Abstract
We have investigated the hitherto unexplored possibility that differences in the catalytic efficiencies of thymidylate synthases ThyX and ThyA, enzymes that produce the essential DNA precursor dTMP, have influenced prokaryotic genome evolution. We demonstrate that DNA replication speed in bacteria and archaea that contain the low-activity ThyX enzyme is up to 10-fold decreased compared with species that contain the catalytically more efficient ThyA. Our statistical studies of >400 genomes indicated that ThyA proteins are preferred for the replication of large genomes, providing further evidence that the thymidylate metabolism is limiting expansion of prokaryotic genomes. Because both ThyX and ThyA participate in frequent reciprocal gene replacement events, our observations indicate that the bacterial metabolism continues to modulate the size and composition of prokaryotic genomes. We also propose that the increased kinetic efficiency of thymidylate synthesis has contributed to extending the prokaryotic evolutionary potential.
Keywords: genome evolution, gene replacement, nucleotide metabolism, PBCV-1, thymidylate synthesis
Contrarily to the deoxyribonucleotides dATP, dCTP, and dGTP, which can be produced directly by ribonucleotide reductase, de novo synthesis of dTTP requires the formation of thymidylate (dTMP). This essential DNA precursor is produced by the methylation of dUMP by thymidylate synthase. The mechanisms for de novo synthesis of DNA precursors were thought to be conserved in all free-living prokaryotes and eukaryotes. However, we identified a second family of thymidylate synthases [ThyX, also known as flavin-dependent thymidylate synthase (FDTS)] that is evolutionarily unrelated to the canonical thymidylate synthases ThyA. Approximately 30% of microbial species (based on completed genome sequences) depend on FDTS ThyX (EC 2.1.1.148) (1), whereas the canonical dTMP-forming enzyme ThyA (EC 2.1.1.45) is present in ≈70% of microorganisms. The key difference in ThyA and ThyX catalysis is related to the reductive mechanisms used for the reduction of the methylene group that serves as carbon source in the reaction. During ThyX catalysis, NAD(P)H is used as reductant, whereas ThyA uses methylenetetrahydrofolate (MTHF) not only as a one-carbon donor, but also as a source of reductive power (reviewed in ref. 2). Consequently, ThyX catalysis leads to the production of reduced tetrahydrofolate (THF), whereas ThyA produces oxidized dihydrofolate. These mechanistic differences of the two thymidylate synthases have clearly raised the possibility that the two enzymes might not be fully interchangeable in vivo (3).
Several arguments suggest that the evolutionary trajectories of ThyX and ThyA proteins differ. For instance, ThyA proteins form a highly conserved protein family, whereas ThyX sequences are much more divergent (1, 2). Notably, the high level of sequence conservation of ThyA proteins does not result from functional constraints because extensive mutagenesis studies performed on ThyA proteins identified only five critical residues for catalytic activity (4). Moreover, the sporadic and almost mutually exclusive phylogenetic distribution patterns of thyX and thyA are indicative of frequent lateral gene transfer and/or nonorthologous gene displacement events (1, 5). Environmental factors might also influence thymidylate synthase utilization because our sequence-similarity searches indicated that thyX genes are over-represented in the genomes of (hyper)thermophilic, microaerophilic, and anaerobic microorganisms (6). However, thyX can also be found in a number of mesophilic and aerobic organisms suggesting that these “environmental” factors are not sufficient to explain the complex distribution patterns of the two thymidylate synthases.
Using experimental and statistical analyses, we have addressed why these two analogous enzyme families appeared, and why both thymidylate synthase families have been maintained in current-day organisms.
Results and Discussion
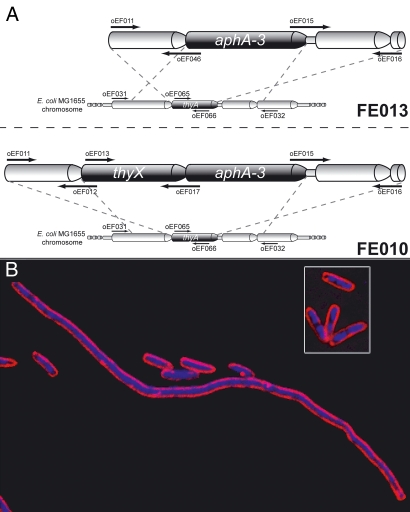
Systematic activity measurements under aerobic and anaerobic conditions using both viral and bacterial ThyX proteins have suggested that flavin adenine dinucleotide (FAD) reduction by NAD(P)H or conformational changes associated with this reductive step limit the catalytic efficiency of ThyX catalysis in vitro compared with ThyA (7–11). This point is illustrated in Table 1 which shows that the catalytic efficiency of the viral ThyX protein is 3–5% of that which has been described for the Lactobacillus casei ThyA enzyme. This observation raised the unexplored possibility that intrinsic biochemical properties of ThyA and ThyX enzymes have influenced thymidylate synthase phylogeny and possibly the dynamics of prokaryotic genome evolution. This could imply that the availability of dTMP in ThyA and ThyX organisms is dramatically different. To test this notion, we first constructed an Escherichia coli strain where the chromosomal copy of thyA was replaced with thyX encoding a highly active viral protein (Materials and Methods). This construct was marked with a nonpolar kanamycin selection marker (Fig. 1A). In the resulting strain FE010 (ΔthyA::thyX), the chromosomal copy of thyX is expressed from the native thyA promoter. In the presence of thymidine, FE010 and the thyA-deleted control strain FE013 grew similarly to the MG1655 WT strain. However, in thymidine-deprived solid and liquid growth media, FE010 grew poorly (0.36 doubling per hour) compared with the WT strain (0.63 doubling per hour), and strain FE013 (ΔthyA) failed to grow. Several experiments established that FE010 is impaired in DNA replication. For instance, we observed that under thymidine deprivation, FE010 cells form long filaments (Fig. 1B), similar to what is often found for E. coli DNA replication mutants. In addition, trichloroacetic acid precipitation measurements of the incorporation rate of radioactively marked thymidine into DNA showed that the relative DNA replication velocities of the WT and FE010 (ΔthyA::thyX) strains were 100% (corresponding to 0.17 fmol/min) and 12% (0.02 fmol/min), respectively. The strains overexpressing thyX from a low copy-number plasmid based on the T5 promoter transcription–translation system grew slowly (10) and had slowed-down DNA replication (0.05 fmol/min for FE023). Finally, the frequency of spontaneous rifampicin-resistant mutants of FE010 was increased by three orders of magnitude compared with the parental strain (data not shown). Altogether, these results indicate that chromosomally or plasmid encoded thyX replacing thyA limits E. coli DNA replication.
Table 1.
Catalytic parameters of PBCV-1 ThyX and Lactobacillus casei ThyA
| Parameter* | PBCV-1 ThyX | L. casei ThyA |
|---|---|---|
| Reductant | NADPH/FAD | THF |
| kcat, s−1 | 0.35 (10%) | 3.6 (100%) |
| Km, mM | ||
| dUMP | 15 | 5 |
| MTHF | 20 | 10 |
| NADPH | 43 | |
| kcat/KmdUMP, s−1·μM−1 | 0.023 (3.2%) | 0.72 (100%) |
| kcat/KmMTHF, s−1·μM−1 | 0.0175 (4.8%) | 0.36 (100%) |
Fig. 1.
Chromosomal replacement of E. coli thyA with thyX from PBCV-1. (A) Schematic representations of the chromosome of E. coli strains FE010 (ΔthyA::thyX) and FE013 (ΔthyA) (see Materials and Methods for details). DNA primers [supporting information (SI) Table S1] used for various PCRs are indicated with arrows. (B) Dual staining of E. coli cells grown under thymidine deprivation. The cell membrane is red (FM 4–64 staining) and DNA is blue (DAPI staining). The figure shows the representative field for the filamentous phenotype observed for FE010 (ΔthyA::thyX), whereas WT strain (Inset) did not filament under the same conditions.
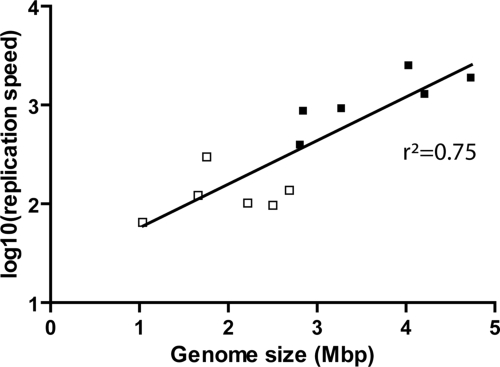
To investigate whether these results also hold true for organisms that naturally contain thyX, we measured the combined fork progression speed during Helicobacter pylori DNA replication. By using quantitative PCR, we determined a replication origin/terminus ratio of 2.7 ± 0.6 in midexponential phase cultures of H. pylori. This ratio corresponds to a replication period (C-period) of 230 min (see Materials and Methods), whereas the corresponding value is ≈40 min in E. coli. Accounting for the differences in genome size (H. pylori ≈1.7 Mbp; E. coli ≈4.6 Mbp), we estimate a mean DNA replication speed of ≈120 bp/s for H. pylori. This value is an order of magnitude lower than what has been measured for E. coli [1,870 bp/s (Table 2)]. Most notably, the low replication speed we have found for H. pylori has been observed in other bacteria and archaea that use ThyX for dTMP synthesis. Indeed, Table 2 shows that the average replication speed in a diverse group of ThyX organisms (135 ± 82 bp/sec) is 10-fold lower than in ThyA-containing species (1,301 ± 762 bp/s). This observation contradicts the current view that “high” and “low” replication speeds in prokarya reflect the differences in the molecular architectures of bacterial and archaeal replisomes, respectively. We propose the alternative explanation that the observed differences in replication speed result, at least in part, from dTMP availability influenced by the use of different thymidylate synthases. Earlier observations have indicated that in E. coli, a decreased replication rate results in a slower growth rate and decreased replicative fitness (12). The above suggestion and the significant growth difference of ThyX- and ThyA-containing species (Fig. 2A) raise the possibility that organisms with highly active ThyA could compete out catalytically less efficient ThyX species in natural populations.
Table 2.
Correlation between thymidylate synthase type and replication speed
| Thymidylate synthase | Organism | Domain* | Genome size, Mbp† | Doubling time, min‡ | C-period, min‡ | Replication speed, bp/s‡ | Ref. |
|---|---|---|---|---|---|---|---|
| ThyX | Chlamydia trachomatis | B | 1.1 | 1,440 | 276 | 64 | 32 |
| Helicobacter pylori26695 | B | 1.7 | 167 | 232 | 120 | This work | |
| Prochlorococcus | B | 1.7 | 1,020 | ≈240–360 | 95 | 33 | |
| Synechococcus | B | 2.4 | 360 | ≈240–360 | 135 | 33 | |
| Pyrococcus abyssi | A | 1.8 | 30 | 100 | 294 | 34 | |
| Sulfolobus acidocaldarius | A | 2.2 | ND | ND | ≈80–110 | 35 | |
| ThyA | Bacillus subtilis | B | 4.2 | 26 | 55 | 1,277 | 36 |
| Brucella abortus | B | 3.3 | 216 | 60 | 913 | 37 | |
| Enteroccocus faecium | B | 2.8 | 90 | 55 | 863 | 38 | |
| Escherichia coli | B | 4.6 | 20 | 41 | 1,870 | 39 | |
| Staphyloccus aureus | B | 2.8 | 120 | 120 | 392 | 40 | |
| Vibrio cholerae | B | 4.0 | 42 | 27 | 2,488 | 41 |
ND, not determined.
*Domain of the species is indicated (B for Bacteria and A for Archaea)
†Genome size data were collected from the IMG database
‡Doubling time, C-period, and replication speed were collected from studies cited.
Fig. 2.
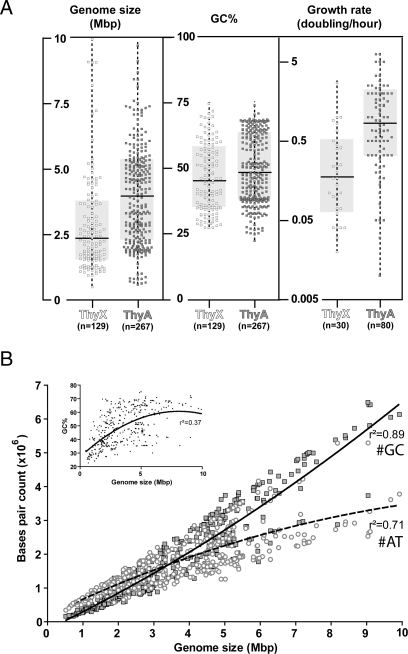
Size distribution and nucleotide counts of thyA- and thyX-containing genomes. (A) Distributions of genome size, GC% and growth rate are shown. In each case, thyX and thyA containing organisms were analyzed individually. (B) A scatter plot demonstrating the relationship between the genome size and the nucleotide counts for 425 prokaryotic genomes is shown [GC count (filled squares); AT counts (open circles)]. The best polynomial nonlinear fits (second order) for GC (solid line) and AT (dotted line) counts are shown. (Inset) Plotting of the relative GC% as a function of genome size.
To address why ThyX enzymes are nevertheless used in a large number of species, we determined the genome size and guanine and cytosine (GC) composition of 425 prokaryotic organisms as a function of their thymidylate synthase use. When these genomic features were separately plotted by using a box-and-whiskers graph (Fig. 2A), we observed only a small difference in GC percentage between ThyX and ThyA-containing organisms. The striking observation is that although the genome sizes of thyA organisms were widely distributed, with a median size of 3.9 Mb, the size distribution of thyX-containing genomes was significantly narrower, with a median size of 2.3 Mb [P < 0.0001 (a nonparametric two-tailed Mann–Whitney test)]. We also plotted the number of different nucleotides as a function of the genome size (Fig. 2B). The nonlinear polynomial fit of the combined guanine and cytosine count, as a function of genome size, had a high goodness-of-fit value of R2 = 0.89 and revealed an obvious gradual increase in all analyzed genomes. Although the counts of adenine and thymidine nucleotides (R2 = 0.70) also showed an initial increase, starting from a genome size of ≈3 Mb, this increase was less important. This observation is unexpected because synthesis of dATP and dTTP precursors requires less energy than the production of dGTP and dCTP precursors (13). Notably, our observations suggest that prokaryotic genomes have enlarged mainly by increasing their GC content (GC%) to overcome the “AT-barrier” and are likely to be at the origin of the much weaker positive correlation that has been observed for the relative GC% and the average genome size (Fig. 2B) (14, 15). Further support for this notion is shown in Fig. 3, which depicts a positive correlation between the replication speed and genome size in prokarya. Altogether these observations predict that organisms with large genomes that thrive in complex and variable environments should show an increased GC content.
Fig. 3.
Relation between mean replication speed (represented by a logarithmic scale) and prokaryotic genome size. ThyX (open squares) and ThyA (filled squares) containing organisms are indicated. A linear regression curve (r2 = 0.75) is plotted. Data were extracted from Table 2.
Although reciprocal gene replacements implicating thyX and thyA are occurring frequently and randomly, in the light of functional constraint imposed by the nature of the thymidylate synthase, the likelihood for observing replacement of thyA by thyX in natural populations is small. This constraint, however, does not operate in species with a relatively small genome, explaining why the catalytically less efficient ThyX proteins are still present in 30% of prokaryotic organisms (based on completed genome sequences). Our work also provides the hypothesis that thymidylate synthase use globally affects prokaryotic genome dynamics. Our data predict that massive gene duplications or lateral gene transfer into thyX-containing species should be counterselected. Moreover, the replacement of thyA by thyX coincides with the well documented case of reductive genome evolution observed in Rickettsia species (16). Even though this correlation does not prove causation, future experimental work will be needed to address the possibility that an analogous increase in mutation rate, as observed for the strain FE010 (ΔthyA::thyX), could accelerate genome reduction as has been proposed for Pelagibacter and Prochlorococcus species that both contain thyX (17). The observation that some cyanobacteria have strikingly large, AT-rich genomes suggests that alternative strategies to enlarge prokaryotic genomes exist.
In conclusion, our combined experimental results and statistical analyses have indicated that metabolic enzymes can shape prokaryotic genomes. Because the work reported here indicates that ThyX enzymes limit DNA replication kinetics in vivo, our observations suggest that the increased catalytic efficiency of ThyA enzymes was necessary to increase the prokaryotic evolutionary potential.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains used in this study are listed in Table S2. E. coli cells were grown at 37°C in either thymidine-containing LB medium or thymidine-free M9 minimal medium supplemented with 0.1 mM CaCl2, 2 mM MgSO4, 0.2% casamino acids (Difco) or thymidine-free L+ medium (18). When necessary, thymidine (50 μg/ml) and isopropyl d-thiogalactopyranoside (1 mM) were added to the growth medium. H. pylori 26695 was grown on either blood agar base two (Oxoid) plates supplemented with 10% defibrinated horse blood (Oxoid) or in liquid culture in brain-heart infusion broth (Oxoid) supplemented with 10% FBS (Gibco BRL). An antibiotics–fungicide mix consisting of vancomycin (final 10 μg/ml), polymyxin B (2.5 units per liter), trimethoprim (5 μg/ml), and fungizone (2.5 μg/ml) was added. Plates were incubated at 37°C in a microaerobic atmosphere in jars by using CampyGen (Oxoid). Liquid cultures were shaken at 175 rpm.
Molecular Biology Techniques.
The replacement of the complete thyA gene of E. coli strain MG1655 by the Paramecium bursaria Chlorella virus 1 (PBCV-1) thyX gene, encoding the most catalytically active ThyX protein isolated to date, was performed as follows. First, thyX and 500-bp DNA fragments flanking thyA were amplified by PCR (primers used are indicated in Table S1). The kanamycin cassette aphA-3 was obtained by SmaI digestion of pUC18K2. The deletion construct (Fig. 1A) was assembled in two sequential PCRs. The obtained PCR product was introduced into the chromosome of E. coli by the use of the lambda-red recombination system (19, 20). This system uses the thermosensitive pKOBEG plasmid that carries the λ phage redγβα operon under the control of a pBAD promotor. Transformants were selected with kanamycin in the presence of thymidine (replacement efficiency ≈7.103 CFU/μg of linear DNA). The thyA-deleted strain, FE013, was constructed as above without insertion of thyX. Both constructs were confirmed by PCR and DNA sequencing (data not shown).
Fluorescence Microscopy.
E. coli strains MG1655 and FE010 were diluted in thymidine-deprived L+ medium to an OD600 ≈ 0.2–0.3 and incubated at 37°C with agitation. One-milliliter culture aliquots were removed and stained as described (21). Briefly, cell membranes were stained with N-(3-triethylammonium-propyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (FM 4-64) at 0.01 μg.μl−1, and the nucleoid was stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI) at 2 μg·ml−1. FM 4-64 stains lipid membranes and emits a red fluorescence (excitation/emission ≈515/640 nm); DAPI is a nucleoid stain with blue fluorescence (excitation/emission ≈350/470 nm). The stained cells were observed by using a Leica DM RXA microscope. Images were captured with a CCD camera 5 MHz Micromax 1300Y (Roper Instruments). The final reconstructed images were obtained by deconvoluting Z-series with Metamorph software (Universal Imaging).
Runout Replication Assay.
E. coli strains were grown in LB medium for 12–14 h and diluted 10-fold by using M9 minimal medium. At the beginning of the exponential phase, rifampicin (50 μg/ml final; Sigma), uridine (50 μg/ml final; Sigma), and 3 μCi of [methyl-3H]-thymidine (84 Ci/mmol; Amersham) were added. Incorporation of radioactive thymidine was measured by scintillation counting after trichloroacetic acid precipitation of whole cells.
Determination of Ori/Ter Ratio and Replication Period (C-Period).
For the experimental quantification of the relative ratios of H. pylori origin and terminus regions [previously identified by using bioinformatic analyses (22)], DNA was extracted from midexponential cultures (OD600 ≈ 0.8) by using QIAamp DNA Mini kit (Qiagen). The primers used (oEF102, oEF103, oEF106, and OEF107) for the quantification reactions are indicated in Table S1. Quantitative PCR was performed in a MiniOpticon real-time PCR detection system (Bio-Rad) by using SYBR green PCR MasterMix Q-PCR (Bio-Rad). Observed cycle thresholds and PCR efficiencies allowed determination of a calibrator-normalized relative quantification of the replication origin over terminus for each sample by using Opticon Monitor Software version 3.1.32 (Bio-Rad). The replication time (C in the following equation) was calculated from the origin/terminus ratio by using the formula O/T = 2τ/C, where τ is the doubling time (23, 24).
Statistical Analysis.
We analyzed several genomic characteristics of 425 complete prokaryotic genomes (Table S3) that were retrieved in March 2006 from the Integrated Microbial Genomes (25) and the Cluster of Orthologous Groups databases (26, 27). Automated analyses were completed and validated by manual investigation. In particular, the presence and/or absence of both thymidylate synthases was confirmed by using BLAST and PSI-BLAST tools (28, 29). The maximum growth rates of prokaryotes used in this study were taken from ref. 30. Statistical analyses were performed by using GraphPad Prism version 4.00 for Windows.
Acknowledgments.
We thank B. Dalmais and E. Bentchikou for technical advice, H. de Reuse and J. M. Ghigo (Pasteur Institute, Paris) for H. pylori strain 26695 and pKOBEG plasmid, respectively. F.E. was supported by the Ministère de la Recherche Française and the Fondation pour la Recherche Médicale. This work was supported by the Centre National de la Recherche Scientifique Program Microbiologie Fondamentale (U.L. and H.M.) and the Institut National de la Santé et de la Recherche Médicale AVENIR program and the Fondation Bettencourt Schuller (H.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801356105/DCSupplemental.
References
- 1.Myllykallio H, et al. An alternative flavin-dependent mechanism for thymidylate synthesis. Science. 2002;297:105–107. doi: 10.1126/science.1072113. [DOI] [PubMed] [Google Scholar]
- 2.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 3.Leduc D, et al. Flavin-dependent thymidylate synthase ThyX activity: Implications for the folate cycle in bacteria. J Bacteriol. 2007;189:8537–8545. doi: 10.1128/JB.01380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finer-Moore JS, Santi DV, Stroud RM. Lessons and conclusions from dissecting the mechanism of a bisubstrate enzyme: Thymidylate synthase mutagenesis, function, and structure. Biochemistry. 2003;42:248–256. doi: 10.1021/bi020599a. [DOI] [PubMed] [Google Scholar]
- 5.Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 6.Leduc D, et al. Two distinct pathways for thymidylate (dTMP) synthesis in (hyper)thermophilic Bacteria and Archaea. Biochem Soc Trans. 2004;32:231–235. doi: 10.1042/bst0320231. [DOI] [PubMed] [Google Scholar]
- 7.Leduc D, et al. Functional evidence for active site location of tetrameric thymidylate synthase X at the interphase of three monomers. Proc Natl Acad Sci USA. 2004;101:7252–7257. doi: 10.1073/pnas.0401365101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graziani S, et al. Functional analysis of FAD-dependent thymidylate synthase ThyX from Paramecium bursaria Chlorella virus-1. J Biol Chem. 2004;279:54340–54347. doi: 10.1074/jbc.M409121200. [DOI] [PubMed] [Google Scholar]
- 9.Graziani S, et al. Catalytic mechanism and structure of viral flavin-dependent thymidylate synthase ThyX. J Biol Chem. 2006;281:24048–24057. doi: 10.1074/jbc.M600745200. [DOI] [PubMed] [Google Scholar]
- 10.Zhong J, Skouloubris S, Dai Q, Myllykallio H, Barbour AG. Function and evolution of plasmid-borne genes for pyrimidine biosynthesis in Borrelia spp. J Bacteriol. 2006;188:909–918. doi: 10.1128/JB.188.3.909-918.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernyshev A, Fleischmann T, Koehn EM, Lesley SA, Kohen A. The relationships between oxidase and synthase activities of flavin dependent thymidylate synthase (FDTS) Chem Commun. 2007;27:2861–2863. doi: 10.1039/b700977a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmstetter C. Timing of synthetic activities in the cell cycle. In: Neidhardt FC, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. Washington, DC: Am Soc Microbiol; 1996. pp. 1627–1639. [Google Scholar]
- 13.Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- 14.Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 15.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 16.Blanc G, et al. Reductive genome evolution from the mother of Rickettsia. PLoS Genet. 2007;3:e14. doi: 10.1371/journal.pgen.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marais GA, Calteau A, Tenaillon O. Mutation rate and genome reduction in endosymbiotic and free-living bacteria. Genetica. 2007 doi: 10.1007/s10709-007-9226-6. [DOI] [PubMed] [Google Scholar]
- 18.Giladi M, Bitan-Banin G, Mevarech M, Ortenberg R. Genetic evidence for a novel thymidylate synthase in the halophilic archaeon Halobacterium salinarum and in Campylobacter jejuni. FEMS Microbiol Lett. 2002;216:105–109. doi: 10.1111/j.1574-6968.2002.tb11422.x. [DOI] [PubMed] [Google Scholar]
- 19.Derbise A, Lesic B, Dacheux D, Ghigo JM, Carniel E. A rapid and simple method for inactivating chromosomal genes in Yersinia. FEMS Immunol Med Microbiol. 2003;38:113–116. doi: 10.1016/S0928-8244(03)00181-0. [DOI] [PubMed] [Google Scholar]
- 20.Chaveroche MK, Ghigo JM, d'Enfert C. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 2000;28:E97. doi: 10.1093/nar/28.22.e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Servant P, et al. The ClpPX protease is required for radioresistance and regulates cell division after gamma-irradiation in Deinococcus radiodurans. Mol Microbiol. 2007;66:1231–1239. doi: 10.1111/j.1365-2958.2007.06003.x. [DOI] [PubMed] [Google Scholar]
- 22.Hallin PF, Ussery DW. CBS Genome Atlas Database: A dynamic storage for bioinformatic results and sequence data. Bioinformatics. 2004;20:3682–3686. doi: 10.1093/bioinformatics/bth423. [DOI] [PubMed] [Google Scholar]
- 23.Riber L, Lobner-Olesen A. Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli. J Bacteriol. 2005;187:5605–5613. doi: 10.1128/JB.187.16.5605-5613.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bremer H, Churchward G. An examination of the Cooper-Helmstetter theory of DNA replication in bacteria and its underlying assumptions. J Theor Biol. 1977;69:645–654. doi: 10.1016/0022-5193(77)90373-3. [DOI] [PubMed] [Google Scholar]
- 25.Markowitz VM, et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusov RL, et al. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 29.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couturier E, Rocha EP. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol. 2006;59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, Santi DV. Asparagine 229 in thymidylate synthase contributes to, but is not essential for, catalysis. Proc Natl Acad Sci USA. 1993;90:8604–8608. doi: 10.1073/pnas.90.18.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambden PR, Pickett MA, Clarke IN. The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology. 2006;152:2573–2578. doi: 10.1099/mic.0.29032-0. [DOI] [PubMed] [Google Scholar]
- 33.Burbage CD, Binder BJ. Relationship between cell cycle and light-limited growth rate in oceanic Prochlorococcus (mit9312) and Synechococcus (wh8103) (cyanobacteria) J Phycol. 2007;43:266–274. [Google Scholar]
- 34.Myllykallio H, et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- 35.Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: Synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharpe ME, Hauser PM, Sharpe RG, Errington J. Bacillus subtilis cell cycle as studied by fluorescence microscopy: Constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol. 1998;180:547–555. doi: 10.1128/jb.180.3.547-555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altenbern RA. Chromosomal mapping of Brucella abortus, strain 19. Can J Microbiol. 1973;19:109–112. doi: 10.1139/m73-016. [DOI] [PubMed] [Google Scholar]
- 38.Higgins ML, Koch AL, Dicker DT, Daneo-Moore L. Autoradiographic studies of chromosome replication during the cell cycle of Streptococcus faecium. J Bacteriol. 1986;168:541–547. doi: 10.1128/jb.168.2.541-547.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meacock PA, Pritchard RH. Relationship between chromosome replication and cell division in a thymineless mutant of Escherichia coli B/r. J Bacteriol. 1975;122:931–942. doi: 10.1128/jb.122.3.931-942.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altenbern RA. Chromosome mapping in Staphylococcus aureus. J Bacteriol. 1968;95:1642–1646. doi: 10.1128/jb.95.5.1642-1646.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26:3124–3131. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]