Abstract
Calcium sensitivity of myosin cross-bridge activation in striated muscles commonly varies during ontogeny and in response to alterations in muscle usage, but the consequences for whole-organism physiology are not well known. Here we show that the relative abundances of alternatively spliced transcripts of the calcium regulatory protein troponin T (TnT) vary widely in flight muscle of Libellula pulchella dragonflies, and that the mixture of TnT splice variants explains significant portions of the variation in muscle calcium sensitivity, wing-beat frequency, and an index of aerodynamic power output during free flight. Two size-distinguishable morphs differ in their maturational pattern of TnT splicing, yet they show the same relationship between TnT transcript mixture and calcium sensitivity and between calcium sensitivity and aerodynamic power output. This consistency of effect in different developmental and physiological contexts strengthens the hypothesis that TnT isoform variation modulates muscle calcium sensitivity and whole-organism locomotor performance. Modulating muscle power output appears to provide the ecologically important ability to operate at different points along a tradeoff between performance and energetic cost.
Striated muscles from a variety of taxa show substantial inter- and intra-specific variation in the sensitivity of myosin cross-bridge activation by calcium (1–6). Within individual animals, calcium sensitivity of muscle activation varies during ontogeny, training, and disease and appears to be one of the primary ways that striated muscles adjust to changes in contractile regimes (7–9). It generally is thought that varying the calcium sensitivity of muscle activation affects recruitment of force-generating cross-bridges during a calcium transient, thereby modulating the rate and amount of force and power output (7–12). However, little is presently known regarding the effects of variation in calcium sensitivity on whole-muscle contractile performance, locomotor ability, and energetics.
Variation in muscle calcium sensitivity often involves changes in the molecular composition of the troponin-tropomyosin complex (7–9, 13). This group of molecules constitutes the molecular “switch” that turns myosin cross-bridge activity on and off in response to neurally induced calcium signals. Troponin T (TnT), one of the components of the troponin-tropomyosin complex, varies in isoform composition during development (14–17), training (18), and human heart failure (19, 20). Changes in TnT isoform composition frequently correlate with variation in the calcium sensitivity of myosin cross-bridge activation (1–9), and experimental manipulations of TnT isoform composition have been shown to affect the calcium sensitivity of actomyosin ATPase (21). Point mutations in human cardiac TnT alter both calcium sensitivity and myosin cross-bridge kinetics (22, 23). The emerging picture is that many regions of TnT interact in functionally significant ways with the other troponins, tropomyosin (7–9, 13), and perhaps with myosin as well.
In both vertebrates and invertebrates, variability in TnT isoform composition arises from alternative splicing of mRNA (24–30). There are evolutionarily conserved features of the splicing pattern at the 5′ end of the molecule (27, 29), thus indicating that isoform variation in TnT has deep evolutionary roots, and perhaps evolutionarily conserved functional effects. Here we test the hypothesis that alternative splicing of TnT affects muscle calcium sensitivity and flight performance of dragonflies. We use a comparative approach that takes advantage of the existence of two morphs of Libellula pulchella dragonflies that follow different ontogenetic trajectories of TnT splicing. This approach allows us to test for associations between TnT transcript variation, muscle calcium sensitivity, and contractile performance that are independent of the developmental and physiological context in which TnT alternative splicing occurs. We relate our results to what is known (31) about the life history, behavior, and ecology of this species.
Methods
L. pulchella dragonflies were collected at Ten-Acre Pond, Centre County, Pennsylvania, during 1998. Adult dragonflies were netted in the field and placed immediately in an insulated cooler at 10–15°C. Dragonflies were transported to the laboratory, and within 3 hr of capture were filmed with high-speed video during free flight (500 frames per sec; Redlake HR1000, Morgan Hill, CA). Before each flight test, the dragonfly was warmed in an incubator at 36°C. After release onto a horizontal platform, dragonflies immediately made vigorous escape flights, which generally took the form of shallow-angled ascents. Three or four flights were filmed for each of 97 dragonflies. A sample recording in QuickTime format is available at http://www.bio.psu.edu/Faculty/Marden/LPulchella.mov.
Wing-beat frequency was measurable from all video records and tended to be relatively invariant within and between flights for an individual. Wing-beat amplitude could be measured only from individuals (n = 57) whose body axis, at an appropriate point in a flight (i.e., nonturning, approximately midway along the flight path), was aligned nearly perpendicular to the film plane of the camera. Recent studies (32, 33) of free flight in another Libellulid dragonfly show that the product of wing angular velocity (i.e., frequency × amplitude) and wing area has a tight linear relationship (r2 = 0.95) with total aerodynamic power output estimated from a quasi-steady-state model. Thus, we used the product of wing angular velocity and area as an index of total aerodynamic power output.
A subset of dragonflies (n = 16) was additionally tested for peak metabolic rate during intermittent flight attempts in a flow-through respirometry system. Dragonflies were placed in a 6-liter jar, through which CO2-free air was passed at a flow rate of 6.7 liter min−1. A subsample of excurrent air from the respirometer was dried by using a small magnesium perchlorate filter, then passed through a LiCor CO2 gas analyzer at a flow rate of 0.1 liter min−1. Output from the flow meter and the CO2 analyzer was digitized and recorded on a computer. Peak metabolic rate during 10 min of intermittent flight was obtained by adjusting the observed maximal rate with a Z-transformation (34) that corrected for nonequilibrium metabolic rates according to the known washout characteristics of the respirometry system. Metabolic rates were estimated based on a respiratory quotient of 1.0 and an energy release of 21.31 J⋅ml−1 CO2 (35).
After flight testing, a subset of dragonflies (n = 38) was euthanized, and samples of their flight muscles were collected. To determine the calcium sensitivity of muscle activation, muscle fibers were dissected, demembranated, and bathed in solutions containing a range of calcium concentrations, as described in ref. 36, except that in the present study, we increased the concentration of creatine phosphate to 20 mM. Fibers were clamped by using a modification of the method described in ref. 37 and were held between an Aurora Scientific 404A force transducer (Aurora, Ontario) and a three-dimensional micromanipulator. The fiber rig was situated on the stage of an inverted microscope (Leica DMIL, Deerfield, IL). Sarcomere length was visualized on a video monitor that was connected to a charge-coupled device camera on the microscope. Sarcomere length was set at 2.3–2.4 μM, which we established from in situ fixation of flight muscles. The calcium concentration producing half-maximal tension (pCa50; the negative log of the calcium concentration) was determined by using an iterative fit of the Hill equation.
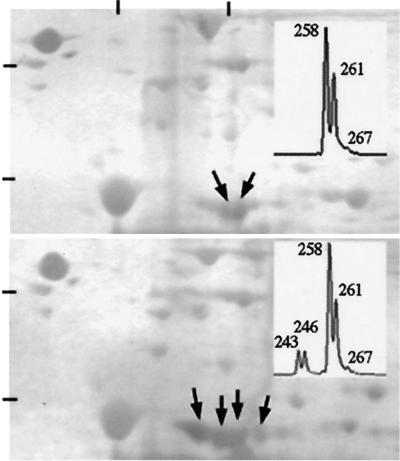
For 22 of the individuals that were tested for calcium sensitivity, we also collected fresh samples of intact flight muscle to isolate mRNA and determine the relative quantity of TnT splice variants. Relative abundances of alternatively spliced TnT transcripts have been shown in previous studies to correspond closely with abundances of the protein isoforms they encode (20, 38), and we found qualitative agreement between variation in transcripts and TnT protein isoforms separated on two-dimensional gels (Fig. 1). Thus, variation in transcript abundance should be a reasonably accurate indication of variation in muscle protein composition.
Figure 1.
Coomassie-stained two-dimensional separation of L. pulchella Tnt isoforms (arrows; identity was confirmed by Western blots of the same gels). (Upper) An individual with two predominant protein isoforms; the same individual showed two predominant mRNA fragment sizes (Inset from genescan analysis). (Lower) An individual with four or perhaps five predominant protein isoforms; this individual showed four predominant mRNA fragment sizes. Bars at top show pH 5.3 (Left) and 5.75 (Right). Bars on the vertical scale show molecular masses of 94 kDa (Upper) and 60 kDa (Lower). TnT isoforms had an apparent molecular mass of 44.3 kDa on these gels. Numbers on genescan insets are defined in Table 1.
Muscle samples were flash-frozen in liquid nitrogen and stored at −70°C for up to 2 weeks until further processing. RNA was recovered by using RNAqueous (Ambion, Austin, TX), and first-strand cDNA synthesis was performed (Boehringer Mannheim). TnT cDNAs were amplified by using the primers TnT Rev (5′-CCTTCCGCTTGGCTTGCTTC-3′) and TnT 5′dUTR (5′-6fam CGCTTCTTTCACTCGTTGTTCAAAC-3′), which was fluorescently labeled with 6fam dye. PCR conditions were as follows: 94°C for 5 min, then 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min, ending with a final incubation at 72°C for 7 min. Extensive amplification and sequencing of L. pulchella TnT cDNAs (39) has shown no evidence for alternative splicing in regions other than what is covered by these two primers (i.e., alternative splicing of TnT in L. pulchella dragonflies has not been observed at the 3′ end, as occurs in vertebrates). The resulting TnT fragments were separated on an automated sequencer (ABI 377), then analyzed with genescan 2.1 software (Applied Biosystems) to determine the size and relative quantity of each fragment. The peak height for each fragment was normalized to the total peak height for all fragments in that lane. For each individual, we ran two samples of the PCR and averaged the results. For five individuals, we performed replicate RNA isolations and all subsequent steps to determine the repeatability of our measures of relative abundance. For those five individuals, the relative quantity estimated for each fragment in the first replicate (n = 50 total fragment relative abundance estimates) explained 97% of the variation in values from the second replicate, i.e., the procedure was highly repeatable.
Relative quantities were arc-sine transformed (40) before statistical analyses. All analyses were performed by using jmp software (SAS Institute, Cary, NC).
Results
Our sample of L. pulchella dragonflies included both newly emerged adults whose body masses ranged from 202 to 342 mg and mature adults that ranged up to 751 mg. Much of the maturational change in body mass is attributable to development of the flight muscles (41), which hypertrophy and increase in aerobic metabolic capacity during adult maturation. Previous studies have shown that the fractional cross-sectional area of mitochondria increases from about 0.15 to 0.46 (31, 41) between emergence and maturity, whereas the specific activity of an enzyme that is an indicator of aerobic metabolic activity (citrate synthase) increases by approximately 1.6-fold (J.H.M. and K. Dennison, unpublished data). Activity of the anaerobic enzyme lactate dehydrogenase falls by 2.8-fold over the same period. Thus, our sample included dragonflies that varied widely in muscle ultrastructure and metabolic physiology.
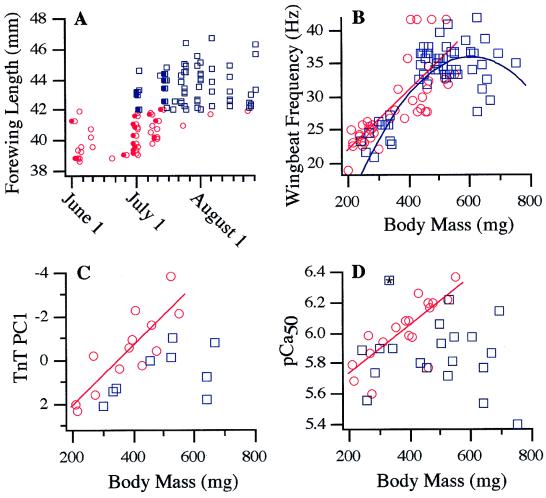
Our study population also showed seasonal variation in overall size, which we characterized by measuring wing length. The wing cuticle hardens at adult emergence and thus provides an age-invariant measure of exoskeletal size. L. pulchella dragonflies that were present from June 1 until approximately mid-July had smaller forewing lengths (Fig. 2A; mean = 40 mm, range = 39–42 mm), whereas dragonflies that began appearing on July 1 were larger (Fig. 2A; mean = 44 mm; range = 42–47 mm). Morphological characteristics other than overall size, including the fine details of wing venation, were indistinguishable between these two groups. Our working hypothesis is that these are two forms of the same species, which we will refer to as the “early” and “late” morph.
Figure 2.
(A) Forewing length of L. pulchella dragonflies during the summer of 1998. A cutoff of 42 mm was used to assign individuals to the early season (circles) or late season (squares) morphs. Filled symbols denote newly emerged adults, none of which were observed after July 13. The small data set for mid-June is the result of a period of cold, rainy weather during which there was little dragonfly activity. (B–D) Wing-beat frequency (B), the first principal component of relative abundances of different-sized TnT transcripts (Table 1) (C), and calcium sensitivity of demembraneated muscle fibers (D) as a function of body mass. The * in D indicates an outlier that may have resulted from an error in the measurement of calcium sensitivity.
Both morphs showed an increase in wing-beat frequency during adult maturation (Fig. 2B), as would be expected when increasingly large muscles drive constant-sized wings. Curiously, however, the late morph showed a significant parabolic trend between wing-beat frequency and body mass (r2 = 0.69, P < 0.0001 for the second-order term in the quadratic fit), with wing-beat frequency becoming much more variable and, on average, declining in the most massive individuals (Fig. 2B).
Eight TnT cDNA fragment sizes, ranging in length from 243 to 270 nt, were produced by our amplification of the alternatively spliced 5′ region of the molecule (Table 1). Six of those fragment sizes were predicted from an extensive survey of cDNAs (37) from flight muscle. The two other fragment sizes were found at very low levels (fragments 255 and 264); they did not correspond to any of our characterized sequences and may be artifacts. Relative abundances of fragment sizes varied widely among individuals (Table 1). For example, fragment size 258 constituted 82% of the TnT transcripts detected from one individual, but only 12% in another individual. Other fragment sizes varied from undetectable levels to as much as 47% of the total.
Table 1.
Fragment sizes and mean relative abundances (% of total TnT transcripts) of the PCR products obtained from amplification of the 5′ alternatively spliced region of L. pulchella TnT cDNA
| cDNA fragment size | Deduced amino acid sequence of variable region | Mean relative abundance (range; SD) |
|---|---|---|
| 243 | MSDEEEYSEEEEEV............................................... | 3.5 (0–10.4; 3.4) |
| 246 | MSDEEEYSEEEEEV......K....................................... | 12.2 (1.8–42.0; 10.5) |
| 258 | MSDEEEYSEEEEEV.................................RPRGK......... | 54.0 (11.9–82.1; 18.4) |
| 261 | MSDEEEYSEEEEEV......K..........................RPRGK......... | 20.7 (0–36.6; 12.3) |
| 267 | MSDEEEYSEEEEEV.............KEPEKKTE.......................... | 7.5 (0–46.5; 12.6) |
| 270 | MSDEEEYSEEEEEV......K......KEPEKKTE.......................... | 1.2 (0–22.5; 4.8) |
| 285 | MSDEEEYSEEEEEV......K......KEPEKKTE............RPRGK......... | Not in flight muscle |
Also shown are the deduced amino acid sequences from characterized cDNAs (39) that correspond to the variable region of these fragments. Putative exons are separated by gaps. A constitutive exon (not shown) begins after the 3′-most base shown for each sequence. The exon structure deduced from cDNAs requires the presence of a micro-exon, such as the one that occurs in Drosophila TnT (30). A fragment size (285) present in other L. pulchella muscles (39) is not found in flight muscle but is shown here for completeness. Two fragment sizes that occur at low levels (255, 264; mean relative abundance = 0.6 and 0.5, respectively) do not correspond to any of our sequenced cDNAs and may be artifacts. The pattern of variation in nucleotides among the cDNAs that we have sequenced (39) is consistent with alternative splicing from a single TnT gene, as occurs in Drosophila (27, 30).
To reduce the dimensionality of the TnT fragment data set, we used principal components analysis to express TnT fragment relative abundances as a single independent variable. The first principal component (TnT PC1) is the linear combination of the original variables (arc-sine-transformed relative abundances of each TnT fragment size) that accounts for the greatest amount of variation. TnT PC1 = 0.0209*AS243 + 0.019*AS246 + 0.100*AS255 + 0.043*AS258–0.031*AS261 + 0.048*AS264 − 0.042*AS267 − 0.060*AS 270 − 1.552 (where ASn refers to the arc-sine-transformed relative abundance of each fragment size). Relative abundances of fragments 261 and 267 accounted for 90% of the variation in TnT PC1.
The early morph showed a tight linear relationship (Fig. 2C; r2 = 0.72, P = 0.0003) between body mass and TnT PC1, whereas the late morph showed no significant trend (Fig. 2C; r2 = 0.24, P = 0.18). The second and third principal components of TnT fragment relative abundance were not correlated with body mass (nor calcium sensitivity; see below) in either morph.
We observed similar maturational trends for calcium sensitivity of skinned fiber activation. The early morph showed a tight, linear increase in muscle calcium sensitivity with increasing body mass (Fig. 2D; r2 = 0.62, P = 0.0001), whereas the late morph showed no significant trend (r2 = 0.02, P = 0.59).
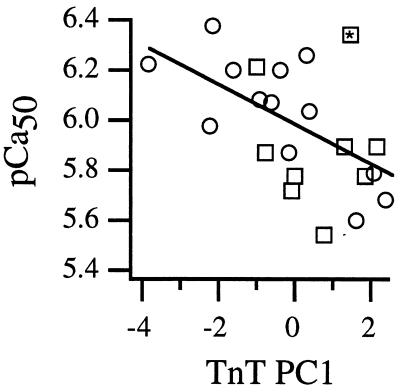
Despite the fact that alternative splicing of TnT followed different ontogenetic patterns in the two morphs, they showed the same relationship between TnT PC1 and muscle calcium sensitivity (r2 = 0.30, P = 0.009; Fig. 3). In a multivariate model that included TnT PC1 (P = 0.03), neither flight muscle mass (P = 0.40), gender (P = 0.88), or morph identity (P = 0.89) had significant effects on muscle calcium sensitivity (total r2 = 0.34). Thus, the relationship between calcium sensitivity and the mixture of TnT transcripts was consistent across wide variation in the developmental and physiological context in which TnT alternative splicing occurred. These results support the hypothesis that TnT isoform variation, either alone or in combination with coregulated molecules, determines calcium sensitivity of muscle activation.
Figure 3.
Calcium sensitivity as a function of the first principal component of relative abundances of different sized TnT transcripts (Table 1). Symbols are as in Fig. 2 B–D. The * indicates a point that was also an outlier for the maturational pattern of calcium sensitivity (Fig. 2D).
Two of the individual transcript size classes (fragments 261 and 267) showed significant positive correlations with calcium sensitivity (r2 = 0.21, 0.19; P = 0.03, 0.04, respectively). Neither of these relationships were affected by morph (P = 0.26–0.40), gender (P = 0.47–0.61), or muscle mass (P = 0.26–0.62).
The largest outlier in the relationship between calcium sensitivity and TnT PC1 (∗ in Fig. 3) was also the largest outlier in the relationship between calcium sensitivity and body mass (∗ in Fig. 2D). This outlier was the only newly emerged individual from either morph that showed a high calcium sensitivity, which suggests a measurement error. When this individual was excluded, TnT PC1 explained 44% of the variation in muscle calcium sensitivity, and the effects of fragment sizes 261, 267, and 258 on calcium sensitivity were significant (r2 = 0.26–0.30, P < 0.02 in each case; fragments 261 and 267 had positive effects, whereas fragment 258 had a negative effect).
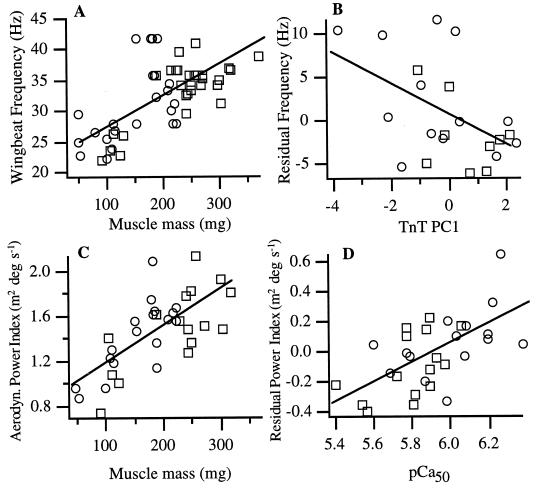
Not surprisingly, dragonflies with larger muscles achieved higher wing-beat frequencies and power outputs (Fig. 4 A and C). The question we were primarily interested in addressing is whether variation in calcium sensitivity and TnT splicing also affect these variables. Thus, we first performed linear regressions of wing-beat frequency and power output index on flight muscle mass, then examined the relationship between the variation not explained by flight muscle mass (the residuals from Fig. 4 A and C) and calcium sensitivity and TnT PC1. Flight muscle mass explained 42% of the variation in wing-beat frequency (Fig. 4A), and both calcium sensitivity (r2 = 0.21; P = 0.005; data not shown) and TnT PC1 (r2 = 0.26; P = 0.01; Fig. 4B) accounted for a significant portion of the residual variation. Wing-beat amplitude was not correlated with muscle mass (r2 = 0.01; P = 0.6; data not shown), but was weakly related to calcium sensitivity (r2 = 0.13; P = 0.046; data not shown). Muscle mass explained 50% of the variation in our index of power output (P < 0.0001; Fig. 4C), with 36% of the residual variation explained by calcium sensitivity (P = 0.0004; Fig. 4D). In a multivariate model using muscle mass, calcium sensitivity, gender, and morph as independent variables, neither morph or gender had significant effects (P = 0.63 and 0.89, respectively) on power output index (P = 0.007 for calcium sensitivity in this model), thus indicating that calcium sensitivity affected performance in a consistent fashion, even though the two morphs showed different ontogenetic trends in calcium sensitivity (Fig. 2D).
Figure 4.
Relationships between total flight muscle mass and wing-beat frequency (A) and relative aerodynamic power output (the product of wing area, wing-beat amplitude, and frequency; C). The first principal component of TnT transcript relative abundances (TnT PC1; B) explains a significant portion of the residual variation in wing-beat frequency after accounting for variation in muscle mass. Calcium sensitivity explains a significant portion of residual variation in relative power output (D). Sample sizes vary because not all of the variables (or components thereof, such as wing-beat amplitude) were measured from all dragonflies. Symbols are as in Fig. 2 B–D.
A striking result is that calcium sensitivity appeared to have nearly as strong an effect as the bulk amount of muscle in determining wing-beat frequency and aerodynamic power output. According to a multivariate fit using calcium sensitivity and muscle mass as independent variables, increasing the pCa50 of muscle activation from 5.4 to 6.4 at the mean flight muscle mass increased wing-beat frequency by a factor of 1.33 and power output index by a factor of 1.58. A tripling of flight muscle mass from 100 to 300 mg at the mean pCa50 was required to increase wing-beat frequency and power output index by comparable factors (1.4- and 1.5-fold, respectively). These factors were calculated from the following regression equations (units in parentheses): wing-beat frequency (Hz) = 9.56*pCa50 + 0.05*flight muscle mass (mg) − 34.24; power output index (m2*degrees*s−1) = 0.62*pCa50 + 0.0032*flight muscle mass (mg) − 2.82.
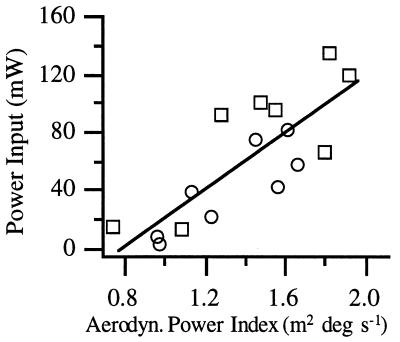
Power output index explained 65% of the approximately 15-fold variation (Fig. 5) in metabolic rate during intermittent flight in the respirometer (P < 0.0001). Thus, dragonflies with larger and more powerful muscles consumed metabolic fuel at a markedly higher rate.
Figure 5.
Metabolic rates of L. pulchella dragonflies during intermittent flight in a respirometer as a function of their relative aerodynamic power output during free flight. Symbols are as in Fig. 2 B–D.
Discussion
We found wide variation in TnT isoform composition in the flight muscles of L. pulchella dragonflies, which was associated with substantial variation in the calcium sensitivity of muscle activation and flight performance. Two individual TnT transcripts were associated with an up-regulation of calcium sensitivity (fragments 261 and 267; Table 1), and another transcript (fragment 258) may encode a down-regulator, although properties of the overall mixture of TnT proteins may be more important than the abundance of individual components. We cannot eliminate the possibility that TnT variation is part of a coregulated complex of molecules that together cause the observed effects; however, no variability in other troponins or tropomyosin is revealed by one-dimensional SDS/PAGE in this species (36). Comparative physiologists working with other taxa, particularly fish, also have begun to focus on troponin isoform variation as a likely cause of differences in contractile properties of muscle (42–47). As this body of work progresses, it will be interesting to see whether TnT splicing has consistent effects on contractile properties and locomotor performance across different types of animals.
The mechanisms by which TnT may affect calcium sensitivity and muscle power output are not yet clear. Point mutations in human cardiac TnT affect both calcium sensitivity and myosin cross-bridge kinetics (22, 23); however, no experiments have yet determined whether alternative splicing causes similar effects. A recent discussion of the relationship between calcium sensitivity and cross-bridge kinetics (48) suggests that the problem is complex, as there is feedback (i.e., positive cooperativity) between calcium binding to thin filament regulatory sites and the rate and extent of additional cross-bridge binding.
For weight-supported flight, insects require at least 12–16% of their body mass to be flight muscle (49). In L. pulchella dragonflies, flight muscle constitutes approximately 40% of body mass at adult emergence, and this ratio increases to 58–63% in mature males. Thus, this species is probably overdesigned for the brief, low-exertion foraging flights used by maturing adults to capture prey, but appropriately designed for the intense aerial interactions that occur during territoriality and mating (50). The significance of being able to modulate muscle power output may be that newly emerged adults can down-regulate contractility and thereby conserve energy while developing a very large flight motor, which then is up-regulated at maturity when maximal flight performance is advantageous for territorial defense and mating. An analogy would be using a governor to restrain power output of the engine of a race car while driving in ordinary traffic, then removing it at the race track.
Given this interpretation, and the previous demonstration that Libellulid dragonflies carrying small experimentally applied loads (5–13% of body weight) fare poorly in territorial competition (41), it is curious that the late-season morph of L. pulchella shows either no increase or perhaps a decrease in muscle calcium sensitivity at maturity (Fig. 2B). One hypothesis for this apparent down-regulation of muscle performance involves seasonal changes in population size and age structure. During the summer of 1998, emergence of new adults ended after mid-July, although mature individuals were present until the end of August (Fig. 2A). Termination of adult emergence may reduce the frequency and intensity of aerial interactions for the late-season morph, because there are few newly matured dragonflies seeking territories. This possibility also may explain the apparently increased longevity of the late-season morph, which unlike the early season morph, persists well beyond the period of new adult emergence (Fig. 2A). Continual pressure on mature adults of the early-season morph from younger individuals may stimulate up-regulation of muscle power, which negatively affects energy balance and longevity. Thus, the contractile differences that we have observed between the two morphs may be a training effect resulting from differences in experience, or they may reflect fixed genetic differences between populations that consistently experience different social environments.
In summary, we propose that dragonflies use alternative splicing of TnT to modulate the calcium sensitivity of their flight muscles, and thereby accomplish ecologically relevant variation in flight performance and energy consumption. This conclusion is similar to the interpretation presented for a recent experiment involving mammalian heart muscle. Rats that were fed high-salt diets to stimulate cardiac hypertrophy and eventual heart failure showed altered expression of cardiac TnT isoforms, which correlated with decreases in an index of power output (the end-systolic pressure-volume relationship), and with increased energy efficiency and economy (51). Thus, there is an emerging understanding that calcium regulatory processes mediate an inverse relationship between performance and energy economy of striated muscles.
Acknowledgments
We thank D. S. Grove for assistance with the genescan procedures and S. Schaeffer for help interpreting cDNA splicing patterns. This project was supported by National Science Foundation Grants IBN-9600840 and IBN-9722196 and the American Heart Association.
Abbreviation
- TnT
troponin T
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF133521).
References
- 1.Schachat F H, Diamond M S, Brandt P W. J Mol Biol. 1987;198:551–554. doi: 10.1016/0022-2836(87)90300-7. [DOI] [PubMed] [Google Scholar]
- 2.Tobacman L S, Lee R. J Biol Chem. 1987;262:4059–4064. [PubMed] [Google Scholar]
- 3.McAuliffe J J, Gao L, Solaro R J. Circ Res. 1990;66:1204–1216. doi: 10.1161/01.res.66.5.1204. [DOI] [PubMed] [Google Scholar]
- 4.Nassar R, Malouf N N, Kelly M B, Oakeley A E, Anderson P A. Circ Res. 1991;69:1470–1475. doi: 10.1161/01.res.69.6.1470. [DOI] [PubMed] [Google Scholar]
- 5.Reiser P J, Greaser M L, Moss R L. J Physiol (London) 1992;449:573–588. doi: 10.1113/jphysiol.1992.sp019102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akella A B, Ding X L, Cheng R, Gulati J. Circ Res. 1995;76:600–606. doi: 10.1161/01.res.76.4.600. [DOI] [PubMed] [Google Scholar]
- 7.Moss R L, Diffee G M, Greaser M L. Rev Physiol Biochem Pharmacol. 1995;126:1–63. doi: 10.1007/BFb0049775. [DOI] [PubMed] [Google Scholar]
- 8.Solaro R J, Rarick H M. Circ Res. 1998;83:471–480. doi: 10.1161/01.res.83.5.471. [DOI] [PubMed] [Google Scholar]
- 9.Moss R L. Circ Res. 1999;84:862–865. doi: 10.1161/01.res.84.7.862. [DOI] [PubMed] [Google Scholar]
- 10.Sweeney H L, Bowman B F, Stull J T. Am J Physiol. 1993;264:C1085–C1095. doi: 10.1152/ajpcell.1993.264.5.C1085. [DOI] [PubMed] [Google Scholar]
- 11.Baylor S M, Hollingworth S J. Gen Physiol. 1998;112:297–316. doi: 10.1085/jgp.112.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry S V. J Muscle Res Cell Motil. 1998;19:575–602. doi: 10.1023/a:1005397501968. [DOI] [PubMed] [Google Scholar]
- 13.Gergely J. Adv Exp Med Biol. 1998;453:169–176. doi: 10.1007/978-1-4684-6039-1_20. [DOI] [PubMed] [Google Scholar]
- 14.Briggs M M, Schachat F. Dev Biol. 1993;158:503–509. doi: 10.1006/dbio.1993.1208. [DOI] [PubMed] [Google Scholar]
- 15.Morgan M J, Earnshaw J C, Dhoot G K. J Cell Sci. 1993;106:903–908. doi: 10.1242/jcs.106.3.903. [DOI] [PubMed] [Google Scholar]
- 16.Wong T S, Ordahl C P. Dev Biol. 1996;180:732–744. doi: 10.1006/dbio.1996.0342. [DOI] [PubMed] [Google Scholar]
- 17.Jin J P, Chen A, Huang Q Q. Gene. 1998;214:121–129. doi: 10.1016/s0378-1119(98)00214-5. [DOI] [PubMed] [Google Scholar]
- 18.Schantz P G, Dhoot G K. Acta Physiol Scand. 1987;131:147–154. doi: 10.1111/j.1748-1716.1987.tb08216.x. [DOI] [PubMed] [Google Scholar]
- 19.Anderson P A, Malouf N N, Oakeley A E, Pagani E D, Allen P D. Circ Res. 1991;69:1226–1233. doi: 10.1161/01.res.69.5.1226. [DOI] [PubMed] [Google Scholar]
- 20.Mesnard-Rouiller L, Mercadier J J, Butler-Browne G, Heimburger M, Logeart D, Allen P D, Samson F. J Mol Cell Cardiol. 1997;29:3043–3055. doi: 10.1006/jmcc.1997.0519. [DOI] [PubMed] [Google Scholar]
- 21.Wu Q L, Jha P K, Du Y, Leavis P C, Sarkar S. Gene. 1995;155:225–230. doi: 10.1016/0378-1119(94)00846-k. [DOI] [PubMed] [Google Scholar]
- 22.Sweeney H L, Feng H S, Yang Z, Watkins H. Proc Natl Acad Sci USA. 1998;95:14406–14410. doi: 10.1073/pnas.95.24.14406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanaga F, Morimoto S, Ohtsuki I. J Biol Chem. 1999;274:8806–8812. doi: 10.1074/jbc.274.13.8806. [DOI] [PubMed] [Google Scholar]
- 24.Breitbart R E, Nguyen H T, Medford R M, Destree A T, Mahdavi V, Nadal-Ginard B. Cell. 1985;1:67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- 25.Briggs M M, Schachat F. J Mol Biol. 1989;206:245–249. doi: 10.1016/0022-2836(89)90538-x. [DOI] [PubMed] [Google Scholar]
- 26.Bucher E A, de la Brousse F C, Emerson C P., Jr J Biol Chem. 1989;264:12482–12491. [PubMed] [Google Scholar]
- 27.Fyrberg E, Fyrberg C C, Beall C, Saville D L. J Mol Biol. 1990;216:657–675. doi: 10.1016/0022-2836(90)90390-8. [DOI] [PubMed] [Google Scholar]
- 28.Nadal-Ginard B, Smith C W, Patton J G, Breitbart R E. Adv Enzyme Regul. 1991;31:261–286. doi: 10.1016/0065-2571(91)90017-g. [DOI] [PubMed] [Google Scholar]
- 29.Briggs M M, Schachat F. Am J Physiol. 1996;270:C298–C305. doi: 10.1152/ajpcell.1996.270.1.C298. [DOI] [PubMed] [Google Scholar]
- 30.Benoist P, Mas J A, Marco R, Cervera M. J Biol Chem. 1998;273:7538–7546. doi: 10.1074/jbc.273.13.7538. [DOI] [PubMed] [Google Scholar]
- 31.Marden J H, Fitzhugh G F, Wolf M R. Am Zool. 1998;38:528–545. [Google Scholar]
- 32.Wakeling J M, Ellington C P. J Exp Biol. 1997;200:557–582. doi: 10.1242/jeb.200.3.557. [DOI] [PubMed] [Google Scholar]
- 33.Wakeling J M, Ellington C P. J Exp Biol. 1997;200:583–600. doi: 10.1242/jeb.200.3.583. [DOI] [PubMed] [Google Scholar]
- 34.Bartholomew G A, Vleck D, Vleck C M. J Exp Biol. 1981;90:17–32. [Google Scholar]
- 35.Harrison J F, Lighton J R. J Exp Biol. 1998;201:1739–1744. doi: 10.1242/jeb.201.11.1739. [DOI] [PubMed] [Google Scholar]
- 36.Fitzhugh G H, Marden J H. J Exp Biol. 1997;200:1473–1482. doi: 10.1242/jeb.200.10.1473. [DOI] [PubMed] [Google Scholar]
- 37.Moss R L. J Physiol (London) 1979;292:177–192. doi: 10.1113/jphysiol.1979.sp012845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan M J, Earnshow J C, Dhoot G K. J Cell Sci. 1993;106:903–908. doi: 10.1242/jcs.106.3.903. [DOI] [PubMed] [Google Scholar]
- 39.Wolf M R. Ph.D. thesis. University Park: Pennsylvania State University; 1999. [Google Scholar]
- 40.Sokal R R, Rohlf F J. Biometry: The Principles and Practice of Statistics in Biological Research. 3rd Ed. New York: Freeman; 1995. [Google Scholar]
- 41.Marden J H. Physiol Zool. 1989;62:505–521. [Google Scholar]
- 42.Crockford T, Wommack K E, Johnston I A, McAndrew B J, Mutungi G, Johnson T P. J Muscle Res Cell Motil. 1991;12:439–446. doi: 10.1007/BF01738328. [DOI] [PubMed] [Google Scholar]
- 43.Rome L C, Syme D A, Hollingworth S, Lindstedt S L, Baylor S M. Proc Natl Acad Sci USA. 1996;93:8095–8100. doi: 10.1073/pnas.93.15.8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnston I, Cole N, Vieira V, Davidson I. J Exp Biol. 1997;200:849–868. doi: 10.1242/jeb.200.5.849. [DOI] [PubMed] [Google Scholar]
- 45.Thys T M, Blank J M, Schachat F H. J Exp Biol. 1998;201:2993–3001. doi: 10.1242/jeb.201.21.2993. [DOI] [PubMed] [Google Scholar]
- 46.Rome L C. Comp Biochem Physiol B. 1998;120:51–72. doi: 10.1016/s0305-0491(98)00023-6. [DOI] [PubMed] [Google Scholar]
- 47.Jackman D M, Pham T, Noel J J, Waddleton D M, Dhoot G K, Heeley D H. Biochim Biophys Acta. 1998;1387:478–484. doi: 10.1016/s0167-4838(98)00165-4. [DOI] [PubMed] [Google Scholar]
- 48.Moss R L. Circ Res. 1999;84:862–865. doi: 10.1161/01.res.84.7.862. [DOI] [PubMed] [Google Scholar]
- 49.Marden J H. J Exp Biol. 1987;130:235–258. [Google Scholar]
- 50.Marden J H, Kramer M G, Frisch J. J Exp Biol. 1996;199:529–535. doi: 10.1242/jeb.199.3.529. [DOI] [PubMed] [Google Scholar]
- 51.Kameyama T, Chen Z, Bell S P, VanBuren P, Maughan D, LeWinter M M. Circulation. 1998;98:2919–2929. doi: 10.1161/01.cir.98.25.2919. [DOI] [PubMed] [Google Scholar]