Abstract
Germinal center (GC) B cells undergo somatic hypermutation, class switch recombination, and rapid clonal expansion to produce high-affinity antibodies. The BCL6 transcriptional repressor facilitates this phenotype because it can repress DNA damage checkpoint genes. GC B and T cells can make transient direct physical contact; T cells were observed to be associated with dead B-cell fragments. We thus hypothesized that one function of CD40 signaling from T cells within this timeframe could be to modulate BCL6 activity. CD40 signaling rapidly disrupts the ability of BCL6 to recruit the SMRT corepressor complex by excluding it from the nucleus, leading to histone acetylation, RNA polymerase II processivity, and activation of BCL6 target genes, such as CD23b, ATR, and TP53. Washout of CD40 to emulate transient T-cell contact permitted BCL6 target gene mRNA levels to return to their repressed levels, demonstrating that this is a reversible process, which could allow centroblasts that pass quality control to either continue proliferation or undergo terminal differentiation. These data suggest that transient CD40 signaling in the GC might allow T cells to weed out heavily damaged centroblasts while at the same time promoting survival of intact B cells, which could undergo differentiation or additional rounds of proliferation.
Introduction
During the humoral immune response, T helper (Th) cells interact with B cells, inducing migration to secondary lymphoid tissues where they proliferate to establish germinal centers (GCs).1,2 The purpose of the GC reaction is to produce clones of B cells expressing high-affinity antibodies. B cells in the dark zone of the GC, known as centroblasts, undergo rapid proliferation and simultaneous somatic hypermutation and class switch recombination.1,3 As centroblasts migrate to the more heterogeneous GC light zone, they encounter and interact with T cells and follicular dendritic cells.1,2 T cells and follicular dendritic cells signal to centrocytes using cytokines and ligands, such as interleukin (IL)–4, IL-2, and CD40.1,2 Measurement of the kinetics of the interaction between T and B cells in live murine GCs demonstrated that many of these interactions are transient and were observed to last up to 30 minutes.1 Centroblasts that have successfully generated high- affinity antibodies differentiate into centrocytes, which can then differentiate into either plasma or memory B cells and exit the GC reaction. The remaining unselected cells undergo cell death and are presumably removed by GC macrophages
Up-regulation of the BCL6 transcriptional repressor is required for B cells to form GCs.4,5 BCL6 contributes to the GC phenotype by repressing multiple DNA damage and cell proliferation checkpoint genes, including ATR (ataxia and telangiectasia, rad3 related), CHEK1,6 TP53 (tumor suppressor p53), and CDKN1A (cyclin-dependent kinase inhibitor 1A).6–10 BCL6-mediated loss of function of these checkpoints results in a potentially hazardous state of physiologic genomic instability, which has the potential to lead B cells toward malignant transformation. Accordingly, constitutive expression of BCL6 in mice can cause diffuse large B-cell lymphoma11,12 and genetic mutations that deregulate BCL6 expression occur frequently in human B-cell lymphomas (reviewed by Ye13). BCL6 also binds and represses multiple genes involved in B-cell maturation, such as CD23bFCER2 (Fc fragment of IgE, low-affinity II receptor),4 and differentiation, such as PRDM1 (PR domain containing 1, with ZNF domain).14–17 Accordingly, BCL6 down-regulation is required for B cells to undergo further differentiation to memory or plasma cells.14,16,18,19
The proper control and timing of BCL6 down-regulation are thus critical for the normal immune response as well as to prevent lymphomagenesis. One important signaling pathway implicated in BCL6 down-regulation is mediated by the CD40 receptor. CD40 stimulation of mouse splenocyte for 24 hours could down-regulate of BCL6 mRNA.20 CD40 stimulation of Ramos B-cell lymphoma cells could down-regulate BCL6 and induce the BCL6 target gene CD23b.21,22 More recently, it was shown that continuous exposure to CD40 signaling leads to NF-κB–mediated induction of IRF4, which binds to an IRF4 responsive element in the BCL6 promoter and represses its transcription. These IRF4 binding sites were shown to be mutated in a subset of patients with diffuse large B-cell lymphoma, possibly contributing to constitutive expression of BCL6.23 In the context of the GC reaction, this has an important biologic implication because T cells signal to B cells through the CD40 receptor. CD40 transcriptional responses were found only in post- and pre-GC populations with the exception of a small subpopulation of GC centrocytes.24
In previous work, we found that a peptide derived from the silencing mediator of retinoid and thyroid receptor (SMRT) corepressor could reactivate BCL6 target genes, such as CD23b, to a similar extent as CD40 signaling, and both could synergize in identical manner with STAT signaling to further induce transcription.22 Therefore, we wondered whether CD40 signaling could disrupt the BCL6-SMRT interaction in a similar manner as the peptide inhibitor. Indeed, we found that CD40 can disrupt the BCL6-SMRT repression complex, causing rapid induction of CD23b and other target genes long before BCL6 becomes transcriptionally down-regulated. The data suggest a biologic mechanism whereby the transient interactions of GC B cells and T cells might allow reversible and temporary reactivation of BCL6 target genes. Reactivation of BCL6 target genes involved in cellular checkpoints could allow B cells damaged during affinity maturation to be rapidly culled from the GC reaction.
Methods
Mammalian cell culture
Ramos cells were grown in medium containing 90% Iscove medium (Cellgro, Manassas, VA), 10% fetal bovine serum (Gemini, Irvine, CA), and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA). To trigger CD40 signaling, cells were incubated with 830 to 1000 ng/mL CD40 ligand (R&D Systems, Minneapolis, MN). For kinase inhibitor studies, cells were pretreated with 50 μM PD98059 and 20 μM UO126 (Sigma-Aldrich, St Louis, MO) or vehicle (dimethyl sulfoxide) for 1 hour then incubated with CD40L for 2 hours. Both PD98059 and UO126 are the inhibitor of MEK1/2.
Isolation of centroblasts from tonsils
The use of discarded tonsillectomy specimens was approved by the Albert Einstein Institutional Review Board and informed consent was obtained in accordance with the Declaration of Helsinki. The tissue was minced and cell pellets obtained using Histopaque 1077. Cell pellets were washed twice with phosphate-buffered saline (PBS) and resuspended in 250 μL PBS plus 0.5% bovine serum albumin (BSA). Cell concentration was determined by staining with trypan blue; 108 cells were incubated with 25 μL CD77 antibody (BD Biosciences, San Jose, CA) for 10 minutes on ice. Cells were washed with PBS by centrifugation for 5 minutes at 240g. The pellet was resuspended in PBS plus 0.5% BSA. For the second staining, cells were incubated with 5 μL MARM-4 antibody for 10 minutes on ice. Cells were washed with PBS by centrifugation for 5 minutes at 240g. The pellet was resuspended in PBS plus 0.5% BSA. For the third staining, cells were incubated with 50 μL rat-antimouse-IgG1a/b microbeads for 20 minutes on ice. Cells were washed with PBS by centrifugation for 5 minutes at 240g. The pellet was resuspended in PBS plus 0.5% BSA; 500 μL of PBS plus 0.5% BSA/108 cells was applied to a magnetic LS column (Miltenyi Biotec, Auburn, CA) followed by the cell sample. The column was washed with PBS plus 0.5% BSA (Miltenyi Biotec), and the sample was eluted outside of the magnetic field with 2 to 5 mL runs of PBS plus 0.5% BSA. The eluted sample was applied to a fresh LS column followed by the addition of PBS until all cells had entered the column. The sample was eluted outside of the magnetic field with 2 to 5 mL runs of PBS plus 0.5% BSA. Purity of the sample was determined by staining with CD38-PE and CD77/MARM-FITC conjugated antibodies and analyzed in an LSRII flow cytometer (BD Biosciences).
Western blots
Whole-cell lysates were prepared using radioimmunoprecipitation assay modified buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid) supplemented with protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN); 5 × 106 cells were resuspended in 10 μL buffer and incubated on ice for 30 minutes and centrifuged for 10 minutes at 9000g at 4°C. Supernatant was subjected to sodium dodecyl sulphate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA), washed in PBS plus 0.1% Tween (PBS-T), and blocked in 5% milk in PBS-T for 30 minutes. The membranes were then incubated for 1 hour at room temperature in primary antibody as follows: anti-BCL6, diluted 1/1000 in PBS-T, 5% milk (Santa Cruz Biotechnology, Santa Cruz, CA); anti-SMRT, diluted 1/500 in PBS-T, 5% milk (Upstate Biotechnology, Charlottesville, VA). Membranes were washed 3 times with PBS-T and incubated in secondary antibody for 1 hour at room temperature. The membranes were washed with PBS-T 3 times, dried and incubated in enhanced chemiluminescence reagent (Santa Cruz Biotechnology) for 1 minute, and visualized in a Fuji Imager (FujiFilm, Tokyo, Japan).
Immunofluorescence microscopy for SMRT localization
Ramos cells (3 × 106) were transfected with 3 μg GFP-SMRT plasmid25 using the Amaxa nucleofector buffer V program O-006. After 24 hours, the cells were treated with 1 μg/mL CD40L for 2 hours, and then the cells were fixed with 1% paraformaldehyde 10 minutes at 37°C. Then cells were collected and seeded onto a CultureWell Chambered Coverglass (Stratech Scientific, Newmarket, United Kingdom), and the fluorescence was monitored by Zeiss AxioSkop II (Carl Zeiss, Thornwood, NY) with optics for phase contrast and epi-fluorescence with 100× objectives and captured using an Olympus digital camera (Olympus, Tokyo, Japan).
Quantitative reverse-transcribed polymerase chain reaction
RNA was extracted from 104 to 5 × 106 cells, using the RNeasy kit (Qiagen, Valencia, CA). cDNA was synthesized using Superscript III First Strand cDNA Synthesis kit (Invitrogen). The mRNA levels of the different genes were detected using the Power SYBR green kit (Applied Biosystems, Foster City, CA) and an Opticon Engine 2 thermal cycler (Bio-Rad). The CT values of the genes of interest were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (ΔCT). ΔCT values of the treated cells were expressed relative to control treated cells using the ΔΔCT method. The fold change in expression of each gene in treated versus control treated cells was determined by the relative quantitation method, 2-ΔΔCT with ΔΔCT + s and ΔΔCT-s where “s” is the standard deviation of the ΔΔCT value for the triplicates. Results were represented as fold expression with standard deviation.
Primers were as follows: GAPDH, F: 5′-CGACCACTTTGTCAAGCTCA-3′; R: 5′-CCCTGTTGCTGTAGCCAAAT-3′; BCL6, F: 5′-AACCTGAAAACCCACACTCG-3′; R: 5′-TTCGCATTTGTAGG-GCTTCT-3′; ATR, F: 5′-TGGGTCCTATGGGAACAGAG-3′; R: 5′-ACGCATCAGCCTCATTGTAA-3′; CCL3, F: 5′-CTTTGAGACGAGCAGCCAGT-3′; R: 5′-ATTTCTGGACCCACTCCTCA-3′; CD23b, F: 5′-ATGAATCCTCCAAGCAGGAG-3′; R: 5′-GACTTGAAGCTGCTCAGATCTGCT-3′; p53, p53-F: 5′-CTTTGAGGTGCGTGTTTGTG-3′; p53-R: 5′-TCTTGCGGAGATTCTCTTCC-3′.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described.17 ChIPs for BCL6, SMRT, and HDAC3 were performed with 10 to 20 × 106 cells each, using 4 μg BCL6 (Santa Cruz Biotechnology), SMRT (Upstate Biotechnology) 4 μg HDAC3y (Santa Cruz Biotechnology), 2 μg RNA polymerase II (POLII) (Santa Cruz Biotechnology) antibodies. ChIPs for histone modifications were performed on 2.5 × 106 cells using 2 μg H4 pan-acetylated or H3 acetylated antibodies (Upstate Biotechnology). An actin antibody (Upstate Biotechnology) was used as negative control. DNA fragments were detected by quantitative polymerase chain reaction (PCR; quantitative ChIP). For QChIP, ΔCT values (Antibody-Input) were expressed relative to control antibody using the ΔΔCT method or as percent of the input according to a standard curve established previously using genomic DNA.
CD23b locus primers were as follows: promoter, F: 5′-CCTTAGCTACTGCCTTTCACC-3′; R: 5′-TCTGTGCCAGGAAAGTCAGTC-3′; exon 1, F: 5′-TTTAGCATAATGAATCCTCCAAG-3′; R: 5′-CCAGACCCCACCCAAAGG-3′; exon 4, F: 5′-CTCCAGTCTCTCAAGTTTCCAA-3′; R: 5′-CTGGGTCCAGGAAGTCATC-3′.
Results
CD40 signaling rapidly up-regulates CD23b expression before down-regulation of BCL6
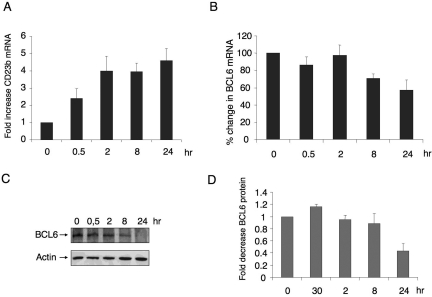
To study the kinetics of CD40-mediated reactivation of the BCL6 target gene CD23b, we treated Ramos B-cell lymphoma cells with 800 ng/mL recombinant CD40 ligand for 0, 0.5, 2, 8, and 24 hours and measured the levels of endogenous CD23b mRNA by quantitative PCR (QPCR). We found that, after 30 minutes of CD40 signaling, the RNA levels of CD23b started to increase, reaching a plateau at 2 hours (Figure 1A). BCL6 protein and RNA levels were observed to decline within 8 hours after treatment (Figure 1B,C) with a 40% decrease in RNA levels at 24 hours (Figure 1B). Densitometry of bands corresponding to the time points of 3 independent Western blots showed that BCL6 protein decrease starts at 8 hours (10% decrease) and was decreased to approximately 40% by 24 hours (Figure 1D). These results demonstrate that reactivation of CD23b is not entirely a consequence of BCL6 down-regulation because CD23b RNA is up-regulated before there is an appreciable decrease in BCL6 protein levels (Figure 1C,D).
Figure 1.
CD40 signaling rapidly up-regulates CD23b expression before down-regulation of BCL6. (A) Ramos cells were exposed to CD40L for the indicated durations, after which QPCR was performed to detect the mRNA abundance of the CD23b gene product. CD23b mRNA levels were first normalized by GAPDH levels and expressed as fold change compared with untreated cells. (B) BCL6 mRNA levels were detected by QPCR in the same cells as in panel A. Panels A and B correspond to the average of 3 independent replicates. (C) Western blots were performed in cells treated as in panels A and B using antibodies for BCL6 and for actin as a loading control. (D) Densitometry of 3 independent BCL6 Western blot replicates. The signal of each BCL6 Western blot was first normalized by actin levels and then expressed in fold compared with untreated cells. Error bars represent standard error of the mean (SEM).
CD40 signaling rapidly ejects corepressors from the BCL6 repression complex on the CD23b promoter
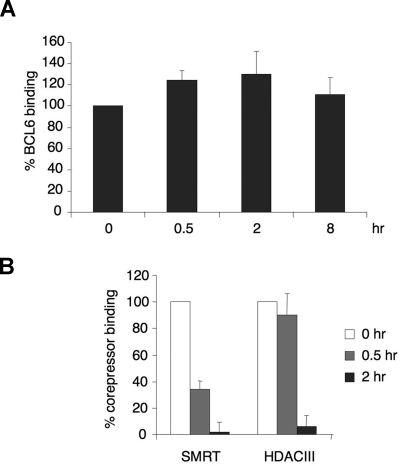
To examine whether BCL6 repression complexes located on the CD23b promoter were affected by CD40 signaling, we treated Ramos cells with CD40L for 0, 0.5, 2, and 8 hours and performed quantitative chromatin immunoprecipitations (QChIP) using BCL6 antibodies. There was no change in BCL6 recruitment to the BCL6 binding site in CD23b (Figure 2A) during the time points of our experiment. A nonspecific IgG did not enrich the CD23b promoter (not shown). These data suggest that, although present at the CD23b promoter, BCL6 may be functionally impaired by CD40 signaling. Because the magnitude of gene reactivation by corepressor blocking peptides was previously shown to be similar to the effect of CD40 on CD23b,22 we hypothesized that CD40 signaling might also disrupt the interaction between SMRT and BCL6. Ramos cells were exposed to CD40L for 0, 0.5, and 2 hours and subjected to QChIP using antibodies for SMRT and HDAC3 (which is a component of the SMRT complex26). In untreated cells, SMRT and HDAC3 were associated with the BCL6 binding region of the CD23b locus (Figure 2B). However, after 30 minutes of CD40 treatment, only approximately 50% of the initial SMRT signal remained, and at 2 hours SMRT was almost fully excluded. HDAC3 was also released after 2 hours of exposure to CD40L (Figure 2B). The apparently slower reduction in HDAC3 could be the result of several factors, such as the inherent difficulty in precisely quantifying protein abundance by ChIP, or residual HDAC3 localization to the promoter despite SMRT ejection. This result demonstrated that SMRT is ejected from BCL6 repression complexes on CD40 stimulation.
Figure 2.
CD40 rapidly ejects corepressors from the BCL6 repression complex on the CD23b promoter. (A) Chromatin immunoprecipitations were performed in Ramos cells at the indicated time points of CD40L exposure using BCL6 antibody. QPCR was performed to measure the enrichment of the CD23b promoter BCL6 binding site compared with input chromatin. The BCL6 enrichment values were first normalized to the input levels and expressed relative to untreated cells. The results correspond to the average of 3 independent replicates. (B) Chromatin immunoprecipitations were performed with SMRT and HDAC3 antibodies at the indicated time points after CD40L exposure to Ramos cells. The enrichment values for the different corepressors were first normalized to input levels and expressed relative to untreated cells. The results correspond to the average of 3 independent replicates. Error bars represent SEM.
CD40 triggers posttranslational modification of SMRT and its translocation to the cytoplasm
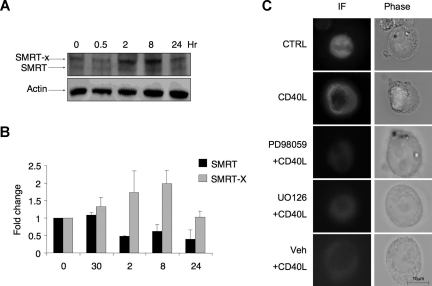
Western blots of Ramos cell lysates yielded 2 bands immunoreactive with SMRT antibodies (Figure 3A,B). The higher-molecular-weight species of SMRT was previously shown to be the result of SMRT phosphorylation.25,27 Treatment of cells with CD40 led to a striking shift of SMRT to the more slowly migrating form as shown in densitometry performed on 3 consecutive Western blots (Figure 3A,B), consistent with the occurrence of posttranslational modifications. We previously observed a similar shift of SMRT mobility after FLT3 singling in myeloid cells.25 SMRT phosphorylation via MAP/MEK kinases was previously shown to result in redistribution into the cytoplasm.25,27 CD40 can signal through MAP/MEK kinase pathways.28 Accordingly, a 2-hour exposure to CD40L was observed to cause transfected SMRT-GFP in Ramos cells to translocate from nucleus to cytoplasm as determined by immunofluorescence microscopy (Figure 3C). Pretreatment with 2 different MEK1/2 inhibitors, PD98059 and U0126 (but not vehicle control), could both abolish the cytoplasmatic redistribution of SMRT (Figure 3C). Our data suggest a physiologic mechanism through which CD40 can rapidly signal to SMRT via a MAP kinase pathway and cause it to translocate to the cytoplasm, thus disrupting the ability of BCL6 to repress target genes via the SMRT complex.
Figure 3.
CD40L triggers posttranslation modification of SMRT and its translocation to the cytoplasm. (A) Western blots were performed using SMRT antibody in Ramos cells exposed to CD40L during the indicated time points. Actin Western blots were performed as a loading control. As previously reported, SMRT is represented by 2 bands, a faster migrating product (SMRT) and a slower band (SMRT-X) indicative of a posttranslational modification. (B) The densitometry of 3 independent SMRT Western blot replicates. The signal of each SMRT Western blot was first normalized by actin levels and then expressed as fold change relative to untreated cells. ■ represents the faster migrating band;  , the slower migrating posttranslationally modified form of SMRT. Error bars represent SEM. (C) Ramos cells were transfected with a GFP-SMRT-expressing plasmid and then exposed to CD40L for 2 hours. Immunofluorescence and phase contrast microscopy were performed to determine the cellular localization of transfected SMRT. Rows 3 to 5 show experiments in which transfected Ramos cells were pretreated with the MEK kinase inhibitors PD98059 and UO126 or vehicle, followed by exposure to CD40L.
, the slower migrating posttranslationally modified form of SMRT. Error bars represent SEM. (C) Ramos cells were transfected with a GFP-SMRT-expressing plasmid and then exposed to CD40L for 2 hours. Immunofluorescence and phase contrast microscopy were performed to determine the cellular localization of transfected SMRT. Rows 3 to 5 show experiments in which transfected Ramos cells were pretreated with the MEK kinase inhibitors PD98059 and UO126 or vehicle, followed by exposure to CD40L.
The chromatin of the CD23b locus is set to a transcriptionally active state by CD40 signaling, leading to recruitment and processivity of POLII
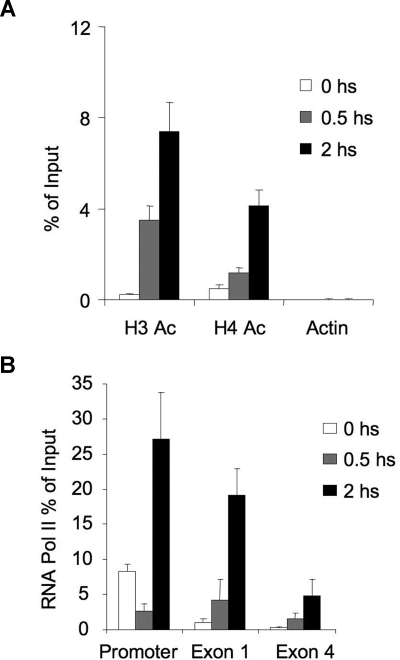
SMRT was previously shown to be an allosteric activator of histone deacetylases.29,30 To determine the effect of CD40L-induced disruption of the BCL6-SMRT-HDAC3 complex, we examined the acetylation status of histones surrounding the BCL6 binding site of the CD23b promoter by QChIP. In the presence of SMRT, the CD23b promoter exhibited low levels of histone acetylation, consistent with its repressed state (Figure 4A). However, exposure to CD40L for 0.5 and 2 hours caused progressive acetylation of both histone 3 and histone 4. This was similar to the kinetics of acetylation change we previously reported for BCl6 target genes after exposure to corepressor inhibitor peptides.22 The greater accessibility of the CD23b locus for transcription was reflected by the progressively greater processivity of RNA POLII observed after CD40L stimulation. This was shown by performing QChIP for RNA POLII at the CD23b gene promoter, exon 1, and exon 4 at different time points, to determine whether its abundance at these sites was increased on CD40L stimulation. Before CD40L stimulation, POLII was primarily associated with the promoter of the gene, but not with exon 1 and exon 4 (Figure 4B). Within 30 minutes of CD40L treatment, there was a reduction in POLII in the promoter and an increase at exon 1. After 2 hours of CD40L treatment, POLII increased by 3-fold at the promoter, 15-fold at exon 1, and 10-fold at exon 4, indicating that POLII was scanning the locus. These results indicated that CD40-mediated disruption of the BCL6-SMRT complex on the CD23b locus results in acetylation of histones 3 and 4 and movement of RNA POLII into the coding region of the gene.
Figure 4.
The chromatin of the CD23b locus is set to a transcriptionally active state by CD40 signaling, leading to recruitment and processivity of POLII. (A) QChIP assays were performed using anti-pan-Histone 3 acetylated, anti-pan-Histone 4 acetylated and antiactin antibodies in Ramos cells treated with CD40L during 0 (□), 30 minutes ( ), and 2 hours (■). QPCR was performed to detect the abundance of CD23b promoter sequence as in Figure 2. The y-axis represents the percentage enrichment by each antibody relative to input. (B) QChIP was performed at the indicated time points using RNA POLII antibodies to detect the abundance of POLII at the CD23b promoter, exon1 and exon 4. The y-axis represents the percentage enrichment of the amplicons relative to input. The result shows increase occupancy by POLII at exon 1 and exon 4 after CD40L exposure, consistent with active transcription of the gene. Each experiment was carried out in duplicate. Error bars represent SEM.
), and 2 hours (■). QPCR was performed to detect the abundance of CD23b promoter sequence as in Figure 2. The y-axis represents the percentage enrichment by each antibody relative to input. (B) QChIP was performed at the indicated time points using RNA POLII antibodies to detect the abundance of POLII at the CD23b promoter, exon1 and exon 4. The y-axis represents the percentage enrichment of the amplicons relative to input. The result shows increase occupancy by POLII at exon 1 and exon 4 after CD40L exposure, consistent with active transcription of the gene. Each experiment was carried out in duplicate. Error bars represent SEM.
CD40 signaling induces several BCL6 target genes in lymphoma cells and primary centroblasts
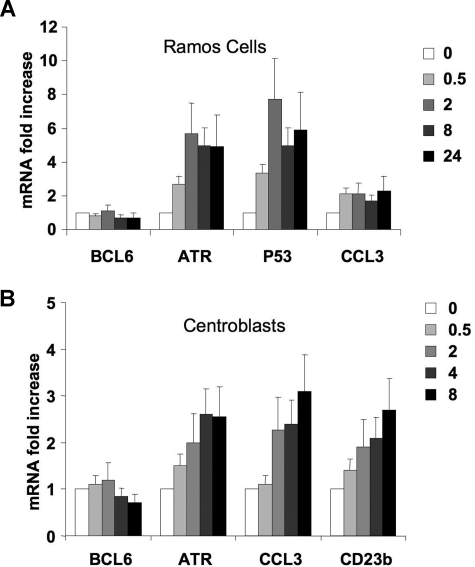
To test whether CD40 signaling could reactivate additional BCL6 targets, Ramos cells were exposed to CD40L, and the mRNA abundance of BCL6 target genes, including TP53, CCL3, and ATR, was measured by QPCR. The ATR mRNA was up-regulated within 30 minutes of CD40 stimulation and reached a plateau 6-fold increase at 2 hours (Figure 5A). TP53 was also rapidly up-regulated by CD40, reaching a 7-fold increase in 2 hours compared with untreated cells (Figure 5A). CCL3 RNA levels increased 2.1-fold after 30 minutes of CD40 stimulation and remained at that level during all time points. As shown in Figure 1B, a decrease in BCL6 mRNA abundance was noted at 8 hours of CD40 treatment (Figure 5A).
Figure 5.
CD40 signaling induces several BCL6 target genes in lymphoma cells and primary centroblasts. (A) Ramos cells were exposed to CD40L for 0, 0.5, 2, 8, or 24 hours, after which QPCR was performed to detect the mRNA abundance of BCL6, ATR, p53, and CCL3. The mRNA levels were first normalized by GAPDH levels and expressed as fold change relative to untreated cells. (B) Purified CD77+ centroblasts were exposed to CD40L for 0, 0.5, 2, 4, 8, or 24 hours after which QPCR was performed to detect the mRNA abundance of BCL6, ATR, CCL3, and CD23b. The mRNA levels were first normalized by GAPDH levels and shown as fold change relative to untreated cells. These experiments were performed in triplicate. Error bars represent SD.
To determine the relevance of this finding in the primary B-cell context, we purified primary human centroblasts from routine tonsillectomy specimens as previously reported.7,31 A purity of 95% was routinely confirmed by flow cytometry using the centroblast markers CD38 and CD77 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). As in the previous experiments with Ramos cells, QChIP assays showed that CD40L induced ejection of SMRT and HDAC3 from the CD23b locus within 30 minutes of exposure and without affecting BCL6 binding. This led to rapid histone 3 and histone 4 acetylation as well as increased occupancy by RNA POLII of exons 1 and 4 (Figure S2). To determine the impact of these CD40L on BCL6 target gene expression, mRNA was collected from centroblasts exposed to CD40L for increasing lengths of time and submitted to QPCR for measurement of BCL6, CD23b, CCL3, and ATR RNA abundance (Figure 5B). We found that the mRNA levels of CD23b increased after 30 minutes of CD40 stimulation and continued to steadily increase during the remainder of the time course, reaching a 2.7-fold increase compared with untreated cells. The other BCL6 target genes, ATR and CCL3, were also found to be up-regulated by CD40L, 3.1-fold for CCL3 and 2.6-fold for ATR. BCL6 mRNA was observed to become down-regulated within 8 hours after CD40 stimulation (Figure 5B). In the context of the GC reaction, these results suggest that T-cell signaling through CD40L to GC B cells can rapidly up-regulate BCL6 target genes before down-regulation of BCL6.
CD40-mediated up-regulation of BCL6 target genes is reversible
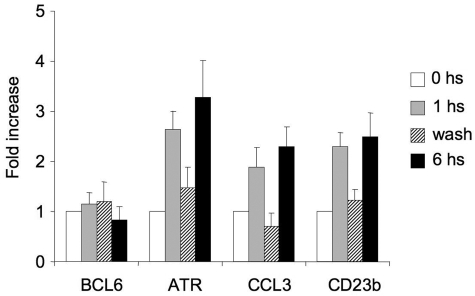
Live imaging of murine GCs has shown that GC B cells shuttle back and forth between the dark and light zones32,33 after transient interaction with T cells,32 which is a source of CD40 signaling and disruption of the BCL6-SMRT complex. Presumably, transcriptional repression of BCL6 target genes, especially those involved in key checkpoints, such as ATR and TP53, would need to be restored for cells to continue to proliferate and undergo further affinity maturation. Therefore, we wondered whether CD40-mediated induction of BCL6 target genes within the temporal time frame of B cell–T cell interactions would be reversible and thus provide a basis for how B cells could recycle back into the dark zone. For this experiment, Ramos cells were exposed to CD40L for 1 hour, after which cells were divided into 3 aliquots. The first aliquot was immediately lysed (1 hour); the second was washed with medium 3 times to remove the CD40L and then left in culture in CD40L-free media for an additional 5 hours (wash); the third fraction was left in CD40L-containing medium for another 5 hours (total exposure to CD40L = 6 hours). mRNA was extracted from all of these cells and examined by QPCR (Figure 6). We found that the BCL6 target genes CCL3, ATR, and CD23b were induced by 1 hour and sustained at 6-hour exposure to CD40L. However, in the washed cells, BCL6 target genes returned to the same levels as untreated cells. BCL6 mRNA levels did not change in any of the time points of the experiment (Figure 6). These results indicated that CD40 reactivation of BCL6 target genes can be a transient event and, in the context of the findings of Allen et al,1,32 imply that GC B cells can be probed by T helper cells in the light zone without necessarily down-regulating BCL6 permanently.
Figure 6.
CD40-mediated up-regulation of BCL6 target genes is reversible. Ramos cells were exposed to CD40L for 1 hour and then split into 3 fractions. The first fraction was immediately lysed ( ), the second was washed to remove the CD40L and then cultured for an additional 5 hours (▨), and the third fraction remained in CD40L containing media for another 5 hours (■). QPCR was performed in untreated cells and all 3 fractions to detect mRNA abundance of BCL6, ATR, CCL3, and CD23b. The mRNA levels of these genes were first normalized to GAPDH mRNA levels and expressed as fold increase relative to untreated cells. The experiment was performed in triplicate. Error bars represent SEM.
), the second was washed to remove the CD40L and then cultured for an additional 5 hours (▨), and the third fraction remained in CD40L containing media for another 5 hours (■). QPCR was performed in untreated cells and all 3 fractions to detect mRNA abundance of BCL6, ATR, CCL3, and CD23b. The mRNA levels of these genes were first normalized to GAPDH mRNA levels and expressed as fold increase relative to untreated cells. The experiment was performed in triplicate. Error bars represent SEM.
Discussion
Imaging studies in murine GCs show that these are dynamic structures in which highly motile centroblasts actively travel from the B cell–rich GC dark zone to the more heterogeneous GC light zone containing T cells, follicular dendritic cells, and macrophages. Presumably, GC dynamics are similar in humans, although this remains to be experimentally proven. Although it was originally thought that centroblasts move unidirectionally toward the light zone, it was recently shown that these B cells actually recycle between these compartments.32,34 B cells in the GC undergo class switch recombination and somatic hypermutation mediated by the enzyme activation-induced cytosine deaminase (AID) while simultaneously undergoing clonal expansion. The purpose of these events is to generate a diversity of mutated immunoglobulin loci and maximize the chance that clones of B cells with high-affinity antibodies will emerge. One reason why B cells may have evolved the ability to recycle back to the proliferative (ie, dark zone) of the GC is to allow further rounds of antibody mutagenesis to occur to maximize the chance of generating a high-quality antibody.
One hazard of the centroblast phenotype is that AID can introduce double-strand breaks and point mutations at transcriptionally active loci throughout the genome in addition to the immunoglobulin loci.35 When such events activate oncogenes or inactivate tumor suppressors, centroblasts are in danger of transforming into B-cell lymphomas.35,36 BCL6 plays a key role in facilitating this phenotype by repressing important DNA damage-sensing proteins and checkpoints, such as ATR and TP53.7,8 The combination of rapid replication and AID-induced DNA mutations could potentially seriously damage B cells. Because BCL6 can physiologically dampen the cell autonomous mechanisms that normally protect against genomic damage, it might be required that these B cells receive help from GC T cells. One way in which this might happen is that GC T cells could rapidly disrupt the actions of BCL6. Accordingly, our data show that the BCL6-SMRT complex is rapidly disrupted in B cells on exposure to CD40L. Under these conditions, SMRT shifts to a higher-mobility form previously shown to be associated with its phosphorylation and becomes delocalized to the cytoplasm. As a consequence, SMRT no longer associates with BCL6 on a target gene promoter. Promoter-associated nucleosomes become acetylated, RNA POLII is able to scan through the coding sequence, and mRNA for genes such as CD23b, ATR, and TP53 is produced. Washout of CD40L from the cells restores the ability of BCL6 to repress its target genes. Presumably, even transient restoration of these genes could allow damaged centroblasts to undergo cell death and is consistent with the finding that GC T cells were observed to be attached to fragments of apoptotic B cells. Along these lines, we previously showed that CD40 signaling can facilitate the death of damaged centroblasts and was associated with induction of ATR.7 In this way, transient interactions between B and T cells would allow the weeding out of damaged B cells from the GC reaction and would allow cells that have passed this inspection to restore BCL6 transcriptional programming and undergo additional cycles of affinity maturation. A recent report suggests that heavily damaged B cells could induce BCL6 protein degradation through an ataxia telangiectasia mutated–dependent pathway.37 How this pathway would connect with the CD40 effects reported herein remains to be determined.
The SMRT corepressor can bind to the BCL6 N-terminal BTB domain through an 18-amino acid BCL6 binding domain motif, which is also highly conserved with the N-CoR corepressor.38 Both N-CoR and SMRT thus bind to BCL6 in an identical manner. A peptide derived from the SMRT BCL6 binding domain was previously shown to disrupt BCL6-mediated transcriptional repression by displacing SMRT and N-CoR from the BCL6 repression complex.22 Both SMRT and N-CoR form HDAC3-containing complexes. Therefore, BCL6 can recruit either SMRT or N-CoR to its target genes, and our data show that, within a population of cells, both corepressors can be found associated with a number of BCL6 target genes and may thus functionally overlap.39 We have shown that CD40 signaling can also disrupt the association between N-CoR and BCL6.7 Collectively, these data suggest that CD40 can eliminate both HDAC3-containing complexes from BCL6 target genes, which we show herein can result in recovery of histone acetylation, RNA POLII processivity, and gene expression. These data are consistent with previous reports showing that SMRT association with the PLZF transcriptional repressor and nuclear hormone receptors can be disrupted by EGFR and FLT3 signaling and results in delocalization of SMRT to the cytoplasm.25,27
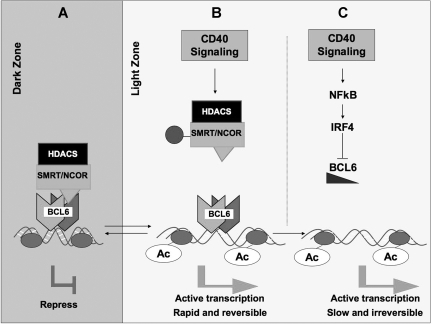
These data are complementary to reports demonstrating that longer exposure to CD40L causes transcriptional down-regulation of BCL6 in B cells.20,23,40,41 Taken together, the data suggest that CD40 stimulation could inhibit BCL6 through 2 separate mechanisms (Figure 7). In the first, transient interaction with T cells temporarily disrupts BCL6 repression of checkpoint genes, allowing B cells to undergo apoptosis if they are damaged or to recycle back for more affinity maturation (Figure 7). However, sustained exposure to CD40L, possibly mediated by more extended contact with follicular dendritic cells, can definitively down-regulate BCL6, at least in part through induction of IRF4, which can directly repress the BCL6 locus23 (Figure 7). This complete and sustained loss of BCL6 in combination with other factors triggered at this stage could facilitate the further differentiation of cells with productive antibody rearrangement into plasma or memory cells. Therefore, differences in the delivery of CD40 signaling could induce 2 alternative regulatory effects on BCL6 with distinct biologic outcomes.
Figure 7.
CD40 signaling can block the function of BCL6 through 2 independent mechanisms. (A) In dark zone centroblasts, BCL6 represses target genes through recruitment of the SMRT and N-CoR corepressors, both of which form histone deacetylase (HDAC) complexes. This facilitates proliferation and immunoglobulin affinity maturation. (B) CD40 signaling in the light zone, for example, by GC T cells, leads to posttranslational modification of these corepressors ( ) and loss of their association with BCL6. This leads to a failure to maintain silencing of these genes, which can now become reactivated even in the presence of BCL6. The double arrow between dark zone and light zone indicates that this is a reversible mechanism. This mechanism is rapid and may allow damaged B cells to be removed from the GC reaction. (C) In the second mechanism, sustained CD40 signaling, for example, by follicular dendritic cells, can activate NFκB, which in turn induces expression of IRF4, which can then directly repress BCL6 mRNA expression leading to down-regulation of BCL6 and up-regulation of its target genes. This mechanism is slower but is irreversible and leads to differentiation of B cells positively selected for high-affinity antibody.
) and loss of their association with BCL6. This leads to a failure to maintain silencing of these genes, which can now become reactivated even in the presence of BCL6. The double arrow between dark zone and light zone indicates that this is a reversible mechanism. This mechanism is rapid and may allow damaged B cells to be removed from the GC reaction. (C) In the second mechanism, sustained CD40 signaling, for example, by follicular dendritic cells, can activate NFκB, which in turn induces expression of IRF4, which can then directly repress BCL6 mRNA expression leading to down-regulation of BCL6 and up-regulation of its target genes. This mechanism is slower but is irreversible and leads to differentiation of B cells positively selected for high-affinity antibody.
Supplementary Material
Acknowledgments
A.M. is supported by National Cancer Institute grant R01 CA104348, the Chemotherapy Foundation, and the Sam Waxman Cancer Research Foundation, and is a Scholar of the Leukemia & Lymphoma Society. J.D.L. and A.M. are supported by National Cancer Institute grant R01 CA59936.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: J.M.P. performed the experiments shown in Figures 1 to 6, helped design the experiments, and wrote the first draft; W.C. performed the experiments shown in Figures 3, 5, and 6 and helped design the experiments; J.D.L. designed the experiments, provided key intellectual contributions and critical reagents, and edited the manuscript; and A.M. designed the experiments, interpreted the results, and edited and wrote the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ari Melnick, Weill Cornell Medical College-C6, 1300 York Avenue, New York, NY, 10021; e-mail: amm2014@med.cornell.edu.
References
- 1.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janeway CA, Jr, Bottomly K. Signals and signs for lymphocyte responses. Cell. 1994;76:275–285. doi: 10.1016/0092-8674(94)90335-2. [DOI] [PubMed] [Google Scholar]
- 3.Berek C, Ziegner M. The maturation of the immune response. Immunol Today. 1993;14:400–404. doi: 10.1016/0167-5699(93)90143-9. [DOI] [PubMed] [Google Scholar]
- 4.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 5.Ye BH, Cattoretti G, Shen Q, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- 6.Ranuncolo SM, Polo JM, Melnick A. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B cells. Blood Cells Mol Dis. 2008 Mar 15; doi: 10.1016/j.bcmd.2008.02.003. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ranuncolo SM, Polo JM, Dierov J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 8.Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- 9.Phan RT, Saito M, Basso K, Niu H, Dalla-Favera R. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat Immunol. 2005;6:1054–1060. doi: 10.1038/ni1245. [DOI] [PubMed] [Google Scholar]
- 10.Polo JM, Juszczynski P, Monti S, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci U S A. 2007;104:3207–3212. doi: 10.1073/pnas.0611399104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baron BW, Anastasi J, Montag A, et al. The human BCL6 transgene promotes the development of lymphomas in the mouse. Proc Natl Acad Sci U S A. 2004;101:14198–14203. doi: 10.1073/pnas.0406138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattoretti G, Pasqualucci L, Ballon G, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell. 2005;7:445–455. doi: 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 13.Ye BH. The role of Bcl-6 in normal lymphoid system and non-Hodgkin's lymphomas. In: Ravid K, Licht JD, editors. Transcription Factors: Normal and Malignant Development of Blood Cells. Vol. 1. New York: John Wiley & Sons; 2001. pp. 271–289. [Google Scholar]
- 14.Shaffer AL, Lin KI, Kuo TC, et al. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 15.Vasanwala FH, Kusam S, Toney LM, Dent AL. Repression of AP-1 function: a mechanism for the regulation of Blimp-1 expression and B lymphocyte differentiation by the B cell lymphoma-6 protooncogene. J Immunol. 2002;169:1922–1929. doi: 10.4049/jimmunol.169.4.1922. [DOI] [PubMed] [Google Scholar]
- 16.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 17.Parekh S, Polo JM, Shaknovich R, et al. BCL6 programs lymphoma cells for survival and differentiation through distinct biochemical mechanisms. Blood. 2007;110:2067–2074. doi: 10.1182/blood-2007-01-069575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reljic R, Wagner SD, Peakman LJ, Fearon DT. Suppression of signal transducer and activator of transcription 3-dependent B lymphocyte terminal differentiation by BCL-6. J Exp Med. 2000;192:1841–1848. doi: 10.1084/jem.192.12.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo TC, Shaffer AL, Haddad J, Jr, Choi YS, Staudt LM, Calame K. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med. 2007;204:819–830. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allman D, Jain A, Dent A, et al. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- 21.Gupta S, Jiang M, Anthony A, Pernis AB. Lineage-specific modulation of interleukin 4 signaling by interferon regulatory factor 4. J Exp Med. 1999;190:1837–1848. doi: 10.1084/jem.190.12.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polo JM, Dell'Oso T, Ranuncolo SM, et al. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med. 2004;10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 23.Saito M, Gao J, Basso K, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 24.Basso K. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi S, McConnell MJ, Harigae H, et al. The Flt3 internal tandem duplication mutant inhibits the function of transcriptional repressors by blocking interactions with SMRT. Blood. 2004;103:4650–4658. doi: 10.1182/blood-2003-08-2759. [DOI] [PubMed] [Google Scholar]
- 26.Guenther MG, Lane WS, Fischle W, Verdin E, Lazar MA, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 27.Hong SH, Privalsky ML. The SMRT corepressor is regulated by a MEK-1 kinase pathway: inhibition of corepressor function is associated with SMRT phosphorylation and nuclear export. Mol Cell Biol. 2000;20:6612–6625. doi: 10.1128/mcb.20.17.6612-6625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li YY, Baccam M, Waters SB, Pessin JE, Bishop GA, Koretzky GA. CD40 ligation results in protein kinase C-independent activation of ERK and JNK in resting murine splenic B cells. J Immunol. 1996;157:1440–1447. [PubMed] [Google Scholar]
- 29.Fischle W, Dequiedt F, Hendzel MJ, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 30.Yu J, Li Y, Ishizuka T, Guenther MG, Lazar MA. A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO J. 2003;22:3403–3410. doi: 10.1093/emboj/cdg326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein U, Tu Y, Stolovitzky GA, et al. Transcriptional analysis of the B cell germinal center reaction. Proc Natl Acad Sci U S A. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007;315:528–531. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 33.Hauser AE, Shlomchik MJ, Haberman AM. In vivo imaging studies shed light on germinal-centre development. Nat Rev Immunol. 2007;7:499–504. doi: 10.1038/nri2120. [DOI] [PubMed] [Google Scholar]
- 34.Schwickert TA, Lindquist RL, Shakhar G, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007;446:83–87. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 35.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Li Z, Naganuma A, Ye BH. Negative autoregulation of BCL-6 is bypassed by genetic alterations in diffuse large B cell lymphomas. Proc Natl Acad Sci U S A. 2002;99:15018–15023. doi: 10.1073/pnas.232581199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phan RT, Saito M, Kitagawa Y, Means AR, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad KF, Melnick A, Lax S, et al. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell. 2003;12:1551–1564. doi: 10.1016/s1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 39.Polo J, Chatzi K, Dell'Oso T, Lev P, Melnick A. BCL6 transcriptionally reprograms the DLBCL genome by forming distinct biochemically active complexes on genes corresponding to different pathways[abstract]. Blood. 2007;118:778a–779a. [Google Scholar]
- 40.Gupta S, Anthony A, Pernis AB. Stage-specific modulation of IFN-regulatory factor 4 function by Kruppel-type zinc finger proteins. J Immunol. 2001;166:6104–6111. doi: 10.4049/jimmunol.166.10.6104. [DOI] [PubMed] [Google Scholar]
- 41.Niu H, Cattoretti G, Dalla-Favera R. BCL6 controls the expression of the B7-1/CD80 costimulatory receptor in germinal center B cells. J Exp Med. 2003;198:211–221. doi: 10.1084/jem.20021395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.