1 and SE proteins act by stimulating correct cleavage, rather than by suppressing incorrect cleavage of the precursor.
By contrast to Drosha, which contains a single dsRBD and cannot form a stable complex with pri-miRNA (56), DCL1 has two dsRBDs and does cleave pri-miR167b in vitro. Moreover, Arabidopsis plants homozygous for the dcl1–9 mutation that disrupts the second dsRBD are defective in miRNA biogenesis (28, 29). It therefore appears likely that the presence of two dsRBDs in DCL1 is required for its ability to carry out at least the first cleavage step from pri-miRNA to pre-miRNA. The Dicer-like activity of DCL1 in the release of mature miRNA from pre-miRNA may not require both dsRBDs because human Dicer, with only one dsRBD at the C terminus, is able to process the dsRNA substrates without other cofactors (52, 53). Furthermore, the Dicer from Giardia intestinalis, which lacks a C-terminal dsRBD, is active on dsRNA substrates (57).
The dsRNA-binding proteins TRBP and PACT are required for miRNA-mediated gene silencing and are believed to help Dicer load mature miRNA into the RISC complex. However, their precise functions remain uncertain because two reports have failed to find differences in in vitro pre-miRNA processing between Dicer and the Dicer-dsRBP complex (21, 23), whereas a third showed that depletion of TRBP reduced the Dicer activity on pre-miRNAs in vitro (22). Another study demonstrated that TRBP and PACT facilitate Dicer in siRNA production in vitro (58). The results of the present study, together with those of earlier genetic studies, establish HYL1 and SE as DCL1 cofactors in pri-miRNA processing to mature miRNA. There is both in vivo and in vitro evidence that DCL1, HYL1, and SE colocalize and interact (43–46). DCL1 has been detected in association with a very large complex (>660 kDa) that may contain DCL1, HYL1 and SE (51). As judged by gel mobility-shift assays, DCL1, HYL1, and SE are each capable of binding pri-miRNAs (data not shown). Hence, HYL1 and SE may function to present the substrate to DCL1 for correct cleavage. Among animal pri-miRNAs, the distance from the bottom of miRNA-containing stem–loop structure within the pri-miRNA to the Drosha cleavage site is consistently ≈11 nt, and the dsRNA-binding protein DGCR8 is believed to measure this distance to determine the Drosha cleavage position on pri-miRNAs in generating pre-miRNA from pri-miRNA (59). The mechanism by which HYL1 and SE define the correct initial cleavage site on the pri-miRNAs for DCL1 is likely to be more complex, because the corresponding distance between the end of the stem and the initial site of cleavage is much more variable in plants than in animals. Henderson et al. (30) reported that small RNAs cloned from the dcl2 dcl3 dcl4 triple mutant form fewer families than those from wild-type plants. In other words, DCL1 tends to cleave its substrates at specific positions compared with the random cleavage by DCL2–4, which suggests that DCL1 cleaves its RNA substrate in a sequence-specific way at some frequency in vivo, as we show that it does in vitro. Nonetheless, our observation that HYL1 and SE promote correct processing of pri-miRNA by DCL1 implies that they provide additional information to stimulate the initial cleavage of pri-miRNA at the correct position for subsequent release of the miRNA by DCL1. Further studies will be required to define the precise substrate sequence and structural requirements as well as to define the nature of the interaction among DCL1, HYL1, SE, and their substrates.
Methods
Preparation of Baculovirus DCL1, HYL1, and SE Expression Cassettes for Insect Cells.
To obtain C-terminal fusions of HA-DCL1-FLAG, oligonucleotide pairs FLAG-AscI-F, FLAG-AscI-R, and HA-NotI-F, HA-NotI-R were annealed and inserted into the AscI site of plasmid pENTR-D-DCL1 and the NotI site of plasmid pENTR-d-DCL1-FLAG. Although the HA tag was not used in the purification, there is an HA tag at the N terminus of the DCL1 ORF. HA-DCL1-FLAG, HYL1, and SE fragments were inserted into pDEST10 vector by LR recombination. The Invitrogen Bac-to-Bac system was used to produce the bacmid to transform the insect cells.
Purification of Recombinant DCL1, HYL1, and SE Proteins from Insect Cells.
The His-HA-DCL1-FLAG recombinant protein was purified by two affinity columns by using Ni/NTA beads, followed by anti-FLAG beads. Recombinant His-HYL1 and His-SE proteins were purified by using Talon (cobalt) resin, followed by NTA/Ni resin.
Activity Assay.
Briefly, the RNA cleavage assays (10 μl) contained 20 mM Tris·HCl (pH 7.0), 50 mM NaCl, 4 mM MgCl2, 5 mM ATP, 1 mM GTP, 2 units of RNase inhibitor (RNaseOUT; Invitrogen), RNA substrate, and recombinant DCL1, HYL1, and SE proteins. After incubation at 37°C, the products were extracted with phenol/chloroform and precipitated. The processing products were fractionated by PAGE in a 12% acrylamide-8 M urea gel.
Cloning of RNA Cleavage Products.
The bands corresponding to oligonucleotides in the vicinity of 21 nt were eluted from gel slices and coprecipitated with glycogen. A 5′ adaptor and a 3′ adaptor were ligated to the RNA cleavage products sequentially. RT-PCR was carried out by using the adaptor-ligated RNA cleavage products. PCR fragments were cloned into the pGEM-T-easy vector (Promega) and sequenced with M13F primer. Sequences were analyzed with Vector NTI software (Invitrogen).
Supplemental Data.
See SI Text and Table S1 for detailed methods, oligonucleotide sequences, in vitro processing of pri-mir171a, and small RNA sequences cloned from in vitro processing of pri-miR167b.
Supplementary Material
Acknowledgments.
We thank M. Axtell, S. Narendra, S. Wang, and C. Vandenberg for the suggestions and discussion. This work was supported by National Science Foundation Grants IBN-0091650 and MCB-0344151 (to N.F.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 9851.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803356105/DCSupplemental.
References
- 1.Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- 2.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 4.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 9.Gregory RI. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 10.Han J. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–2167. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 13.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grishok A. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 16.Hutvagner G. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 17.Ketting RF. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forstemann K, et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chendrimada TP, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haase AD, et al. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y, et al. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The Mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie Z, et al. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch J, et al. Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 2006;140:1192–1204. doi: 10.1104/pp.105.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henderson IR, et al. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- 31.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 36.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 38.Song L, Han MH, Lesicka J, Fedoroff N. Arabidopsis primary microRNA processing proteins HYL1 and DCL1 define a nuclear body distinct from the Cajal body. Proc Natl Acad Sci USA. 2007;104:5437–5442. doi: 10.1073/pnas.0701061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez F, Gasciolli V, Crete P, Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 41.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 42.Lobbes D, Rallapalli G, Schmidt DD, Martin C, Clarke J. SERRATE: A new player on the plant microRNA scene. EMBO Rep. 2006;7:1052–1058. doi: 10.1038/sj.embor.7400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hiraguri A, et al. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 46.Fang Y, Spector DL. Identification of nuclear dicing bodies containing proteins for microRNA biogenesis in living Arabidopsis plants. Curr Biol. 2007;17:818–823. doi: 10.1016/j.cub.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu F, et al. The N-terminal double-stranded RNA binding domains of Arabidopsis HYPONASTIC LEAVES1 are sufficient for pre-microRNA processing. Plant Cell. 2007;19:914–925. doi: 10.1105/tpc.106.048637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu B, et al. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang Z, Ebright YW, Yu B, Chen X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 2006;34:667–675. doi: 10.1093/nar/gkj474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS. Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:3691–3696. doi: 10.1073/pnas.0405570102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 52.Provost P, et al. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gustafson AM, et al. ASRP: The Arabidopsis Small RNA Project Database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macrae IJ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 58.Kok KH, Ng MH, Ching YP, Jin DY. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem. 2007;282:17649–17657. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 59.Ha, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.