Abstract
To gain insight into the interaction of intracellular pathogens with host innate immune pathways, we performed an unbiased genetic screen of Listeria monocytogenes mutants that induced an enhanced or diminished host innate immune response. Here, we show that the major facilitator superfamily of bacterial multidrug resistance transporters (MDRs) controlled the magnitude of a host cytosolic surveillance pathway, leading to the production of several cytokines, including type I IFN. Mutations mapping to repressors of MDRs resulted in ectopic expression of their cognate transporters, leading to host responses that were increased up to 20-fold over wild-type bacteria, and a 20-fold decrease in bacterial growth in vivo. Mutation of one of the MDRs, MdrM, led to a 3-fold reduction in the IFN-β response to L. monocytogenes infection, indicating a pivotal role for MdrM in activation of the host cytosolic surveillance system. Bacterial MDRs had previously been associated with resistance to antibiotics and other toxic compounds. This report links bacterial MDRs and host immunity. Understanding the mechanisms through which live pathogens activate innate immune signaling pathways should lead to the discovery of adjuvants, vaccines, and perhaps new classes of therapeutics. Indeed, we show that the mutants identified in this screen induced vastly altered type I IFN response in vivo as well.
Keywords: bacterial genetic screen, immune response, interferon-beta, intracellular pathogen
Intracellular pathogens have evolved exquisite mechanisms that lead to their compartmentalization and replication within host cells (1). Conversely, the mammalian innate immune system has evolved to recognize microbial infection within different cellular compartments by using a variety of surface, vacuolar, and cytosolic receptors that recognize conserved molecules of microbial origin (2, 3). Not surprisingly, pathogens acquired counter measures to avoid and/or exploit the host innate immune system to promote their pathogenesis (2, 4). Although the field of innate immunity has received enormous attention from those interested in infectious diseases, inflammation, and adaptive immunity, much of our current understanding is still derived from studying the interaction of host cells with purified components derived from microorganisms. How host cells distinguish and respond to live, replicating, pathogenic microorganisms and nonvirulent microbes is still unclear and is central to the understanding of host–pathogen interactions.
Intracellular pathogens fall into two broad classes: those living in vacuole-like compartments and those living directly in the host cell cytosol. Listeria monocytogenes is an example of the latter that has been used as a model organism for decades to study basic aspects of both innate and acquired immunity. On entry into the host cytosol, wild-type (w.t.) strains of L. monocytogenes activate a MyD88/TRIF-independent, IRF-3/TBK1-dependent host transcriptional response that leads to transcription of dozens of genes, including robust expression and synthesis of the cytokine IFN-β (5–8). L. monocytogenes mutants that fail to access the cytosol do not activate the cytosolic surveillance pathway and do not induce IFN-β (5–8). Virulent strains of other intracellular bacterial pathogens, such as Mycobacterium tuberculosis, Legionella pneumophila, Brucellae and Francisella tularensis, also activate this pathway, whereas, in each case, avirulent mutants do not (9–12). Currently, the nature of the bacterial ligand(s) or the host receptor(s) that are involved in recognition of bacteria in the host cytosol are unknown, although nucleic acids, as well as flavone-related compounds, can activate a similar host response (9, 13, 14). To begin to understand how pathogenic bacteria activate the cytosolic surveillance pathway and its role during infection and immunity, we performed an unbiased forward genetic screen and identified L. monocytogenes mutants that induced either elevated or diminished host responses to replicating cytosolic bacteria. We found that expression of L. monocytogenes multidrug resistance transporters (MDRs) of the major facilitator superfamily of bacterial MDRs controlled activation of the host cytosolic surveillance pathway both in vitro and in vivo.
Results and Discussion
Identification of L. monocytogenes Mutants That Differentially Induce IFN-β in Macrophages.
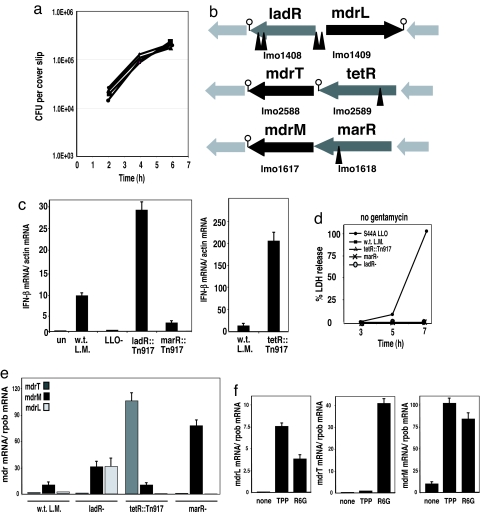
To address how L. monocytogenes activates the host cytosolic surveillance system, we screened a L. monocytogenes Tn917 transposon library (15) for mutants that exhibited an enhanced or diminished type I IFN response upon infection of macrophages. Approximately 5,000 L. monocytogenes::Tn917 mutants were used to infect bone marrow-derived macrophages (BMM) in 96-well plates. The amount of type I IFN (i.e., IFN-β and/or IFN-α) secreted by macrophages during infection was measured by transferring macrophage culture supernatant onto a type I IFN reporter cell line that produces luciferase in response to type I IFN (16). We identified 17 mutants that induced altered induction of type I IFN compared with w.t. bacteria. Among these, six mutants, with transposon insertions located in three different genes, behaved like w.t. in their ability to infect, escape from a vacuole, and grow inside macrophages as shown by their intracellular growth curves (Fig. 1a). Transposon insertions in these mutants were located in genes encoding predicted transcription regulators (Fig. 1b): ladR, previously shown to be a negative regulator of its adjacent multidrug resistance transporter, MdrL (17); lmo2589 encoding a TetR-like protein; and lmo1618 encoding a MarR-like protein (18). Real-time qRT-PCR analysis of IFN-β induction in macrophages infected with these mutants confirmed that the ladR mutant induced 3-fold more IFN-β, the tetR mutant induced 20-fold more IFN-β, and the marR mutant induced 3-fold less IFN-β compared with the level of IFN-β induced by w.t. bacteria [Fig. 1c and supporting information (SI) Fig. S1]. Although the three mutants affected the level of IFN-β in macrophages, none of them induced macrophage cell death as shown by a lactate dehydrogenase (LDH) release assay (Fig. 1d).
Fig. 1.
L. monocytogenes strains with mutations in regulators of multidrug resistance transporters induce altered host IFN-β responses. (a) Intracellular growth curves of w.t. L. monocytogenes (filled square), ladR::Tn917 (filled triangle), tetR::Tn917 (large filled circle), and marR::Tn917 (small filled circle), in bone marrow-derived macrophages (BMMs) (31). (b) Schematic presentation of site of transposon insertions (marked with triangles), mapped to genes predicted to be transcription regulators. (c) Quantitative RT-PCR (qRT-PCR) analysis of IFN-β gene induction in BMM in response to infection with w.t. L. monocytogenes, ladR::Tn917, tetR::Tn917, and marR::Tn917. (d) Lactate dehydrogenase (LDH) release assay was performed on macrophages infected with w.t. L. monocytogenes, tetR::Tn917, marR−, and ladR− mutants at various time points postinfection. L. monocytogenes cytotoxic LLO mutant S44A (32, 33) was used as a positive control. (e) qRT-PCR analysis of L. monocytogenes MDR transporters expression in w.t. bacteria, ladR−, tetR::Tn917, and marR− mutants grown to midlog in BHI broth. (f ) qRT-PCR analysis of MDRs expression by w.t. L. monocytogenes in the presence of the toxic compounds tetraphenylphosphonium (TPP) or rhodamine 6G (R6G). All error bars represent one standard deviation; n = 2 or 3.
LadR, TetR, and MarR Are Repressors of MDRs.
This is the first description of the tetR and marR genes in L. monocytogenes. Interestingly, like the ladR transcription regulator, the tetR and marR regulators were located adjacent to putative multidrug resistance transporters of the major facilitator superfamily, named here mdrT and mdrM, respectively (lmo2588, lmo1617) (Fig. 1b). Among the three MDRs, MdrM and MdrT are highly similar (46% amino acid identity and 64% similarity) and share similarity with the well studied multidrug efflux transporter system, QacA-QacR, of Staphylococcus aureus (18). In S. aureus, QacR represses expression of the MDR qacA. To study the regulation of mdrL, mdrT, and mdrM expression by their adjacent regulators and their effect on the cytosolic innate immune response, we generated a series of in-frame deletions (19) of the regulator genes, the MDR genes, and a double deletion of each MDR-regulator set of genes (Table 1). Unfortunately, for reasons that are still not clear, we were unable to generate in-frame deletions of the tetR gene or the double tetR-mdrT genes; thus, for the rest of the study, we used the original transposon tetR::Tn917 mutant. The expression level of each MDR was analyzed by real-time qRT-PCR from bacteria grown in broth. We found that w.t. L. monocytogenes did not express mdrL or mdrT, but expressed a measurable level of mdrM (Fig. 1e). In the ladR mutant the multidrug transporter, mdrL, was highly induced (≈30-fold) (17). In addition, mutation in the ladR gene resulted in ≈3-fold induction of mdrM, compared with its basal level of expression (Fig. 1e). In the tetR::Tn917 mutant, the adjacent multidrug transporter, mdrT, was specifically and highly induced (≈100-fold) (Fig. 1e). In the case of the marR regulator, the mdrM gene was located downstream of marR, and both genes were predicted to be part of an operon (Fig. 1b). Although mdrM was not expressed in the original marR::Tn917 mutant (data not shown), it was highly induced in the marR in-frame deletion (≈70-fold) (Fig. 1e), suggesting that the transposon insertion blocked the expression of both genes because of polarity. These results clearly demonstrated that LadR, TetR, and MarR are negative regulators of the putative MDRs MdrL, MdrT, and MdrM, respectively.
Table 1.
Listeria monocytogenes strains generated in this study
| L. monocytogenes strain | Description | IFN-β induction/w.t. L. monocytogenes |
|---|---|---|
| 10403S | Wild type | 1 |
| DP-L5396 | ladR::Tn917 | 3 |
| DP-L5418 | marR::Tn917 | 0.3 |
| DP-L5397 | tetR::Tn917 | 20 |
| DP-L5523 | w.t. 10403S + pLIV2:mdrT | 3.5 |
| DP-L5441 | ladR− | 3 |
| DP-L5445 | marR− | 6 |
| DP-L5444 | mdrM− | 0.3 |
| DP-L5516 | mdrM− + pLIV2:mdrM | 1 |
| DP-L5446 | mdrT− | 1 |
| DP-L5442 | mdrL− | 1 |
| DP-L5448 | marR−/mdrM− | 0.3 |
| DP-L5443 | ladR−/mdrL− | 3 |
Listed for each strain is the relative level of IFN-β induced by host macrophages, compared with the level of IFN-β induced by w.t. L. monocytogenes.
One common property of MDRs is that their expression is often induced by the presence of their cognate drug substrates (18, 20). For example, in the QacA-QacR system, the repression of qacA imposed by QacR is relieved when QacR binds toxic drugs, leading to induction of qacA expression (18, 20). When w.t. L. monocytogenes was grown in the presence of the commonly used toxic drugs, tetraphenylphosphonium (TPP) or rhodamine 6G (R6G) (18), the transcription of the three MDRs was highly induced (Fig. 1f), suggesting that the regulator genes identified in this screen were involved in the regulation of MDR transporters.
Expression of L. monocytogenes Multidrug Resistant Transporters Controls the Induction of IFN-β in Macrophages.
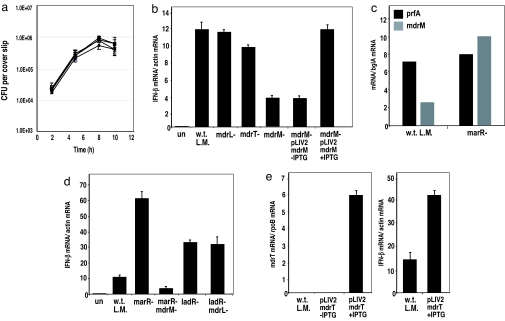
To evaluate the role of each MDR in the induction of type I IFN, we infected macrophages with w.t. L. monocytogenes and the MDR mutants, and analyzed the induction of IFN-β at 4 h postinfection by real-time qRT-PCR. The results clearly demonstrated that among the three MDRs, MdrM was the only one necessary for induction of IFN-β, because this mutant induced only a third of the IFN-β induced by w.t. bacteria (Fig. 2b). This was not due to a growth defect, because all 3 MDR deletion mutants were able to infect and replicate within macrophages like w.t. bacteria (Fig. 2a). Complementation of mdrM expression with an IPTG-inducible expression system (pLIV2 integration vector) (21) restored the induction of IFN-β to the level induced by w.t. L. monocytogenes (Fig. 2b). Further analysis of mdrM expression during infection revealed that mdrM is expressed intracellularly, but, as in broth, its expression is well below the level of the marR deletion mutant (Fig. 2c). These results suggested that this basal expression of mdrM accounted for the majority of IFN-β expression.
Fig. 2.
Role of the L. monocytogenes multidrug resistance transporters mdrL, mdrT, and mdrM in the induction of IFN-β in macrophages. (a) Intracellular growth curves of w.t. L. monocytogenes (filled circle) and the deletion mutants: mdrL− (filled square), mdrT− (×), and mdrM− (filled diamond) in BMMs (31). (b) qRT-PCR analysis of IFN-β induction in BMMs infected with w.t. L. monocytogenes, mdrL−, mdrT−, mdrM−, and a complemented mdrM− strain expressing mdrM from the IPTG-inducible vector pLIV2 (21). (c) qRT-PCR analysis of MdrM expression by w.t. L. monocytogenes and marR mutant intracellularly at 4 h postinfection. (d) qRT-PCR analysis of IFN-β induction in BMMs infected with ladR−, marR− or the double deletions of ladR−/mdrL− or marR−/mdrM−. (e) qRT-PCR analysis of mdrT expression level in w.t. L. monocytogenes bacteria containing IPTG-inducible plasmid pLIV2::mdrT, and analysis of IFN-β induction by this strain in infected BMMs. All error bars represent one standard deviation; n = 2 or 3.
Overexpression of mdrM, or its related MDR, mdrT, caused massive expression of IFN-β. The marR deletion mutant, which overexpressed mdrM (Fig. 1e), induced 6-fold more IFN-β than w.t. bacteria (Fig. 2d). This level of IFN-β induction was completely dependent on mdrM expression because it was not observed with the marR-mdrM double-deletion mutant, which induced the same level of IFN-β as the mdrM mutant alone, thereby excluding a potential role for other MarR-inducible genes (Fig. 2 b and d). Further support for the role of MdrM in IFN-β induction came from infecting macrophages with the ladR− mutant. As shown in Fig. 1e, LadR also repressed the expression of mdrM, although to a lesser extent than the MdrM repressor, MarR. Infecting macrophages with the ladR− mutant resulted in 3-fold higher induction of IFN-β than with w.t. bacteria; however, infection with the double-deletion ladR-mdrL mutant still induced 3-fold more IFN-β then w.t. bacteria, suggesting that this induction was not due to the expression of mdrL (Fig. 2d). Microarray analysis comparing total gene expression of w.t. bacteria versus the ladR− mutant revealed that, besides mdrL, mdrM was the most differentially expressed gene in the ladR− mutant (Table S1 and SI Methods). Because mdrM overexpression in the marR− mutant resulted in enhanced host IFN-β expression (Figs. 1e and 2d), it is most likely that the induction of IFN-β by the ladR− mutant was because of overexpression of mdrM and not mdrL. Overall, these results demonstrated a direct role for MdrM in activation of IFN-β in response to L. monocytogenes infection. Interestingly, w.t. bacteria expressing IPTG-inducible MdrT (MdrM homolog) also resulted in increased induction of IFN-β in infected macrophages (Fig. 2e). These observations strongly suggest that the induction of IFN-β was not restricted to MdrM, but could be recapitulated by expression of homologous MDRs, likely with similar substrate specificity.
L. monocytogenes MDRs Control the Magnitude of the Host Cytosolic Response.
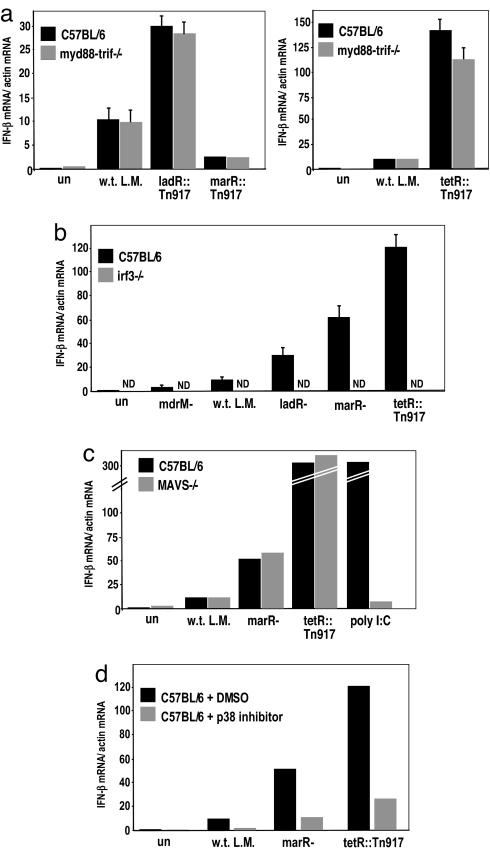
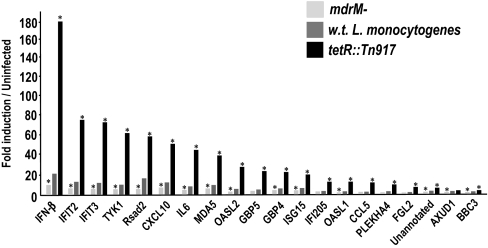
How immune cells recognize intracellular pathogens such as L. monocytogenes is not fully understood. The cytosolic innate immune response to L. monocytogenes is generally described as independent of Toll-like receptors (TLRs) and their signaling adaptors, Myd88 and Trif, and dependent on p38 MAPK signaling and the IFN regulatory factor 3 (IRF-3) (5, 7). To test whether induction of IFN-β by the mutants identified in this screen activated the same pathway, we infected Myd88-Trif double-knockout macrophages, IRF-3 deficient (22) macrophages, and macrophages treated with a p38 MAPK inhibitor with ladR, marR, and tetR mutants. We found that the increased induction of IFN-β by these mutants was almost entirely independent of TLRs, largely dependent on p38 MAPK, and absolutely dependent on IRF-3 (Fig. 3a, b, and d). One well characterized cytosolic pathway that leads to IRF-3 activation and IFN-β expression depends on the cytosolic receptors RIG-I and MDA-5 and their adaptor, MAVS (23–25). We infected MAVS-deficient macrophages with w.t. L. monocytogenes, marR−, and tetR::Tn917 mutants, and the induction of IFN-β by w.t. L. monocytogenes and the mutants was independent of MAVS (Fig. 3c) (25, 26). These results are consistent with the hypothesis that w.t. L. monocytogenes and the mutants induced altered levels of activation of the same host cytosolic surveillance pathway. To gain further insight into the host pathways and downstream genes activated by bacterial MDRs, we compared the macrophage response to infection with w.t. L. monocytogenes, mdrM− and tetR::Tn917 mutants by using microarray analysis. We used type I IFN receptor minus (IFNαβR−/−) macrophages to avoid the complication of IFN-β autocrine signaling. Macrophages infected with the mdrM− mutant, which induced a 3-fold lower host IFN-β response, had altered expression of only 16 genes (by SAM analysis), all of which were diminished compared with macrophages infected with w.t. L. monocytogenes. Macrophages infected with the tetR::Tn917 mutant, which induced a 20-fold higher IFN-β response, had strongly increased induction (by SAM and at least 4-fold) of 13 genes, compared with macrophages infected with w.t. L. monocytogenes. Interestingly, the genes whose expression was affected by mdrM− and tetR::Tn917 mutants largely overlapped and are presented in Fig. 4. Moreover, the vast majority of these genes were previously identified as “cytosolic response genes” (i.e., genes that are induced only by w.t. L. monocytogenes in the cytosol) and included IFN-β, IL-6, CCL5, and CXCL10 (13). Like IFN-β, the expression of these genes appears to be almost entirely IRF-3-dependent (Fig. S2). These experiments provided further evidence that bacterial MDR expression specifically controlled the magnitude of the host cytosolic response to L. monocytogenes, which includes a range of host genes, in addition to IFN-β.
Fig. 3.
Induction of IFN-β by ladR, tetR, and marR mutants is independent of MyD88/Trif and MAVS but dependent on IRF-3 and p38 MAPK. qRT-PCR analysis of IFN-β induction in C57BL/6 w.t. vs. myd88/trif−/− BMMs (a), and C57BL/6 w.t. vs. irf3−/− BMMs (22) (b) infected with w.t. L. monocytogenes, and ladR, marR, tetR::Tn917 regulator mutants. (c) qRT-PCR analysis of IFN-β induction in C57BL/6 BMMs vs. MAVS−/− BMMs (25) infected with w.t. L. monocytogenes, marR−, tetR::Tn917, or transfected with poly[I:C] as a positive control. (d) qRT-PCR analysis of IFN-β induction in C57BL/6 BMMs treated with either DMSO or 10 μM SB202190 (p38 MAPK inhibitor) (5). All error bars represent one standard deviation; n = 2 or 3.
Fig. 4.
L. monocytogenes MDR expression determines the magnitude of the host immune response. Genes identified by microarray analysis as having lower expression in mdrM− infected IFNαβR−/− macrophages (by SAM) or higher expression in tetR::Tn917 infected IFNαβR−/− macrophages (by SAM and at least 4-fold higher than w.t.), as compared with their expression in w.t. L. monocytogenes infected IFNαβR−/− macrophages. All data are represented as fold induction over uninfected macrophages and are the average of two experiments. * indicates that these values are significantly different from macrophages infected with w.t. L. monocytogenes, by SAM analysis with a false discovery rate of 10%.
In Vivo Analysis of L. monocytogenes MDR Mutants.
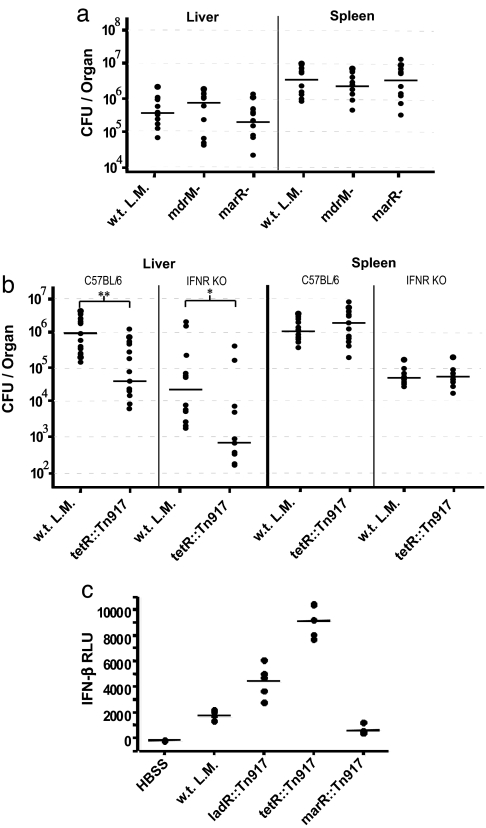
To test the role of the cytosolic surveillance response in the host's defense to L. monocytogenes infection, we infected mice with w.t. L. monocytogenes, mdrM−, marR−, and tetR::Tn917 mutants. Mice infected with the mutant that induced 20 times more IFN-β, tetR::Tn917, had 20-fold lower bacterial loads in the liver, whereas the other mutants had w.t. levels of bacteria, suggesting that only a drastic change in activation of the cytosolic surveillance pathway had a measurable effect on host resistance (Fig. 5 a and b). Although the tetR::Tn917 mutant induced stronger activation of the entire cytosolic response (Fig. 4 and Fig. S2), we specifically tested the role of IFN-β in the increased clearance of the tetR::Tn917 mutant by using IFNαβR−/− mice. Interestingly, IFNαβR−/− mice were still more resistant to tetR::Tn917 than w.t. L. monocytogenes, suggesting that the increased host resistance may not be solely because of the high expression of IFN-β. Instead, the increased host resistance is likely due to a combination of genes in the enhanced cytosolic response. Although previous studies suggested that host induction of IFN-β might be beneficial to L. monocytogenes (22, 27), the results in this study are consistent with a model in which L. monocytogenes avoids excessive activation of the host cytosolic surveillance system.
Fig. 5.
Effect of MDR mutations on in vivo induction of IFN-β and L. monocytogenes virulence. (a) C57BL/6 mice infected with 1 × 104 (0.1 LD50) of w.t., mdrM−, or marR− L. monocytogenes. Organs were collected 48 h postinfection, and bacterial numbers are represented as colony forming units (cfu) per organ, n = 10 mice per strain. (b) C57BL/6 or IFNαβR−/− mice were infected with 1 × 104 of w.t. or tetR::Tn917 L. monocytogenes and processed and analyzed as described for a, n = 15 (C57BL/6) or 12 (IFNαβR−/−) mice per strain. (c) Detection of type I IFN levels in serum of Balb/C mice infected i.v. (1 × 104 bacteria) with Hank's Buffered Salt Solution (HBSS), w.t. L. monocytogenes, ladR::Tn917, tetR::Tn917, or marR::Tn917 for 24 h. Units are presented as relative light units (RLU), detected by luciferase reporter ISRE-L929 cell line assay (16), n = 5 mice per strain. All median values are represented by horizontal lines. Statistical significance was determined by nonparametric Mann–Whitney test. **, P = 0.001; *, P = 0.007.
This report demonstrates a role for bacterial MDR transporters in the activation of a host immune response. MDRs are known to bind and transport a broad range of structurally unrelated compounds. We propose that MDR-mediated transport of bacterial ligands to the host cytosol triggers the host cytosolic surveillance system. However, we cannot rule out that the MDR proteins by themselves are the stimulatory ligands for the host immune system, although we have shown that high expression of one MDR, mdrL, had no effect on the activation of the cytosolic surveillance system (Fig. 2d). The results of this study indicate that the host immune system can detect a live, virulent intracellular pathogen by recognition of the pathogen's own defense mechanism to toxic molecules. Interestingly, other intracellular bacterial pathogens, including Mycobacterium tuberculosis, Brucella, and Legionella pneumophila, also activate a similar host response (9, 10, 12), although for these pathogens, activation requires an auxiliary secretion system. Perhaps L. monocytogenes MDRs and these auxiliary secretion systems release the same or related molecules into the host cytosol, resulting in activation of the host cytosolic surveillance pathway.
We generated L. monocytogenes strains that vary by 60-fold in the amount of IFN-β and >5- or 10-fold in the amount of IL-6, CCL5, and CXCL10 induced in infected macrophages, because of their levels of MDR expression (Figs. 3b and 4; Fig. S2). Importantly, the activation of the cytosolic surveillance system in infected animals, as measured by IFN-β in the serum, recapitulated the results observed in tissue culture (Fig. 5c). Type I interferons have wide ranging effects on innate and adaptive immune responses, and are used to treat multiple sclerosis, hepatitis C, and some malignancies (28, 29). The strains generated in this study may provide insight into the role of IFN-β and the cytosolic surveillance pathway in linking innate and adaptive immunity, thereby leading to the development of adjuvants and vaccines and, perhaps, to the discovery of new therapeutics.
Materials and Methods
Bacterial Genetic Screen.
A total of 5,000 individual L. monocytogenes Tn917-LTV3 transposon insertion mutants (15) were grown on BHI media in 96-well plates overnight at 30°C. Bone marrow-derived macrophages from C57BL/6 mice were plated on 96-well plates, 4 × 104 cells per well, and infected with 2 × 106 bacteria. 30 min postinfection, macrophages were washed and gentamicin was added (50 μg/ml) to prevent extracellular growth of bacteria. At 6 h postinfection 100 μl of macrophage culture media was frozen at −80°C. The amount of type I IFN in the media was detected by using a reporter cell line, ISRE-L929 (16). ISRE-L929 cells were grown in 96-well plates and incubated with 40 μl of infected macrophage culture media for 4 h. Cells were lysed and luciferase activity was detected by using Bright Glow Assay (Promega, E-2620) and measured with a luminescence counter (VICTOR3, PerkinElmer).
Infections and Analysis of Gene Expression in Macrophages.
RNA was collected from infected macrophages at 4 h postinfection, and induction of IFN-β was analyzed by qRT-PCR, as described (30). Where indicated, 10 μM p38 MAPK inhibitor SB202190 (Calbiochem) was added to cells 30 min before infection, and kept on for the duration of the infection (5).
L. monocytogenes Gene Expression.
Expression of MDR genes by L. monocytogenes growing in BHI broth or intracellularly was analyzed by using real-time qRT-PCR analysis (30). Level of gene expression was normalized to the level of expression of the rpoB gene or bglA gene. To test for expression of MDR genes after treatment with toxic drugs, tetraphenylphosphonium (50 μM, Sigma) or rhodamine 6G (50 μM, Sigma) were added for 1 h at log phase, then total bacteria RNA was extracted and analyzed by qRT-PCR (30).
Microarray Analysis.
Microarray analysis of IFNαβR−/− macrophages infected with w.t., mdrM−, or tetR::Tn917 L. monocytogenes was done as described in refs. 13 and 30, with several modifications detailed in the SI Methods.
In Vivo L. monocytogenes Infections.
C57BL/6 or IFNαβR−/− mice were infected with 0.1 LD50 (1 × 104) of w.t. L. monocytogenes, mdrM−, or tetR::Tn917 mutants, and organs were harvested 48 h postinfection, as described (27).
Supplementary Material
Acknowledgments.
We thank Nicole Meyer-Morse for help with animal experiments, Bruce Beutler (The Scripps Research Institute, La Jolla, CA) for the ISRE-L929 IFN reporter cell line and femurs from MyD88Trif−/− mice, Genhong Cheng (University of California, Los Angeles) for femurs from IRF3−/− mice, Zhijian Chen (University of Texas Southwestern Medical School, Dallas, TX) for femurs form MAVS−/− mice, and Hiroshi Nikaido for helpful discussion. This work was supported by National Institutes of Health Grant P01 AI063302. D.A.P. is a Senior Scholar awardee at the Ellison Medical Foundation. A.A.H is a postdoctoral fellow of the Miller Institute of Basic Science, University of California, Berkeley.
Footnotes
Conflict of interest statement: Daniel A. Portnoy has a consulting relationship with and a financial interest in Anza Therapeutics Inc, which stands to benefit from commercialization of the results of this research. Peter Lauer and Tom W. Dubensky Jr., Anza Therapeutics Inc, declare competing financial interests.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804170105/DCSupplemental.
References
- 1.Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 4.Roy CR, Mocarski ES. Pathogen subversion of cell-intrinsic innate immunity. Nat Immunol. 2007;8:1179–1187. doi: 10.1038/ni1528. [DOI] [PubMed] [Google Scholar]
- 5.O'Riordan M, Yi CH, Gonzales R, Lee KD, Portnoy DA. Innate recognition of bacteria by a macrophage cytosolic surveillance pathway. Proc Natl Acad Sci USA. 2002;99:13861–13866. doi: 10.1073/pnas.202476699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCaffrey RL, et al. A specific gene expression program triggered by Gram-positive bacteria in the cytosol. Proc Natl Acad Sci USA. 2004;101:11386–11391. doi: 10.1073/pnas.0403215101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perry AK, Chen G, Zheng D, Tang H, Cheng G. The host type I interferon response to viral and bacterial infections. Cell Res. 2005;15:407–422. doi: 10.1038/sj.cr.7290309. [DOI] [PubMed] [Google Scholar]
- 8.Stockinger S, et al. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J Immunol. 2002;169:6522–6529. doi: 10.4049/jimmunol.169.11.6522. [DOI] [PubMed] [Google Scholar]
- 9.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Stanley SA, Johndrow JE, Manzanillo P, Cox JS. The Type I IFN response to infection with Mycobacterium tuberculosis requires ESX-1-mediated secretion and contributes to pathogenesis. J Immunol. 2007;178:3143–3152. doi: 10.4049/jimmunol.178.5.3143. [DOI] [PubMed] [Google Scholar]
- 11.Henry T, Brotcke A, Weiss DS, Thompson LJ, Monack DM. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J Exp Med. 2007;204:987–994. doi: 10.1084/jem.20062665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux CM, et al. Brucella requires a functional Type IV secretion system to elicit innate immune responses in mice. Cell Microbiol. 2007;9:1851–1869. doi: 10.1111/j.1462-5822.2007.00922.x. [DOI] [PubMed] [Google Scholar]
- 13.Leber JH, et al. Distinct TLR- and NLR-mediated transcriptional responses to an intracellular pathogen. PLoS Pathog. 2008;4:e6. doi: 10.1371/journal.ppat.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts ZJ, et al. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J Exp Med. 2007;204:1559–1569. doi: 10.1084/jem.20061845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camilli A, Portnoy A, Youngman P. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J Bacteriol. 1990;172:3738–3744. doi: 10.1128/jb.172.7.3738-3744.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang Z, et al. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6:565–570. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 17.Huillet E, Velge P, Vallaeys T, Pardon P. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol Lett. 2006;254:87–94. doi: 10.1111/j.1574-6968.2005.00014.x. [DOI] [PubMed] [Google Scholar]
- 18.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camilli A, Tilney LG, Portnoy DA. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol Microbiol. 1993;8:143–157. doi: 10.1111/j.1365-2958.1993.tb01211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schumacher MA, Brennan RG. Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol Microbiol. 2002;45:885–893. doi: 10.1046/j.1365-2958.2002.03039.x. [DOI] [PubMed] [Google Scholar]
- 21.Fischetti VA. Gram-Positive Pathogens. Washington, DC: ASM Press; 2006. [Google Scholar]
- 22.O'Connell RM, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 24.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Soulat D, Bauch A, Stockinger S, Superti-Furga G, Decker T. Cytoplasmic Listeria monocytogenes stimulates IFN-beta synthesis without requiring the adapter protein MAVS. FEBS Lett. 2006;580:2341–2346. doi: 10.1016/j.febslet.2006.03.057. [DOI] [PubMed] [Google Scholar]
- 27.Auerbuch V, Brockstedt DG, Meyer-Morse N, O'Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 29.Maher SG, Romero-Weaver AL, Scarzello AJ, Gamero AM. Interferon: Cellular executioner or white knight? Curr Med Chem. 2007;14:1279–1289. doi: 10.2174/092986707780597907. [DOI] [PubMed] [Google Scholar]
- 30.Herskovits AA, Auerbuch V, Portnoy DA. Bacterial ligands generated in a phagosome are targets of the cytosolic innate immune system. PLoS Pathog. 2007;3:e51. doi: 10.1371/journal.ppat.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Portnoy DA, Jacks PS, Hinrichs DJ. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decatur AL, Portnoy DA. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science. 2000;290:992–995. doi: 10.1126/science.290.5493.992. [DOI] [PubMed] [Google Scholar]
- 33.Schnupf P, et al. Regulated translation of listeriolysin O controls virulence of Listeria monocytogenes. Mol Microbiol. 2006;61:999–1012. doi: 10.1111/j.1365-2958.2006.05286.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.