Abstract
HMGA1 is a member of a small family of architectural transcription factors involved in the coordinate assembly of multiprotein complexes referred to as enhanceosomes. In addition to their role in cell proliferation, differentiation, and development, high-mobility group proteins of the A type (HMGA) family members behave as transforming protoncogenes either in vitro or in animal models. Recent reports indicated that HMGA1 might counteract p53 pathway and provided an interesting hint on the mechanisms determining HMGA's transforming potential. HMGA1 expression is deregulated in a very large array of human tumors, including cervical cancer, but very limited information is available on the molecular mechanisms leading to HMGA1 deregulation in cancer cells. Here, we report that HMGA1 expression is sustained by human papilloma virus (HPV) E6/E7 proteins in cervical cancer, as demonstrated by either E6/E7 overexpression or by repression through RNA interference. Knocking down HMGA1 expression by means of RNA interference, we also showed that it is involved in cell proliferation and contributes to p53 inactivation in this type of neoplasia. Finally, we show that HMGA1 is necessary for the full expression of HPV18 E6 and E7 oncoproteins thus establishing a positive autoregulatory loop between HPV E6/E7 and HMGA1 expression.
Introduction
High-mobility group proteins of the A type (HMGA) are a small family of nonhistonic architectural transcription factors, comprising at least three major members: HMGA1a, HMGA1b, and HMGA2 [1]. Their ability to bind the minor groove of DNA and to interact with a large number of proteins allow them to coordinate the assembly and disassembly of multiprotein transcription complexes called enhanceosomes and to control transcription of an ample number of genes [1–5]. HMGA family members are strongly expressed in early developmental stages, where they are involved in controlling cell proliferation and differentiation. Indeed, inactivation of HMGA2 leads to the mouse “pigmy” phenotype [6], whereas overexpression of an active HMGA2 confers a giant phenotype in transgenic animals [7]. HMGA expression declines to very low levels in most adult differentiated tissues, but it is deregulated in human tumors of both mesenchymal and epithelial origin. HMGA1, in particular, is overexpressed in carcinomas of the thyroid [8], colon [9–11], prostate [12,13], pancreas [14], ovary [15], and breast [16] and in neuroblastoma [17]. Its increased expression may be associated to more aggressive and/or metastatic stages [12,16,18,19]. HMGA proteins possess transforming activity, both in vitro and in animal models [20–26]. This might be related to their role in gene transcription [1] but might also involve the inhibition of oncosuppressive pathways. Indeed, HMGA1 binds and counteracts p53 [27,28]. Furthermore, it binds and, if in molar excess, delocalizes to the cytoplasm HIPK2 [29], one of the kinases responsible for p53 serine 46 phosphorylation committing p53 toward an apoptotic response [30]. Relatively little is known on the mechanisms leading to HMGA deregulation in human tumors. Oncogenic pathways, such as those sustained by MYCN, c-myc and ras might be relevant in specific neoplasia [22,31,32].
Cervical cancer is the second most common cancer in women. Its epidemiology is strongly linked to human papilloma virus (HPV) infection that is necessary, but not sufficient, for the development of virtually all cervical squamous cell carcinomas and for approximately 95% of the adenocarcinomas. Human papilloma virus infection is a very common phenomenon, but in 80% of the cases, the infection is quickly cleared. Persistence of the infection of the “high-risk” strains (HPV16 and 18) is frequently associated to the development of noninvasive squamous precursor lesions called cervical intraepithelial neoplasia (CIN) [33–35]. Cervical intraepithelial neoplasia might vary from CIN-1 to CIN-3, mostly depending on the grading of dysplasia. E6 and E7HPV proteins are necessary for the induction and maintenance of the transformed phenotype, mostly through the inactivation of p53 (through E6-mediated degradation) and RB (by means of E7 binding) pathways [36,37].
The Notch pathway is an important tuner of cell proliferation and differentiation whose deregulation contributes to human carcinogenesis. In cervical cancer, conflicting reports described either Notch1 increased expression and functional cooperation with HPV oncoproteins [38–41] or the requirement for Notch1 signaling downmodulation to maintain the transformed phenotype and HPV E6/E7 expression [42,43]. The levels of Notch1 overexpression achieved in the different experimental settings might be at least partially responsible for these contrasting evidences [44]. Nevertheless, Notch1 is an oncosuppressor in the epithelial context, in vivo [45]. In addition, because p53 might directly induce Notch1 expression [46–48], its repression by HPV oncoproteins might be responsible for the impairment of Notch1 oncosuppression in cervical carcinogenesis [47].
HMGA1 expression in cervical cancer has been previously reported [49], but the mechanism and the consequences of this have not been addressed. By using Notch1 as a tool to convey growth inhibitory signals, we show that HMGA1 expression in cervical cancer cells is sustained by HPV E6/E7 proteins. We also show that HMGA1 repression through RNA interference inhibits cell proliferation and contributes to p53 reactivation leading to sensitization to DNA-damaging drugs in cooperation with Notch1. Finally, we show that HMGA1 is necessary for the full expression of HPV18 E6 and E7 oncoproteins, thus establishing an autoregulatory loop between HPV E6/E7 transcription and HMGA1 itself.
Materials and Methods
Cells and Viruses
Human cervical cancer cells (HeLa, SiHa, and Caski) cells were maintained in standard conditions in Dulbecco's modified Eagle's medium. HeLa E6/E7 cells (expressing the HPV16 E6/E7 transcript under the control of an heterologous promoter) were kindly provided by Dr. D. DiMaio and grown as indicated [50]. Recombinant adenoviruses expressing the constitutively active form of human Notch1 and the green fluorescent protein (GFP) were previously described [43]. Viruses were used at a multiplicity of infection of 100.
Plasmids, Transfections, and Luciferase Assay
The expression plasmids for activated Notch1 and the HPV18 promoter luciferase reporter construct (pGL3-HPV18LCR) were previously described [43]. Transient transfections were achieved by means of the Lipofectamine 2000 Reagent (Invitrogen, San Diego, CA) according to the manufacturer's instructions. PG13 p53 luciferase reporter construct was stabilized in HeLa cells by a short cycle of puromicin selection. Luciferase reporter construct assays were performed using a Luciferase Reporter Assay System (Promega Corporation, Madison, WI). Cells were lysed, and luciferase activity was determined using a Veritas automatic dual-injector luminometer (Turner Biosystems, Sunnyvale, CA). Data were normalized on a Renilla luciferase reporter construct and/or on protein content. All conditions were tested in duplicate samples, and experiments were repeated at least three times.
RNA Preparation and Real-Time Quantitative Polymerase Chain Reaction Analysis
Total RNA was extracted using the RNeasy system (Qiagen, Hilden, Germany) or the TRIzol reagent (Invitrogen). For quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis, total RNA (1 µg) was reverse-transcribed using GeneAmp-Gold RNA PCR Core Kit (Applied Biosystems, Warrington, United Kingdom). One-twentieth of this reaction was employed for real-time quantitative PCR using either SYBR Green or TaqMan PCR Master Mixes (Applied Biosystems). The analysis of human HMGA1, HES1, p21, β-actin mRNA, and that of HPV16 and 18 E6/E7 transcripts was performed with designed (HMGA1, E6/E7) and commercial ABI reagents using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems). All amplification reactions were done in triplicate, and the averages of the threshold cycles were used to interpolate standard curves and to calculate transcript amounts using SDS version 1.7a software (Applied Biosystems). Results were normalized on endogenous controls, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and β-actin.
Western Blot
Protein extracts were made in Laemmli buffer and were separated on SDS-PAGE, blotted onto nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany), blocked in 10% nonfat dry milk, and probed with various antibodies [goat anti-Notch1 (C-20), rabbit antip21, mouse anti-p53, goat anti-β-actin, rabbit anti-ERK1/2, and mouse anti-phospho-ERK1/2 (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-CREB (kindly provided by Luca Canettieri); and rabbit anti-HMGA1 (kindly provided by Dr. G. Manfioletti)]. HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology. Immunoreactive bands were visualized by enhanced chemiluminescence (Pierce Chemical Co., Rockford, IL).
RNA Interference and Cell Proliferation Assays
RNA interference on HPV16 E6/E7 transcript was achieved by means of the small interfering RNA (siRNA) duplex HPV16 E7 (sense seq: GCATGGAGATACTCCTACA) previously described [51], synthesized by Dharmacon Res., Inc. (Lafayatte, CO), and transfected in SiHa cells with Dharmafect 1 according to the manufacturer's instructions. RNA interference on HMGA1 was achieved by means of siRNA duplexes si-A1-pool or si-A1–4 (a pool of four different siRNA against HMGA1, respectively, with one of them used singly, produced by Dharmacon Res., Inc.) or PLKO vectors expressing shRNA (Sigma-Aldrich Co., St. Louis, MO). HeLa cells were plated in multiwell 24 plates and were transfected either with (5 nM) A1-pool or A1-4 or nonspecific siRNA (NT) siRNA duplexes, or vehicle only, by using the Hiperfect reagent (Qiagen), according to the manufacturer's instructions. Three days after plating, cells were detached by trypsinization and split in six replicate wells or plated in chamber slides for 5′-bromo-2′-deoxyuridine (BrdU) labeling assay. The following day, cells were retransfected as previously mentioned. At day 6 after initial plating, cells were processed for RNA and protein extraction. For BrdU incorporation assay, at day 6 from initial plating and after 11 hours of pulse with BrdU, cells were processed by means of a BrdU labeling and detection kit (Roche Diagnostics, Indianapolis, IN).
For long-term colony growth assays, 2 x 106 HeLa cells were transfected with 8 µg of either PLKO A1–49, A1–51, or A1–53 plasmids (targeting different regions of HMGA1 transcript) or with the notarget NT PLKO construct. After a short (3 days) cycle of puromicin selection, cells were monitored for HMGA1 mRNA expression or replated in duplicates for colony formation assay. After 2 weeks, colonies were stained with Coomassie solution (50% methanol, 10% acetic acid, 0.05% Coomassie blue R-250 powder) and were scored.
Results
HPV E6/E7 Oncoproteins Sustain HMGA1 Expression in Human Cervical Cancer Cells
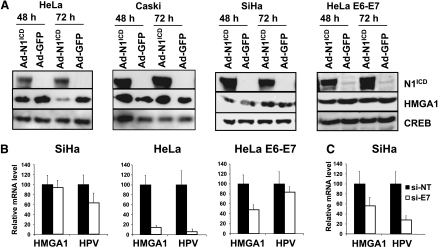
Invasive uterine cervical carcinomas are characterized by high HMGA1 expression, a condition reproduced by a number of cervical cancer cell lines, in vitro [49]. However, the mechanisms through which HMGA1 expression is deregulated in these tumors have not been fully elucidated. Therefore, we sought to study whether growth inhibitory and oncosuppressive stimuli, such as those conveyed by Notch1 signaling, could affect HMGA1 expression in these settings. Adenoviral delivery of the activated form of Notch1 (Ad-N1ICD) clearly repressed HMGA1 protein expression in HeLa cells (Figure 1A) but failed to do so in Caski and SiHa cells (Figure 1A). As previously reported [42], Notch1 activated the ERK pathway and increased Hes1 expression in the three cell lines (not shown). At the mRNA level, again Notch1 repressed HMGA1 expression in HeLa but not in SiHa cells (Figure 1B). This result is reminiscent of Notch1's ability to efficiently repress E6/E7 expression in HeLa but not in SiHa and Caski cells, as we previously observed (Figure 1B and Talora et al. [42]). Together with the observation of a slower kinetics of HMGA1 repression compared to E6/E7 transcript repression on Ad-N1ICD infection (not shown), these data suggest an association between the effects of Notch1 on E6/E7 and the levels of HMGA1 expression. To directly test the contribution of HPV oncoproteins to maintaining HMGA1 expression in human cervical cancer cell lines, we used HeLa E6–E7 cells, where the HPV16 E6/E7 transcript is controlled by a heterologous promoter [50]. In these cells, Ad-N1ICD infection resulted in the repression of the endogenous HPV18 E6/E7 transcript (not shown) but left the expression of the exogenous HPV16 E6/E7 transcript almost unaffected (Figure 1B). Under these conditions, repression of HMGA1 mRNA was far more modest compared to control HeLa cells and did not result in any significant protein change (Figure 1, A and B) suggesting that HPV E6/E7 oncoproteins might be directly involved in stimulating HMGA1 expression. To confirm the role of HPV oncoproteins in sustaining HMGA1 expression in cervical cancer, we knocked down E6/E7 transcript in the SiHa cell line by means of an interfering RNA known to inhibit both E6 and E7 expression [51]. Consistent with the hypothesis, E6/E7 repression resulted in approximately 55% reduction of HMGA1 expression also in SiHa cells (Figure 1C). Overall, these results indicate that HMGA1 expression is directly sustained by HPV E6 and/or E7 protein expression in cervical cancer cells.
Figure 1.
Notch1 represses HMGA1 expression in HeLa cervical cancer cells via HPV E6/E7 downmodulation. (A) Total proteins extracted from cells infected with an adenovirus expressing Notch1 constitutively active intracellular domain (Ad-N1ICD) or a control adenovirus (Ad-GFP) were analyzed for HMGA1 expression. (B) Total RNA extracted from cells infected with Ad-GFP (black bars) or Ad-N1ICD (white bars) were analyzed for the expression of HMGA1 and HPV E6/E7 transcripts (HPV18 E6/E7 for HeLa cells and HPV16 E6/E7 for SiHa and HeLa E6–E7 cells) by quantitative PCR. (C) SiHa cells transfected with the si-NT (black bars) or the si-E7 (white bars) described in the Materials and Methods section were analyzed by quantitative PCR for the expression of HMGA1 and HPV16 E6/E7 transcripts. For (B and C), results were normalized on GAPDH mRNA levels, and expressed as percentage relative to the control samples.
HMGA1 Knock Down Impairs Cervical Cancer Cell Proliferation and Cooperates with Notch1 to Increase p53 Activity and to Sensitize Cells to Cisplatinum
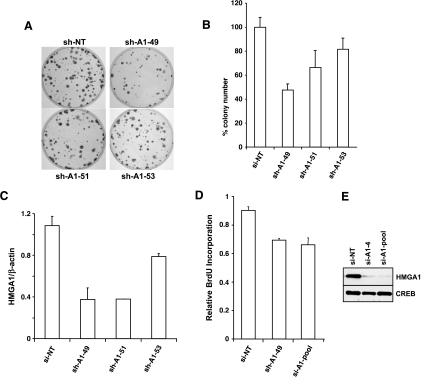
HMGA1 has been described as a potential oncogene whose expression is inappropriately increased in several cancer types. To test whether and how it could contribute to cervical carcinogenesis, we investigated on the consequences of HMGA1 depletion in HeLa cells, by means of short hairpin (sh-) or small interfering (si-) RNA. Transfection of three distinct sh-vectors against human HMGA1 leads to reduced colonyforming capability in HeLa cells (Figure 2, A and B), with an efficiency proportional to their effect on the HMGA1 transcript expression (Figure 2C). These results were consistent with a reduction in the BrdU incorporation observed in HeLa cells transiently transfected with siRNA against HMGA1, which efficiently reduced HMGA1 transcript (not shown) and protein expression (Figure 2, D and E).
Figure 2.
HMGA1 knock down impairs cell growth in HeLa cervical cancer cells. (A) The colony-forming ability of HeLa cells is reduced by transfection of three different PLKO sh-expressing vectors against HMGA1. (B) Graphical representation of the results in (A). (C) Effects of the three different PLKO vectors on HMGA1 expression as measured by quantitative PCR. (D) DNA synthesis of HeLa cells measured by BrdU incorporation is inhibited by cell transfection with two si-RNA oligonucleotides against HMGA1. (E) Effects of the two si-RNA oligonucleotides on HMGA1 protein expression.
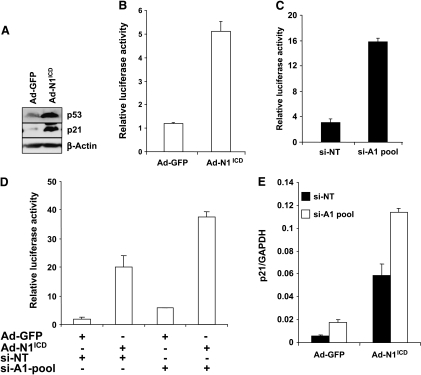
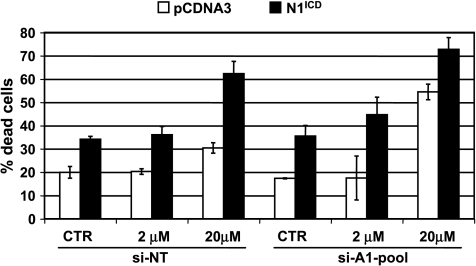
In cervical cancer cells, activation of Notch1 signaling has been associated to cell growth inhibition dependent on E47 degradation and/or on p53 accumulation [42]. The latter condition specifically occurs in HeLa cells on E6 repression due to a Notch1-dependent inhibition of the AP1 transcriptional activity on the E6/E7 promoter [43]. HMGA1 was recently reported to counteract p53-oncosuppressive activities [27–29]. Because p53 inactivation by HPV is a fundamental step in cervical carcinogenesis, we hypothesized that the HPV-dependent HMGA1 expression might contribute to a stronger inactivation of p53 in cervical cancer. To test this hypothesis, we used again Ad-N1ICD-infected HeLa cells, where the reduced expression of the HPV18 E6 protein resulted in p53 accumulation (Figure 3A and Talora et al. [43]) and a five-fold increase in the luciferase activity of the PG13 p53 reporter construct (Figure 3B). Knocking down HMGA1 expression through siRNA also resulted in an increase in the PG13 reporter activity (Figure 3C) and potentiated the effects of Ad-N1ICD infection (Figure 3D). The expression of an endogenous p53 target gene, such as p21, also increased by either Ad-N1ICD infection or HMGA1 RNA interference, and the two conditions resulted in a synergic effect (Figure 3E). These results collectively indicate that Notch1 infection and HMGA1 depletion cooperate in raising p53 activity. Increased p53 activity is expected to result in increased cell death especially on treatment with therapeutic DNA-damaging drugs, such as cisplatinum (CDDP). Indeed, transfection of a plasmid vector expressing N1ICD in HeLa cells increased the rate of cell death compared with control-transfected cells (Figure 4). In addition, it sensitized HeLa cells to the effect of CDDP, raising to more than 60% the rate of dead cells on 20-µM CDDP treatment. Interestingly, HMGA1 knock down also sensitized cells to the effect of CDDP at the highest concentration and cooperated with Notch1 to further increase cell death in response to the two drug concentrations (Figure 4).
Figure 3.
Notch1 and HMGA1 knock down independently and cooperatively downmodulate p53 activity. (A) Total proteins extracted from cells infected with Ad-N1ICD or Ad-GFP were analyzed for p53 and p21 expression. (B) HeLa cells transfected with the PG13 p53 reporter construct were then infected for 24 hours with either Ad-N1ICD or Ad-GFP and analyzed for luciferase activity. (C) HeLa cells transfected with the PG13 p53 reporter construct were transfected with the si-A1-pool or the si-NT as described in the Materials and Methods section and analyzed for luciferase activity. (D) HeLa cells transfected with the PG13 p53 reporter construct were transfected with the si-A1-pool or the si-NT as described in the Materials and Methods section and then infected for 24 hours with either Ad-N1ICD or Ad-GFP and analyzed for luciferase activity. (E) Quantitative PCR analysis of the expression of the endogenous p21 transcript in HeLa cells transfected with si-NT (black bars) or si-A1-pool (white bars) infected by either Ad-N1ICD or Ad-GFP.
Figure 4.
Notch1 and HMGA1 knock down independently and cooperatively sensitize HeLa cells to cisplatinum. HeLa cells were transfected with the si-A1-pool as described in the Materials and Methods section and then transfected with either N1ICD (black bars) or pCDNA3 control vector (white bars), before being treated with the indicated concentrations of cisplatinum. After 48 hours, cells were harvested and counted in a hemocytometer.
HMGA1 Knock Down Impairs HPV18 E6/E7 Expression
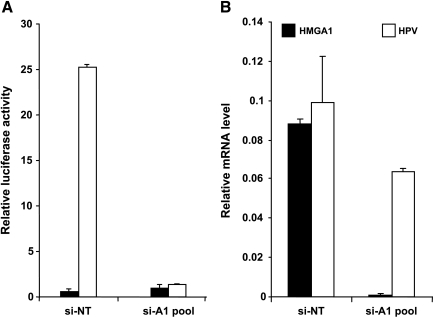
An additional aspect of cervical carcinogenesis deals with the observation that HPV18 enhanceosome contains partially overlapping AP1 and HMGA1 binding sites, both of which were shown to be important for HPV18 transcriptional activity [52,53]. This implicates that HMGA1 increased expression might also be relevant for the overt E6/E7 expression at least in HPV18-infected cells. To test this hypothesis, we looked at the effect of HMGA1 knock down on the transcription of the pGL3-HPV18LCR reporter construct. Indeed, we observed a very strong decrease in luciferase activity on HMGA1 RNA interference compared to control HeLa cells (Figure 5A). Consistent with these data, we could also reproducibly detect a 40% reduction in the HPV18 E6/E7 endogenous transcript (Figure 5B), confirming the relevant role of sustained HMGA1 expression for HPV genome expression.
Figure 5.
HMGA1 knock down impairs HPV E6/E7 transcription. (A) HeLa cells transfected with the si-A1-pool and the si-NT as described in the Materials and Methods section were then retransfected with the PGL3 vector (black bars) or the pGL3-HPV18LCR reporter construct (white bars) and analyzed for luciferase activity. (B) HeLa cells transfected with the si-A1-pool and the si-NT as described in the Materials and Methods section were then analyzed by quantitative PCR for the expression of HMGA1 (black bars) or HPV18 E6/E7 (white bars) transcripts normalized on GAPDH.
Discussion
Despite the largely confirmed association between HMGA1 deregulation and cancer, much less is known about the molecular events leading to this phenomenon. c-myc and MYCN can increase HMGA1 expression through a direct transcriptional regulation in Burkitt lymphoma and neuroblastoma, respectively [22,32]. Ras pathway might also be involved in HMGA1 deregulation in certain neoplasia [31]. Here, we investigated on the molecular mechanisms of HMGA1 deregulation in cervical cancer and showed that HMGA1 expression is sustained by HPV E6 and/or E7 proteins. Indeed, by using Notch1 as a mean to convey growth inhibitory and oncosuppressive stimuli we observed that HMGA1 down-regulation occurs only in HeLa cells, where E6/E7 repression also occurs because of a reduced AP1 transcriptional activity [43]. Notch1-dependent HMGA1 decrease is prevented by the overexpression of the HPV16 E6 and E7 proteins in HeLa cells. Conversely, E6/E7 knockdown through RNAi impairs HMGA1 expression in SiHa cells. Together with the observation that HPV16 E6 protein induced HMGA1 expression in murine fibroblasts [54], our data clearly establish a link between HPV E6/E7 proteins and HMGA1-deregulated expression in cervical cancer. Although this regulation is likely to occur at the transcriptional level, we were unable to show any effect of Notch1 in repressing the activity of the HMGA1 promoter reporter construct we recently described [32] (not shown). Further efforts will be necessary to better characterize the molecular details of this regulation.
HMGA1 clearly possess transforming capabilities both in vitro and in animal models, and this might be related to its ability to modify gene expression [20–26] and to counteract p53. Indeed, HMGA1 can bind p53 and impair its transcriptional activity either directly [27,28] or through the cytoplasmic delocalization and inhibition of HipK2 [29], one of the kinases phosphorylating p53 on serine 46 and turning on its proapoptotic potential [30]. In this article, we also addressed the issue of HMGA1 contribution to cervical carcinogenesis. Our data clearly show that interfering with HMGA1 expression cause reduced BrdU incorporation and impaired colony-forming ability in HeLa cells, confirming the involvement of HMGA1 in cell proliferation in these settings. In addition, we observed that HMGA1 interference was sufficient to induce p53 activity measured either through a p53 luciferase reporter construct or through endogenous p21 induction. In HPV-infected cervical cancer cells, p53 is largely inactivated due to its binding to the E6 protein that targets it for degradation. Notch1 represses E6/E7 expression in AP-1-competent cells, such as HeLa cells, thus leading to p53 accumulation [43]. Indeed, we could measure increased p53 activity both in the p53 luciferase reporter assay and in terms of p21 expression. Notch1 effects could be potentiated by HMGA1 knock down, in keeping with its role in repressing p53 activity. Furthermore, we observed that Notch1 expression could sensitize HeLa cervical cancer cells to DNA-damaging agents such as cisplatinum. Again, HMGA1 knockdown also displayed a similar consequence and further exacerbated Notch1 effects. Therefore, our data support the hypothesis that HMGA1 can contribute to cervical carcinogenesis also through its ability to impair p53 function, in addition to its ability to directly influence gene expression.
An additional aspect addressed in our article deals with the regulation of HPV E6/E7 expression. During the initial phase of the infection by the high-risk HPV strains (such as HPV16 and 18), E6 and E7 expression in cervical cells remains relatively low and reaches higher levels only in differentiated squamous epithelial cells committed to shedding [55]. In contrast, E6/E7 expression reaches very high levels in dysplastic proliferating cells of CIN-3 lesions [56,57], suggesting that some kind of genetic or epigenetic switch occurred. The precise mechanism governing the expression switch is far from being clear. Intriguingly, HMGA1 has been reported to be essential for the formation of an enhanceosome driving the expression of HPV18 E6/E7 transcript [52,53]. In keeping with this, HMGA1 knockdown inhibited HPV18 E6/E7 promoter activity in luciferase reporter assay and reduced E6/E7 expression in HeLa cells. These results point to the existence of a positive feedback loop between E6/E7 and HMGA1 expression. It is worth mentioning that higher HMGA1 levels are detectable in proliferating dysplastic cervical cells of higher-grade (CIN-3) lesions, where the E6/E7 expression switch has occurred, rather than in infected differentiated cells [49]. Whether this loop contributes to the previously mentioned E6/E7 expression switch needs to be specifically addressed.
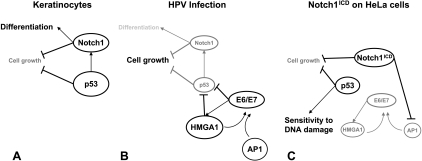
Recent literature has depicted a complex interplay between Notch1 and p53, each of which may apparently work upstream of the other depending on the cell/tissue context [46–48]. On the basis of observation specifically made on keratinocytes, however, we can put forward the following model (Figure 6). p53 directly controls Notch1 expression in both normal and neoplastic keratinocytes, thus contributing to the inhibition of cell growth and induction of cell differentiation [47]. In infected keratinocytes, HPV oncoproteins reduce p53 expression and, by inducing HMGA1, further decrease its activity leading to Notch1 repression, inhibition of cell differentiation, and ablation of growth inhibitory signals, thus providing a strong contribution to carcinogenesis. Overexpression of Notch1 intracellular domain in this context restores the original circuit. It represses E6/E7 expression through AP-1 downmodulation [43]. As a consequence, HMGA1 expression declines, and both conditions favor the accumulation of active p53, which in turn activates a strong oncosuppressive pathway and increases the sensitivity of cervical cancer cells to chemotoxic drugs. HMGA1 decrease further impairs E6/E7 enhanceosome and contributes to the negative control of the expression of the HPV oncogenes.
Figure 6.
Notch1, p53, and HMGA1 interplay in normal and HPV-infected keratinocytes. p53 directly controls Notch1 expression in normal keratinocytes, thus contributing to the inhibition of cell growth and induction of cell differentiation (A). In infected keratinocytes (B), HPV oncoproteins, whose expression is sustained by AP1 and HMGA1, reduce p53 expression, and by inducing HMGA1, further decrease its activity leading to Notch1 repression, inhibition of cell differentiation, and ablation of growth inhibitory signals, thus providing a strong contribution to carcinogenesis. Overexpression of Notch1 intracellular domain in this context (C) impairs AP1-dependent E6/E7 transcription, and as a consequence, HMGA1 levels decline, further reducing E6/E7 enhanceosome activity and HPV oncoproteins' expression. Both conditions favor the accumulation of active p53, the induction of its strong oncosuppressive pathway, and the increase in the sensitivity of cervical cancer cells to chemotoxic drugs.
In conclusion, we showed that the increased HMGA1 expression in cervical carcinoma is directly sustained by HPV infection and is relevant for cervical carcinogenesis.
Acknowledgments
The authors thank A. Gismondi, S. Soddu, G. Canettieri, D. DiMaio, C. Gaetano, and G. Manfioletti for providing materials and for their helpful discussion.
Footnotes
This work was supported by the Associazione Italiana per la Ricerca sul Cancro, Telethon Grant GGP07118, the Ministry of University and Research and National Research Council, the Ministry of Health, the Center of Excellence for Biology and Molecular Medicine, Pasteur Institute, Cenci Bolognetti Foundation, University La Sapienza, Roma, and the Rome Oncogenomic Center.
References
- 1.Reeves R. HMGA proteins: isolation, biochemical modifications, and nucleosome interactions. Methods Enzymol. 2004;375:297–322. doi: 10.1016/s0076-6879(03)75020-4. [DOI] [PubMed] [Google Scholar]
- 2.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 3.Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science. 2001;293:1133–1136. doi: 10.1126/science.293.5532.1133. [DOI] [PubMed] [Google Scholar]
- 4.Munshi N, Merika M, Yie J, Senger K, Chen G, Thanos D. Acetylation of HMG I(Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol Cell. 1998;2:457–467. doi: 10.1016/s1097-2765(00)80145-8. [DOI] [PubMed] [Google Scholar]
- 5.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376:771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 7.Battista S, Fidanza V, Fedele M, Klein-Szanto AJ, Outwater E, Brunner H, Santoro M, Croce CM, Fusco A. The expression of a truncated HMGI-C gene induces gigantism associated with lipomatosis. Cancer Res. 1999;59:4793–4797. [PubMed] [Google Scholar]
- 8.Chiappetta G, Bandiera A, Berlingieri MT, Visconti R, Manfioletti G, Battista S, Martinez-Tello FJ, Santoro M, Giancotti V, Fusco A. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene. 1995;10:1307–1314. [PubMed] [Google Scholar]
- 9.Abe N, Watanabe T, Sugiyama M, Uchimura H, Chiappetta G, Fusco A, Atomi Y. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 1999;59:1169–1174. [PubMed] [Google Scholar]
- 10.Chiappetta G, Manfioletti G, Pentimalli F, Abe N, Di Bonito M, Vento MT, Giuliano A, Fedele M, Viglietto G, Santoro M, et al. High mobility group HMGI(Y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int J Cancer. 2001;91:147–151. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1033>3.3.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Fedele M, Bandiera A, Chiappetta G, Battista S, Viglietto G, Manfioletti G, Casamassimi A, Santoro M, Giancotti V, Fusco A. Human colorectal carcinomas express high levels of high mobility group HMGI(Y) proteins. Cancer Res. 1996;56:1896–1901. [PubMed] [Google Scholar]
- 12.Bussemakers MJ, van de Ven WJ, Debruyne FM, Schalken JA. Identification of high mobility group protein I(Y) as potential progression marker for prostate cancer by differential hybridization analysis. Cancer Res. 1991;51:606–611. [PubMed] [Google Scholar]
- 13.Tamimi Y, van der Poel HG, Denyn MM, Umbas R, Karthaus HF, Debruyne FM, Schalken JA. Increased expression of high mobility group protein I(Y) in high grade prostatic cancer determined by in situ hybridization. Cancer Res. 1993;53:5512–5516. [PubMed] [Google Scholar]
- 14.Abe N, Watanabe T, Izumisato Y, Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A, Fujioka Y, Atomi Y. Diagnostic significance of high mobility group I(Y) protein expression in intraductal papillary mucinous tumors of the pancreas. Pancreas. 2002;25:198–204. doi: 10.1097/00006676-200208000-00015. [DOI] [PubMed] [Google Scholar]
- 15.Masciullo V, Baldassarre G, Pentimalli F, Berlingieri MT, Boccia A, Chiappetta G, Palazzo J, Manfioletti G, Giancotti V, Viglietto G, et al. HMGA1 protein over-expression is a frequent feature of epithelial ovarian carcinomas. Carcinogenesis. 2003;24:1191–1198. doi: 10.1093/carcin/bgg075. [DOI] [PubMed] [Google Scholar]
- 16.Liu WM, Guerra-Vladusic FK, Kurakata S, Lupu R, Kohwi-Shigematsu T. HMG-I(Y) recognizes base-unpairing regions of matrix attachment sequences and its increased expression is directly linked to metastatic breast cancer phenotype. Cancer Res. 1999;59:5695–5703. [PubMed] [Google Scholar]
- 17.Giannini G, Di Marcotullio L, Ristori E, Zani M, Crescenzi M, Scarpa S, Piaggio G, Vacca A, Peverali FA, Diana F, et al. HMGI(Y) and HMGI-C genes are expressed in neuroblastoma cell lines and tumors and affect retinoic acid responsiveness. Cancer Res. 1999;59:2484–2492. [PubMed] [Google Scholar]
- 18.Chiappetta G, Tallini G, De Biasio MC, Manfioletti G, Martinez-Tello FJ, Pentimalli F, de Nigris F, Mastro A, Botti G, Fedele M, et al. Detection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinoma. Cancer Res. 1998;58:4193–4198. [PubMed] [Google Scholar]
- 19.Giannini G, Kim CJ, Marcotullio LD, Manfioletti G, Cardinali B, Cerignoli F, Ristori E, Zani M, Frati L, Screpanti I, et al. Expression of the HMGI(Y) gene products in human neuroblastic tumours correlates with differentiation status. Br J Cancer. 2000;83:1503–1509. doi: 10.1054/bjoc.2000.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21:575–594. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood LJ, Maher JF, Bunton TE, Resar LM. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000;60:4256–4261. [PubMed] [Google Scholar]
- 22.Wood LJ, Mukherjee M, Dolde CE, Xu Y, Maher JF, Bunton TE, Williams JB, Resar LM. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol. 2000;20:5490–5502. doi: 10.1128/mcb.20.15.5490-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, Kenyon L, Visone R, De Martino I, Ciarmiello A, Arra C, et al. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene. 2005;24:3427–3435. doi: 10.1038/sj.onc.1208501. [DOI] [PubMed] [Google Scholar]
- 24.Xu Y, Sumter TF, Bhattacharya R, Tesfaye A, Fuchs EJ, Wood LJ, Huso DL, Resar LM. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004;64:3371–3375. doi: 10.1158/0008-5472.CAN-04-0044. [DOI] [PubMed] [Google Scholar]
- 25.Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, et al. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell. 2006;9:459–471. doi: 10.1016/j.ccr.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Baldassarre G, Fedele M, Battista S, Vecchione A, Klein-Szanto AJ, Santoro M, Waldmann TA, Azimi N, Croce CM, Fusco A. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc Natl Acad Sci USA. 2001;98:7970–7975. doi: 10.1073/pnas.141224998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frasca F, Rustighi A, Malaguarnera R, Altamura S, Vigneri P, Del Sal G, Giancotti V, Pezzino V, Vigneri R, Manfioletti G. HMGA1 inhibits the function of p53 family members in thyroid cancer cells. Cancer Res. 2006;66:2980–2989. doi: 10.1158/0008-5472.CAN-05-2637. [DOI] [PubMed] [Google Scholar]
- 28.Pierantoni GM, Rinaldo C, Esposito F, Mottolese M, Soddu S, Fusco A. High mobility group A1 (HMGA1) proteins interact with p53 and inhibit its apoptotic activity. Cell Death Differ. 2006;13:1554–1563. doi: 10.1038/sj.cdd.4401839. [DOI] [PubMed] [Google Scholar]
- 29.Pierantoni GM, Rinaldo C, Mottolese M, Di Benedetto A, Esposito F, Soddu S, Fusco A. High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J Clin Invest. 2007;117:693–702. doi: 10.1172/JCI29852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat Cell Biol. 2002;4:11–19. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 31.Cleynen I, Huysmans C, Sasazuki T, Shirasawa S, Van de Ven W, Peeters K. Transcriptional control of the human high mobility group A1 gene: basal and oncogenic Ras-regulated expression. Cancer Res. 2007;67:4620–4629. doi: 10.1158/0008-5472.CAN-06-4325. [DOI] [PubMed] [Google Scholar]
- 32.Giannini G, Cerignoli F, Mellone M, assimi I, Ambrosi C, Rinaldi C, Dominici C, Frati L, Screpanti I, Gulino A. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res. 2005;65:8308–8316. doi: 10.1158/0008-5472.CAN-05-0607. [DOI] [PubMed] [Google Scholar]
- 33.Munger K, Baldwin A, Edwards KM, Hayakawa H, Nguyen CL, Owens M, Grace M, Huh K. Mechanisms of human papillomavirus-induced oncogenesis. J Virol. 2004;78:11451–11460. doi: 10.1128/JVI.78.21.11451-11460.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 36.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 37.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 38.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair P, Somasundaram K, Krishna S. Activated Notch1 inhibits p53-induced apoptosis and sustains transformation by human papillomavirus type 16 E6 and E7 oncogenes through a PI3K-PKB/Akt-dependent pathway. J Virol. 2003;77:7106–7112. doi: 10.1128/JVI.77.12.7106-7112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rangarajan A, Syal R, Selvarajah S, Chakrabarti O, Sarin A, Krishna S. Activated Notch1 signaling cooperates with papillomavirus oncogenes in transformation and generates resistance to apoptosis on matrix withdrawal through PKB/Akt. Virology. 2001;286:23–30. doi: 10.1006/viro.2001.0867. [DOI] [PubMed] [Google Scholar]
- 41.Veeraraghavalu K, Pett M, Kumar RV, Nair P, Rangarajan A, Stanley MA, Krishna S. Papillomavirus-mediated neoplastic progression is associated with reciprocal changes in JAGGED1 and manic fringe expression linked to notch activation. J Virol. 2004;78:8687–8700. doi: 10.1128/JVI.78.16.8687-8700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Talora C, Cialfi S, Segatto O, Morrone S, Kim Choi J, Frati L, Paolo Dotto G, Gulino A, Screpanti I. Constitutively active Notch1 induces growth arrest of HPV-positive cervical cancer cells via separate signaling pathways. Exp Cell Res. 2005;305:343–354. doi: 10.1016/j.yexcr.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 43.Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 2002;16:2252–2263. doi: 10.1101/gad.988902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lathion S, Schaper J, Beard P, Raj K. Notch1 can contribute to viral-induced transformation of primary human keratinocytes. Cancer Res. 2003;63:8687–8694. [PubMed] [Google Scholar]
- 45.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 46.Lefort K, Mandinova A, Ostano P, Kolev V, Calpini V, Kolfschoten I, Devgan V, Lieb J, Raffoul W, Hohl D, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKα kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yugawa T, Handa K, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alimirah F, Panchanathan R, Davis FJ, Chen J, Choubey D. Restoration of p53 expression in human cancer cell lines upregulates the expression of Notch1: implications for cancer cell fate determination after genotoxic stress. Neoplasia. 2007;9:427–434. doi: 10.1593/neo.07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bandiera A, Bonifacio D, Manfioletti G, Mantovani F, Rustighi A, Zanconati F, Fusco A, Di Bonito L, Giancotti V. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 1998;58:426–431. [PubMed] [Google Scholar]
- 50.Goodwin EC, DiMaio D. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc Natl Acad Sci USA. 2000;97:12513–12518. doi: 10.1073/pnas.97.23.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sima N, Wang W, Kong D, Deng D, Xu Q, Zhou J, Xu G, Meng L, Lu Y, Wang S, et al. RNA interference against HPV16 E7 oncogene leads to viral E6 and E7 suppression in cervical cancer cells and apoptosis via upregulation of Rb and p53. Apoptosis. 2008;13:273–281. doi: 10.1007/s10495-007-0163-8. [DOI] [PubMed] [Google Scholar]
- 52.Bouallaga I, Massicard S, Yaniv M, Thierry F. An enhanceosome containing the Jun B/Fra-2 heterodimer and the HMG-I(Y) architectural protein controls HPV 18 transcription. EMBO Rep. 2000;1:422–427. doi: 10.1093/embo-reports/kvd091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouallaga I, Teissier S, Yaniv M, Thierry F. HMG-I(Y) and the CBP/p300 coactivator are essential for human papillomavirus type 18 enhanceosome transcriptional activity. Mol Cell Biol. 2003;23:2329–2340. doi: 10.1128/MCB.23.7.2329-2340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kinoshita T, Shirasawa H, Shino Y, Shimizu K, Moriya H, Simizu B. Human papillomavirus type 16 E6 protein up-regulates the expression of the high mobility group protein HMG-I(Y) gene in mouse 10T1/2 cells. Virus Res. 1996;42:119–125. doi: 10.1016/0168-1702(96)01303-2. [DOI] [PubMed] [Google Scholar]
- 55.Chow LT, Broker TR. Papillomavirus DNA replication. Intervirology. 1994;37:150–158. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 56.Durst M, Glitz D, Schneider A, zur Hausen H. Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology. 1992;189:132–140. doi: 10.1016/0042-6822(92)90688-l. [DOI] [PubMed] [Google Scholar]
- 57.Stoler MH, Rhodes CR, Whitbeck A, Wolinsky SM, Chow LT, Broker TR. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum Pathol. 1992;23:117–128. doi: 10.1016/0046-8177(92)90232-r. [DOI] [PubMed] [Google Scholar]