Abstract
Combinatorial assembly of protein domains plays an important role in the evolution of proteins. There is also evidence that protein domains have come together from stable subdomains. This concept of modular assembly could be used to construct new well folded proteins from stable protein fragments. Here, we report the construction of a chimeric protein from parts of a (βα)8-barrel enzyme from histidine biosynthesis pathway (HisF) and a protein of the (βα)5-flavodoxin-like fold (CheY) from Thermotoga maritima that share a high structural similarity. We expected this construct to fold into a full (βα)8-barrel. Our results show that the chimeric protein is a stable monomer that unfolds with high cooperativity. Its three-dimensional structure, which was solved to 3.1 Å resolution by x-ray crystallography, confirms a barrel-like fold in which the overall structures of the parent proteins are highly conserved. The structure further reveals a ninth strand in the barrel, which is formed by residues from the HisF C terminus and an attached tag. This strand invades between β-strand 1 and 2 of the CheY part closing a gap in the structure that might be due to a suboptimal fit between the fragments. Thus, by a combination of parts from two different folds and a small arbitrary fragment, we created a well folded and stable protein.
Keywords: chimeric protein, enzyme evolution, flavodoxin-like fold, protein design, TIM-barrel
Protein domains are classified into folds based on the spatial arrangement of their major secondary structure elements and their topological connections (1). However, many proteins of different folds share fragments that are structurally similar. This has led to the proposition that protein domains evolved by combinatorial assembly of smaller gene fragments, which encode intrinsically stable subunits (2, 3). Combinatorial shuffling of such subunits could have led to the diversification of domain architecture and the generation of new folds. Similar ideas have been used in an approach to build new proteins by combination of a defined protein fragment and peptides encoded by random pieces of Escherichia coli DNA (4, 5). Here, we address the question of fold similarities and modular assembly by comparing and combining protein parts of two major folds: the (βα)8-barrel and the (βα)5-flavodoxin-like fold.
The (βα)8- or TIM-barrel is a common fold among enzymes and consists of a closed eight-stranded parallel β-sheet, which forms the central barrel, surrounded by eight α-helices (6). A remarkable twofold symmetry was revealed by the x-ray structures of the N′-[(5′-phosphoribosyl)formimino]-5-aminoimidazole-4-carboxamide-ribonucleotide isomerase (HisA) and imidazole glycerol phosphate synthase (HisF) enzymes from Thermotoga maritima, which led to the hypothesis about their evolution from an ancestral “half-barrel” by gene duplication and fusion (7, 8). Construction of a (βα)8-barrel from a duplicated and fused C-terminal half of HisF and through the combination of HisA and HisF halves further suggests the evolution and diversification of (βα)8-barrel enzymes by the exchange of half-barrel domains with distinct functional properties (9, 10).
The flavodoxin-like fold is a (βα)5-fold found both as an isolated polypeptide chain and as a part of multidomain proteins. This three-layered fold is made up of a parallel five-stranded β-sheet flanked by two α-helices on one, and three on the other side. The response regulator CheY from T. maritima is one such single-domain flavodoxin-like protein whose three-dimensional structure is known (11, 12).
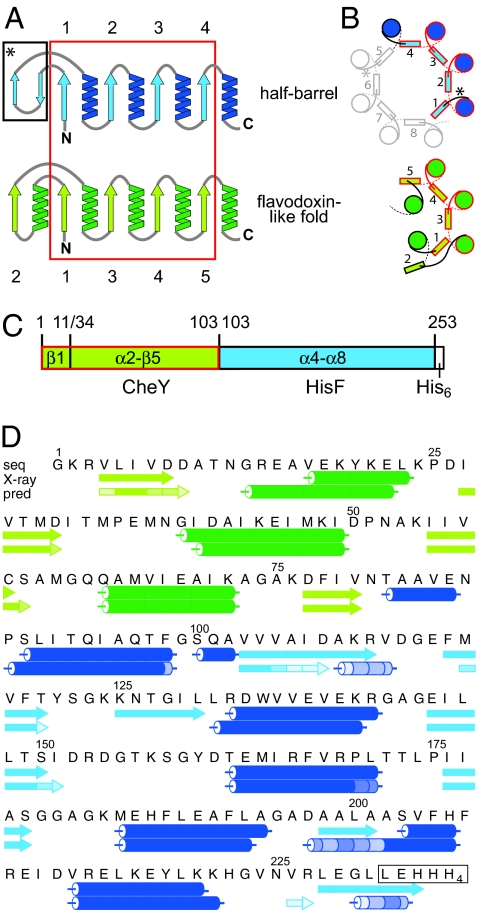
We had detected a strong structural similarity between proteins of both folds (13). When comparing the halves of HisF with all known structures in the protein data bank, we found similarities to proteins described as flavodoxin-like proteins. In particular, four of the five (βα)-elements in the flavodoxin-like fold are topologically equivalent to the half of the (βα)8-barrel fold, but the fifth element (α1β2) aligns with an antiparallel double-stranded β-sheet that is found in the loops of HisA and HisF and is not part of the central barrel (Fig. 1A). The spatial arrangement of the four (βα)-elements is similar apart from the α-helix of the last module: In the half-barrel, the α-helix stays on the same side of the β-sheet outside of the closed barrel, whereas in the flavodoxin-like fold the α-helix (α5) turns to the other side and, together with α1, shields the hydrophobic interior of the curved β-sheet (Fig. 1B).
Fig. 1.
Structural comparison of half-barrels with proteins of the flavodoxin-like fold and construction of the CheYHisF chimera. (A) Topological diagram of the half-barrels [(βα)1–4, blue] and the flavodoxin-like (βα)5-fold (green) (13). The structurally superimposable parts are encircled in red. β-strands are numbered. (B) Comparison of the spatial arrangement of the main secondary structural elements in (βα)8-barrel and flavodoxin-like proteins (top view). α-Helices are depicted as circles, and β-strands are shown as rectangles. Colors and numbering are as in A. The extra two-stranded β-sheets in the (βα)8-barrel are omitted for clarity; their positions are indicated by asterisks. (C) Construction of the CheYHisF chimera. The fragments originating from CheY (residue 1–11 including β1 and 34–103 including α2-β5) are depicted in green, and the fragment originating from HisF (residue 103 to 253 including α4-β8) is depicted in blue. (D) Amino acid sequence and secondary structure of the CheYHisF construct with the attached tag (boxed). Underneath the secondary structure elements are shown as observed in the crystal structure and as predicted by the program psipred (19). The lighter the shade of color in each element the smaller is the confidence of the prediction.
Based on these structural similarities, we constructed a chimeric protein from fragments of the two folds, testing our ability to rationally build a (βα)8-barrel through modular assembly. We decided to use HisF from T. maritima based on our knowledge of the biochemistry of its fragments obtained in previous evolutionary studies, and CheY from T. maritima because it is a single-domain protein in contrast to other flavodoxin-like proteins that had been found in our search. The C-terminal (βα)4-half of HisF with the preceding helix was fused with the parts of CheY (β1 and α2-β5) that show maximum structural similarity to the HisF structure (Fig. 1C). The resulting chimeric protein CheYHisF is a stable monomer that unfolds cooperatively. The biophysical properties of the chimera were analyzed and its structure determined at 3.1 Å resolution. The structure confirms a barrel-like fold with some distinct differences from the originating domains. This study shows how protein folds can evolve and develop anew from a combination of protein fragments in distantly related folds and how this could be exploited for protein design.
Results and Discussion
Production of a Stable Chimeric Protein by Fusing Fragments from the Flavodoxin-Like Protein CheY and the (βα)8-Barrel HisF.
The part of the cheY gene from T. maritima, encoding for β-strand 1 and the sequence from α-helix 2 to β-strand 5 (residues 1–11 and 34–103), was cloned upstream of the part of the hisF gene from T. maritima encoding for the sequence from α-helix 4 through to α-helix 8 (residues 103–253), yielding the chimeric gene cheYhisF (Fig. 1 C and D). The fragment from CheY was selected to substitute for the N-terminal part of HisF based on its remarkable structural similarity (13). The fragment boundaries were set to where the structures deviate from one another in the structural superposition. After heterologous expression of the His6-tagged chimera CheYHisF in E. coli, most of the protein was found in the insoluble fraction of the cell homogenate but could be refolded with high yields after solubilization in guanidinium chloride. Small amounts of CheYHisF could be purified from the soluble fraction via metal affinity and subsequent gel filtration chromatography, however, for higher yields the protein was purified via refolding followed by a gel filtration chromatography. All results presented here have been obtained with refolded CheYHisF.
The wild-type CheY and HisF proteins were also expressed and purified to compare the parent proteins to the chimera. His6-tagged CheY was solubly expressed in E. coli and could be purified in high yields via metal affinity and subsequent gel filtration chromatography. HisF was expressed and purified as described in ref. 14.
Biophysical Characterization of the Chimera in Comparison with Its Parent Proteins.
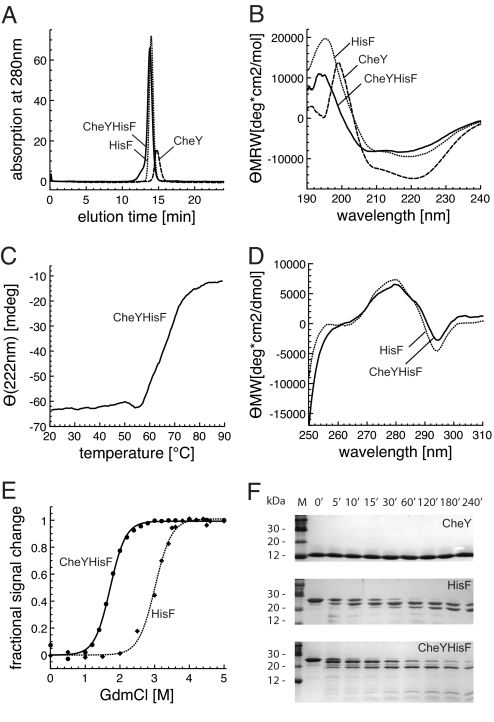
To check the oligomerization state of the chimera in solution, analytical gel filtration experiments were conducted with equal protein concentrations. CheYHisF, HisF, and CheY eluted as sharp peaks with apparent molecular masses of 30.7, 27.9, and 18.6 kDa, respectively, which correspond to the calculated molecular mass of monomeric proteins (26.5, 27.7, and 14.2 kDa, respectively) (Fig. 2A). In the CheYHisF chromatogram a small shoulder at higher molecular weight can be observed indicating the existence of minor amounts of higher association states. The weak absorbance of CheY is due to the lack of tryptophans in its sequence.
Fig. 2.
Characterization of the fold chimera CheYHisF compared with its parent proteins CheY and HisF. (A) Association states measured by analytical gel filtration. Equal amounts of each protein were loaded (0.23 mg). The main peaks of CheY, HisF, and CheYHisF correspond to molecular masses of 18.6, 27.9, and 30.7 kDa, respectively, which are equivalent to monomeric proteins. The small peak of CheY is due to its low molar extinction coefficient. (B) Secondary structure evaluation by far-UV CD spectroscopy. Protein concentrations are 0.2 mg/ml (d = 0.1 cm). The shown spectra are the mean of 10 individual spectra. (C) Stability measured by thermal denaturation. 0.05 mg/ml protein in 10 mM Tris (pH 7.5) was incubated at increasing temperatures while monitored at 222 nm in a 1-cm cuvette. (D) Tertiary structure evaluation by near-UV CD-spectroscopy. Protein concentrations are 3.4 mg/ml for HisF and 3.6 mg/ml CheYHisF (d = 1 cm). The shown spectra are the mean of 10 individual spectra. (E) Stability measured by GdmCl-induced denaturation. Proteins at concentration of 0.1 mg/ml were incubated with the given concentrations of GdmCl at room temperature. The loss of tertiary structure was followed by recording the decrease of the fluorescence emission at 324 nm (CheYHisF) and 323 nm (HisF) after excitation at 280 nm. The lines were drawn as a visual aid. All measurements were recorded in 10 mM Tris (pH7.5) at 25°C. (F) Stability measured by resistance to proteolysis. The proteins (300 μg) were incubated for the indicated time intervals with trypsin (3 μg) in 1 ml of 10 mM Tris (pH 7.5) at 37°C. The analysis was performed on 20% acrylamide Tris-Tricine gels.
The secondary structure content of the proteins was measured and compared by far-UV circular dichroism (CD) and Fourier-transform infrared (FTIR) spectroscopy. The CD spectra of all three proteins are indicative for significant α-helical content. Comparison of the CheYHisF spectrum with the spectra obtained from HisF and CheY suggests a slightly lower amount of α-helical structure in CheYHisF (Fig. 2B). A complementary quantitative estimate of the secondary structure content was derived from FTIR spectra (data not shown), using a multivariant pattern recognition method (15). We compared these values to the secondary structural contents calculated from the crystal structures of HisF and CheY, using the DSSP algorithm (16), and from this deduced an expectance value for the CheYHisF chimera. This data also indicates that CheYHisF has a lower α-helical but a similar β-sheet content than what we expect from a theoretical model based on the two parent structures [supporting information (SI) Table S1].
Tertiary structure was tested by using fluorescence and near-UV CD spectroscopy. CheYHisF and HisF contain the same single tryptophan residue in α-helix 5, which provides a convenient way to compare their tertiary structures. CheY does not contain tryptophan and therefore was not included in the comparison. The fluorescence spectra of both proteins are almost identical (data not shown), the emission maximum of CheYHisF occurs at 324 nm, which is equivalent to that of HisF (323 nm). The near-UV CD spectra of both proteins are very distinct and virtually identical (Fig. 2D). These two results indicate that the indole chromophore of the tryptophan is in a comparably asymmetric environment, and is as well shielded from solvent in CheYHisF as it is in HisF (17).
Reversible unfolding by guanidinium chloride revealed that CheYHisF is less stable than HisF, but it unfolds as cooperatively (Fig. 2E), indicating that it has a compact structure. This is in accordance with the sharp elution peak observed in the analytical gel filtration. A cooperative unfolding behavior was also observed during thermal denaturation, followed by CD at 222 nm. Although the thermal stability of CheYHisF is lower compared with the parental proteins CheY and HisF (both do not melt entirely up to 95°C), it still retains a remarkable resistance to increasing temperatures with a thermal melting point at 65°C (Fig. 2C).
Furthermore, resistance to proteolytic degradation was used to test the stability of the chimera. CheYHisF, CheY and HisF were incubated with trypsin for different periods of time, and the progress of their degradation was analyzed on Tris-N-tris(hydroxymethyl) methylglycine (Tricine) gels containing 20% acrylamide. CheYHisF was only slowly degraded in a time-dependent manner with a stable fragment of ≈20 kDa similar to HisF, whereas CheY was not cleaved at all after a 4-h treatment (Fig. 2F).
Taken together, the biophysical data shows that the CheYHisF protein has adopted a native fold that strongly resembles the sum of the individual fragments of the originating proteins.
X-Ray Crystal Structure of CheYHisF.
Protein crystals of CheYHisF were obtained in 0.1 M Mes (pH 6.5) with 1.6 M ammonium sulfate and 10% dioxane within 5 days. Freezing the crystals with 15% butanediol allowed collection of a dataset with 3.1 Å resolution with cell dimensions of a = 109, c = 80.5, and α = 90° obtained in the tetragonal space group P41212. The CheYHisF crystals contained one protein molecule in the asymmetric unit, yielding a VM of 4.6.
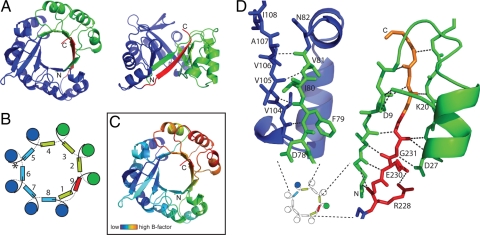
For molecular replacement, the part of the HisF crystal structure (PDB entry 1THF), which corresponds to CheYHisF (namely residues 103–253), was used as a search model. In a second step, residues 1–11 and 34–103 of CheY (PDB entry 1TMY) were placed by molecular replacement. To our surprise, we observed an additional strand within the barrel arrangement, which stems from residues of the C-terminal extension of HisF (residues 229–232) and the attached histidine-tag (residues 233–235) that invade the barrel-structure between β-strand 1 and 2 (Fig. 3 A and B). Its N-terminal end mainly interacts with β-strand 1, and its C-terminal end interacts with β-strand 2 (corresponding to β-strand 3 in wild-type CheY, Fig. 3D), completing a nine-stranded parallel barrel. Even though the structure confirms that CheYHisF is a monomeric barrel-like protein, this additional ninth strand is a feature in this construct that we had not expected. Such an arrangement has not, to our knowledge, been seen in any (βα)8-barrel-like protein, nor have more than eight parallel β-strands been observed in a βα-barrel-protein.
Fig. 3.
The x-ray crystal structure of CheYHisF at 3.1 Å resolution. (A) Ribbon diagram of the full barrel, top and side view. The part originating from HisF is shown in blue and the part originating from CheY is shown in green, and the new β-strand formed by residues of the C terminus of HisF is shown in red. (B) Schematic diagram of the secondary structure arrangement in CheYHisF. α-Helices are depicted as circles and β-strands are depicted as squares. Colors are as in A. (C) Ribbon diagram colored by B factor (dark blue to green, lower; yellow to red, higher). The diagram is shown in the same orientation as in A. The part with the highest B factor corresponds to the part originating from CheY and the new β-strand, whereas the HisF-half is less flexible in comparison. (D) Close-up of the structure at the interfaces between the fragments from CheY and HisF. The positions of the interfaces are indicated in a small copy of the schematic diagram shown in B. On the left, β-strands 4 and 5 are shown as sticks, and the connecting loops and α-helix 4 are shown as a ribbon. On the right, β-strands 1, 2, and 9 are shown as sticks, and the loops and α-helix 1 connecting strand 1 and 2 are depicted as a ribbon. In this figure only the side chains involved in hydrogen bond interactions are shown and labeled. Broken lines indicate hydrogen bonding. Green, CheY; blue, HisF; red, ninth strand; orange, tag.
The contacts between the β-strands at both CheY and HisF interfaces are typical hydrogen-bonding interactions of backbone atoms (Fig. 3D). Apart from the backbone, the side chain of residue D27 at the beginning of β-strand 2 forms a hydrogen bond with the nitrogen of G231 in β-strand 9. Furthermore, a hydrogen-bonding interaction is observed between the side chains of D9 and K20 connecting β-strand 1 and α-helix 1. These side chains partially shield a hydrophobic patch in the middle of β-strand 9. At the C terminus of β-strand 9, the loop connecting β-strand 1 and α-helix 1 further acts as a shield reducing the exposure of hydrophobic β-sheet surfaces (Fig. 3D). To test the importance of the ninth β-strand for the fold, we prepared a shortened construct with a stop codon after residue 226, thereby eliminating the strand forming residues. This variant could be purified via refolding but formed higher oligomers in contrast to the monomeric CheYHisF (data not shown). Most likely, this variant cannot form a fully closed barrel anymore, exposing more of the inner hydrophobic surface area, which leads to the observed aggregation. When comparing β-strands 1 and 2 in CheYHisF with the equivalent strands in the CheY structure, it becomes apparent that their positions are shifted toward each other in CheYHisF; this could indicate a strain somewhere in the structure that prevents the shortened CheYHisF variant from closing the barrel properly.
Another obvious feature of the structure is the high overall B factor, both in the experimental data (Wilson B factor 95.9) and the model data (average B factor of Cα-atoms 102), which indicates a high flexibility within the protein potentially due to fragile crystal packing. Altogether, the part originating from HisF is more rigid than the part originating from CheY (Fig. 3C). The highest B factors, however, are observed around the ninth β-strand and the first and second α-helices.
CheYHisF Structure in Comparison with the Structures of Its Parent Proteins.
(βα)8-barrel proteins have been described by a number of geometric parameters: They have a strand number of n = 8 and a characteristic shear number, which defines the stagger of the β-sheet, of S = 8 (18). Even though the strand number in CheYHisF is n = 9, its stagger remains S = 8 so that the tilt of the strands to the axis of the barrel is the same as in (βα)8-barrel proteins. The almost round barrel of CheYHisF has a radius of ≈7.6 Å, slightly bigger than the radius of 7.2 Å of an idealized (βα)8-barrel (18). The barrel shape of HisF is somewhat more ellipsoid than that of CheYHisF (Fig. 4). However, the curvature of the β-sheet of the HisF part in CheYHisF is exactly as in HisF. It appears that the CheY and HisF parts retain their structures but do not fit as closely together making the barrel shape rounder and leaving space for the insertion of an additional β-strand.
Fig. 4.
Comparison of the CheYHisF structure to the structures of the parent proteins. HisF (PDB entry 1THF) is shown on the left, and CheY (PDB entry 1TMY) is show on the right. The parts equivalent to CheYHisF are colored in marine blue and olive green, respectively. The remainder of the folds are in gray. The superposition of CheYHisF (colors as in Fig. 3) with the parts from HisF and CheY (colored accordingly) in the center shows remarkable structural similarities, the rms deviations are indicated at the arrows. Two close-up views are shown: the lower left shows the phosphate binding sites of HisF with PO4 and of CheYHisF filled by SO4, and the lower right shows how β-strand 9 of CheYHisF invades between β-strands 1 and 2 sitting at the original position of β-strand 1 of CheY.
Overall, the structures of the fused fragments are remarkably similar to the structures seen in the parental proteins. The HisF part in particular superimposes with an rmsd of 0.95 Å over 146 Cα atoms. This part harbors the single tryptophan residue, which we used in near-UV CD and fluorescence spectroscopy measurements to confirm tertiary structure formation. Also, the phosphate binding site is retained: A sulfate is bound in the crystal structure equivalent to the phosphate bound in HisF (Fig. 4), which shows that the part of the binding site that interacts with one phosphate moiety of the substrate is still functional. The CheY part superimposes with an rmsd of 1.11 Å over 66 Cα atoms. Only the β-strand 1, which is displaced by the C terminus of HisF, does not superimpose well (Fig. 4). This new feature could not have been predicted from our spectroscopic data. The overall amount of secondary structure does not change enough to make a measurable contribution to the far-UV CD and FTIR spectra. Further, these spectra rather point to a decreased α-helical content (Fig. 2B and Table S1), which could also be due to slight structural differences in the solution structure compared with the crystal structure.
Implications for Protein Fold Evolution.
Although a simple introduction of structurally similar parts from CheY into HisF did not lead to the expected (βα)8-barrel fold (Fig. S1), the chimera did fold into a similar, yet unobserved structure with the help of a few extra amino acid residues. The HisF and the CheY parts adopt the same spatial arrangements as within their parent proteins, behaving as independent building blocks. The interfaces fit well enough so that they also interact to form a β-sheet. However, there appears to be a constraint in CheYHisF that keeps the fragments from forming a full eight-stranded circular barrel. In the structure, we have found no obvious clash that keeps the fragments from fitting more closely, indicating that it could be a delocalized effect. In any case, the recruitment of the C-terminal residues Lys-Glu-Gly-Leu-Leu-Glu-His rescued the fold. This is especially interesting because these residues do not form a typical β-strand pattern; programs such as psipred (19) do not predict a β-strand at this position (Fig. 1D). The residues therefore have only a low strand-forming propensity. In nature, such recruitment of a small flexible fragment could have stabilized a combinatorial product and thus given it the advantage needed to evolve into a stable fold.
Because gene-recombination events happen frequently in evolving microorganisms, the question arises why we have not observed a nine-stranded βα-barrel structure like CheYHisF in nature. Possibly the geometry of a classical (βα)8-barrel structure is advantageous in terms of stability and functionality. Thus, variations of the structure might have been sampled in the course of evolution but were either outcompeted or gradually evolved into the frequently observed eight-stranded barrel.
Implications for Protein Design.
The experiment shows a potential for the design of new proteins from folding fragments of different protein folds. The protein parts were chosen solely based on structural similarity, no sequence optimization was carried out. The fragments did retain their structures within the new protein context. When comparing the geometry of CheYHisF with other (βα)8-barrel proteins, it appears that the fitting of the CheY and HisF fragments is only slightly off. How many changes are needed to convert it into a proper (βα)8-barrel will have to be investigated. However, the additional strand clearly demonstrates the difficulty in predicting the interactions in detail. Additional examples of chimeras need to be tested to elicit the general potential of this approach for the rational design of proteins. It would be interesting to shift the boundaries and to increase the amount of flavodoxin-like protein in the fold chimera to test the limits of fragment insertion. If this phenomenon is generalizable, (βα)8-barrel proteins could not only be build by the combination of half-barrels (9) but from a large repertoire of natural protein fragments each bringing their own properties to the designed chimera.
Materials and Methods
Cloning of cheY and cheYhisF.
The cheY gene was amplified from T. maritima genomic DNA, using 5′-AGC CAT ATG GGA AAG AGA GTT TTG AT-3′ with a NdeI site (in bold) as the 5′ primer and 5′-GCC CTC GAG CTT CGA AAC CTT GTT G-3′ with a XhoI site (in bold) as the 3′ primer. The gene was cloned into pET21a yielding the construct pET21a-cheY.
The cheYhisF gene was cloned in several steps. The hisF-part (fragment 1) was amplified by using the plasmid pET11c-thisF (14) as template, 5′-GAC TTC ATT GTG AAC ACG GCG GCT GTG GAG-3′ (oligo 1) as the 5′ primer, and 5′-GTG CTC GAG CAA CCC CTC CAG TCT CAC GTT-3′ with a XhoI site (in bold) as the 3′ primer. The cheY-part was constructed the following way: First, the larger part of cheY was amplified from T. maritima genomic DNA, using 5′-TAG TCG ATG ATG CAA CAA ACG GTC GTG AAG CCG-3′ as the 5′ primer and 5′-AGC CGC CGT GTT CAC AAT GAA GTC TTT CGC-3′ as the 3′ primer. A second round of amplification, using 5′-AGC CAT ATG GGA AAG AGA GTT TTG ATA GTC GAT GAT GCA ACA AA- 3′ (oligo 2) with a NdeI site (in bold) as the 5′ primer, led to the shortened cheY-construct, which lacks the nucleotides coding for residues 12 to 33 (fragment 2). The two fragments were then mixed and amplified by using oligos 1 and 2. The resulting gene cheYhisF was cloned into pET21a yielding the construct pET21a-cheYhisF. The constructs were sequenced entirely to exclude any inadvertent PCR mutations.
Heterologous Expression and Purification of Proteins.
The HisF protein was expressed and purified as described in ref. 14. The CheY and CheYHisF proteins, which both carry a His6-tag at their C termini, were produced in E. coli BL21(gold). The cells were grown at 37°C in Luria broth supplemented with 100 μg/ml ampicillin for maintenance of the plasmid. At an OD600 of 0.6, expression was induced by adding isopropyl-β-thiogalactoside to a final concentration of 1 mM, and growth was allowed for another 5 h. Cells were harvested by centrifugation, washed with 10 mM Tris (pH 7.5), and centrifuged again. CheY was mainly found in the soluble fraction of the cell extract and purified from there, whereas CheYHisF was found in the soluble and the insoluble fraction and purified from both. The cells were resuspended in 20 ml of the same buffer and lysed by sonification (Branson Sonifier W-250, 2 × 2 min, Output 5, 50% pulse, on ice), and the resulting homogenate was centrifuged (15000 rpm, 30 min, 4°C). For purification from the soluble extract, the supernatant was filtered and loaded onto a NiNTA column (Amersham Pharmacia) equilibrated with 100 mM Tris and 150 mM KCl (pH 7.5). The protein was eluted with an increasing concentration of imidazole. Fractions with the highest content of protein were dialyzed extensively against 50 mM Tris and then loaded onto a Superdex HiLoad 26/60 column (320 ml, Amersham Pharmacia), which was equilibrated with 50 mM Tris and 300 mM KCl (pH 7.5). Elution was performed in the same buffer at a flow rate of 1.0 ml/min. The eluted protein was extensively dialyzed against 10 mM Tris (pH 7.5) and used as such for characterization. CheYHisF protein found in the insoluble fraction of the cell extract was solubilized and refolded as described in ref. 7, but using Tris instead of potassium phosphate. The almost pure refolded protein was loaded onto a Superdex HiLoad 26/60 and then treated as described for the soluble fraction of the cell extract. Protein yields of CheYHisF were at least 20-fold higher when purified from the insoluble fraction. Because the protein purified from the soluble and the insoluble fraction of the cell extract behaved the same in gelfiltration, circular dichroism (CD) and fluorescence spectroscopy all other analysis was performed with refolded CheYHisF.
Analytical Methods.
Purification of the proteins was checked by electrophoresis on 15% polyacrylamide gels, using the system of Lämmli (20) and staining with Coomassie blue. Protein concentrations were determined by using molar extinction coefficients calculated from the amino acid sequence. Analytical gel filtration was performed by using a calibrated Superose 12 column (Amersham Pharmacia). The proteins (0.23 mg) were eluted at a flow rate of 0.8 ml/min in 50 mM Tris and 300 mM KCl (pH 7.5). CD spectra were recorded with a JASCO model J-810 spectropolarimeter. Fourier transform infrared (FTIR) spectra were recorded with a Bruker Tensor27 spectrophotometer equipped with an ultrathin AquaSpec cuvette. The samples were dialyzed overnight against 10 mM Tris (pH 7.5) at a protein concentrations of >3 mg/ml, and the buffer from dialysis was used for background measurements. The spectra were analyzed by using a multivariant pattern recognition method supplied by Bruker Optics (15). The fluorescence measurements were carried out with a JASCO FP-6500 spectrofluorometer. Protein unfolding induced by guanidinium chloride was followed by the decrease of the fluorescence signal at 324 nm (CheYHisF) and 323 nm (HisF) after excitation at 280 nm. The proteins were incubated with different concentrations of the denaturant, and the signals were recorded after different time intervals until no further change was observed. Temperature-induced unfolding was analyzed by following the far-UV CD signal at 222 nm at slowly increasing temperatures (at 1 and at 2 K/min). Limited proteolysis was performed in 10 mM Tris (pH 7.5) at 37°C, containing 0.003 mg/ml trypsin and 0.3 mg/ml of the test protein. The reaction was stopped after various time intervals by adding SDS/PAGE sample buffer. The time course of proteolysis was followed on 20% polyacrylmide Tris-Tricine gels (21).
Crystallization, Data Collection, and Refinement.
Crystals of native CheYHisF were initially obtained by screening against 1,200 crystallization conditions. Small crystals were obtained and further refined by the hanging drop vapor diffusion method at 18°C. Drops contained 2 μl of the protein solution mixed with 2 μl of 0.1 M Mes at pH 6.5 with 1.6 M ammonium sulfate and 10% dioxane (vol/vol) and were equilibrated against 500 μl of reservoir buffer. After short transfer into crystallization buffer with 15% butanediol, the crystals were flash frozen in liquid nitrogen. Data of single crystals was collected at the synchrotron beamline PXII (Swiss Light Source, Villigen PSI) at 100K, and single one-degree images were recorded on a MarCCD 225-mm detector. Data were indexed, integrated, and scaled with XDS and converted with XDSCONV (23). Molecular replacement searches were performed with PHASER (24), using the coordinates of the HisF part (residues 103–253 of PDB entry 1THF) and the CheY parts (residues 1–11 and 34–103 of PDB entry 1TMY).
Model building was performed with the program Coot (25), and refinement was performed with REFMAC 5.2.0019 (26), resulting in final Rcryst and Rfree values of 22.4% and 25.6%, respectively. The data collection and refinement statistics are summarized in Table S2.
Structural Superimpositions.
Structural superimpositions were performed by using the program STAMP (22).
Supplementary Material
Acknowledgments.
We thank Dr. Sergej Djuranovic for valuable suggestions, Drs. Andrei Lupas and Reinhard Sterner for comments on the manuscript, and Sooruban Shanmugaratnam for technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grant HO 4022/1-1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3CWO).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802202105/DCSupplemental.
References
- 1.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Bogarad LD, Deem MW. A hierarchical approach to protein molecular evolution. Proc Natl Acad Sci USA. 1999;96:2591–2595. doi: 10.1073/pnas.96.6.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Söding J, Lupas AN. More than the sum of their parts: On the evolution of proteins from peptides. BioEssays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 4.Riechmann L, Winter G. Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc Natl Acad Sci USA. 2000;97:10068–10073. doi: 10.1073/pnas.170145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riechmann L, Winter G. Early protein evolution: Building domains from ligand-binding polypeptide segments. J Mol Biol. 2006;363:460–468. doi: 10.1016/j.jmb.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Sterner R, Höcker B. Catalytic versatility, stability, and evolution of the (βα)8-barrel enzyme fold. Chem Rev. 2005;105:4038–4055. doi: 10.1021/cr030191z. [DOI] [PubMed] [Google Scholar]
- 7.Höcker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R. Dissection of a (βα)8-barrel enzyme into two folded halves. Nat Struct Biol. 2001;8:32–36. doi: 10.1038/83021. [DOI] [PubMed] [Google Scholar]
- 8.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 9.Höcker B, Claren J, Sterner R. Mimicking enzyme evolution by generating new (βα)8-barrels from (βα)4-half-barrels. Proc Natl Acad Sci USA. 2004;101:16448–16453. doi: 10.1073/pnas.0405832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seitz T, Bocola M, Claren J, Sterner R. Stabilisation of a (βα)8-barrel protein designed from identical half barrels. J Mol Biol. 2007;372:114–129. doi: 10.1016/j.jmb.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 11.Swanson RV, Sanna MG, Simon MI. Thermostable chemotaxis proteins from the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol. 1996;178:484–489. doi: 10.1128/jb.178.2.484-489.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Usher KC, et al. Crystal structures of CheY from Thermotoga maritima do not support conventional explanations for the structural basis of enhanced thermostability. Protein Sci. 1998;7:403–412. doi: 10.1002/pro.5560070221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Höcker B, Schmidt S, Sterner R. A common evolutionary origin of two elementary enzyme folds. FEBS Lett. 2002;510:133–135. doi: 10.1016/s0014-5793(01)03232-x. [DOI] [PubMed] [Google Scholar]
- 14.Beismann-Driemeyer S, Sterner R. Imidazole glycerol phosphate synthase from Thermotoga maritima. Quaternary structure, steady-state kinetics, and reaction mechanism of the bienzyme complex. J Biol Chem. 2001;276:20387–20396. doi: 10.1074/jbc.M102012200. [DOI] [PubMed] [Google Scholar]
- 15.Fabian H, Schultz CP. In: Encyclopedia of Analytical Chemistry. Meyers RA, editor. Chichester, UK: Wiley; 2000. p. 5799. [Google Scholar]
- 16.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 17.Schmid FX. In: Protein Structure: A Practical Approach. Creighton TE, editor. Oxford: IRL; 1997. pp. 259–295. [Google Scholar]
- 18.Murzin AG, Lesk AM, Chothia C. Principles determining the structure of beta-sheet barrels in proteins. I. A theoretical analysis. J Mol Biol. 1994;236:1369–1381. doi: 10.1016/0022-2836(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 19.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Russell RB, Barton GJ. Multiple protein sequence alignment from tertiary structure comparison: Assignment of global and residue confidence levels. Proteins. 1992;14:309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W. Automatic indexing of rotation diffraction patterns. J Appl Crystallogr. 1988;21:67–72. [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.