Abstract
The Arabidopsis bas1-D mutation suppresses the long hypocotyl phenotype caused by mutations in the photoreceptor phytochrome B (phyB). The adult phenotype of bas1-D phyB-4 double mutants mimics that of brassinosteroid biosynthetic and response mutants. bas1-D phyB-4 has reduced levels of brassinosteroids and accumulates 26-hydroxybrassinolide in feeding experiments. The basis for the mutant phenotype is the enhanced expression of a cytochrome P450 (CYP72B1). bas1-D suppresses a phyB-null allele, but not a phyA-null mutation, and partially suppresses a cryptochrome-null mutation. Seedlings with reduced BAS1 expression are hyperresponsive to brassinosteroids in a light-dependent manner and display reduced sensitivity to light under a variety of conditions. Thus, BAS1 represents one of the control points between multiple photoreceptor systems and brassinosteroid signal transduction.
Because they are fixed in space, plants need to be flexible in their response to external stimuli. Light has an important role, being used by plants both for photosynthesis and as a developmental cue. The quality, quantity, direction, and duration of light are assessed by a variety of photoreceptors. In Arabidopsis, there are five red/far-red absorbing phytochromes (phyA–phyE), two blue/UVA absorbing cryptochromes (cry1 and cry2), and the less understood UVB photoreceptors. All affect gross morphological changes in the seedling as it makes the transition from heterotrophic growth in the dark to photoautotrophic growth in the light (1). Genetic analyses demonstrate a complex web of interactions between these photoreceptor signaling pathways (2–5).
Light regulates photomorphogenesis by interactions with endogenous developmental programs involving multiple phytohormones. For example, genetic analysis between gibberellin (GA)-deficient and phytochrome mutants points to interactions between these two signal transduction systems (6), which may be caused, in part, by phytochrome's regulation of GA biosynthesis genes (7). Genetic analysis also implicates auxin in light signal transduction. The shy2 mutation, a suppressor of mutations affecting phyB (8, 9), resides in the auxin-induced gene IAA3 (10). Brassinosteroid (BR) mutants have been identified in genetic screens for plants that develop as light-grown plants in the absence of the light cue (reviewed in ref. 11). As adults, these mutants are essentially the opposite of mutants lacking phyB, being dark green, slow-growing dwarfs with epinastic leaves, short stems and petioles, and delayed senescence (6).
Genetic screens for loss-of-function mutations have identified >40 loci thought to be involved in photomorphogenesis (1). Despite this large number, it is likely that these screens have missed important light signaling components that are either redundant members of a gene family or are essential for survival. The role of such genes might be uncovered in screens for gain-of-function mutations. One way to target gain-of-function mutations is through extragenic suppressor analysis (12), an approach used successfully in Arabidopsis to identify mutations involved in light signal transduction (9, 13, 14). To facilitate the cloning of dominant suppressor mutations, we have utilized the technique of activation tagging in mutant backgrounds. Activation tagging is a modification of T-DNA tagging that specifically targets gain-of-function mutations. Multimerized copies of enhancer elements from the cauliflower mosaic virus (CaMV) 35S promoter are incorporated near the right border of a T-DNA, which when inserted near a gene may cause enhanced transcription, resulting in a dominant, tagged mutation (15).
In this paper, we report the isolation of activation-tagged suppressors of the missense mutation phyB-4. We describe the identification of bas1-D (phyB activation-tagged suppressor1-dominant) caused by the amplified expression of the cytochrome P450: CYP72B1. We show that this mutant has no detectable brassinolide (BL), the most active BR, and accumulates 26-hydroxybrassinolide (26-OHBL) in feeding experiments. Transgenic lines with reduced expression of this gene have hypocotyls with enhanced responses to BL and reduced responses to light. Crosses with photoreceptor-null mutations place bas1-D downstream of phyA and cry1, making this a bypass suppressor of phyB alleles. We propose that this gene is a control point between multiple photoreceptor signal transduction pathways and BR signaling.
Materials and Methods
Mutant Screen and Genetic Analysis.
The bas1-D phyB-4 mutant was identified as having a shorter hypocotyl than phyB-4 in the following screen. The phyB-4 mutation was originally isolated in the La-er genetic background (16, 17). To improve transformation efficiency, this mutation was introgressed into the Col-0 genetic background six times. Polymorphic markers between La-er and Col-0 were used to confirm introgression (18). phyB-4 mutants were transformed with the activation-tagging construct pSKI074 containing four copies of enhancer elements from the CaMV 35S promoter and conferring resistance to the antibiotic kanamycin in plants (Igor Kardailsky and Detlef Weigel, personal communication). Plants were transformed with the floral dip technique using the Agrobacterium strain GV3101 (19).
Seeds were sterilized and plated on standard growth medium (3, 20). In all experiments, plates with kanamycin (30 μg/L) or gentamicin (60 μg/L) had 0.8% phytagar (Life Technologies, Grand Island, NY). Plates without antibiotics had 1.0% phytagel (Sigma). A 4-day dark treatment at 4°C synchronized germination. Seedlings were grown for 6 days at 20°C in 150 μEm−2s−1 of continuous white light (3). Candidate suppressors having shorter hypocotyls than phyB-4 were analyzed for the phyB-4 mutation by PCR amplification with gene-specific primers (5′-CTGTCGTGGAAAGTGTGAGG-3′ and 5′-GAACCTTGACGCTTGAGG-3′) and digestion with the restriction endonuclease NlaIII.
bas1-D phyB-4 plants were crossed with the null photoreceptor mutants phyB-5, phyA-201, and hy4–2.23N(cry1). For the bas1-D phyB-4 phyA-201 mutant, kanamycin-resistant F2 plants having long hypocotyls after 6 days in far-red light were genotyped for the phyB-4 and phyA-201 mutations (21). For the bas1-D phyB-4 cry1 mutant, kanamycin-resistant F2 plants having long hypocotyls after 6 days in blue light were genotyped for the phyB-4 and cry1 mutations (3). bas1-D phyB-4 was isolated in a Col-0 ecotype background, the photoreceptor-null mutants in the La-er ecotype. We examined F3 populations homozygous for phyB-4 and phyA or cry1 yet segregating the bas1-D mutation, allowing us to test the effect of these photoreceptors in the presence or absence of the bas1-D mutation while controlling for variations caused by the different ecotypes. To test the effect of the bas1-D mutation in a phyB-null mutant background, F3 seedlings heterozygous for bas1-D and segregating phyB-5/phyB-4 were grown. From >200 F3 seedlings, no kanamycin-resistant plants with long hypocotyls in white light were found, showing that bas1-D suppressed a phyB-null mutation.
Cloning of BAS1.
For Southern blot analysis and plasmid rescue, plant DNA was prepared from 1–2 g (fresh weight) of tissue with the PhytoPure plant DNA extraction kit (Nucleon Biosciences, Coatbridge, U.K.). The 7.3-kb HindIII rescued plasmid (pBAS1H) was sequenced with a primer 3′ of the HindIII site in the T-DNA (5′-GCTCTCTCGAGGTCGACGG-3′). A blast search (22) identified genomic sequence encoding CYP72B1, the expressed sequence tag (EST) T04442 (GenBank accession no. T04442) and the bacterial artificial chromosome (BAC) F18A8 (GenBank accession no. AC003105). The PCR primer (5′-GCTTGCTGGACTATTTGAGC-3′) and T7 primer sequence were used to amplify the junction of insertion in the bas1-D phyB-4 mutant and the two rescued plasmids, showing that all three shared the same architecture of insertion and that all four copies of the enhancer elements were intact. The KpnI rescued plasmid (pBAS1K) was 13.67 kb and contained the entire gene encoding CYP72B1 plus 6.33 kb of 3′ genomic sequence.
The EST T04442 is a complete cDNA for BAS1 encoding the CYP72B1 protein. Sequencing uncovered an error in the database entry for BAC F18A8 where nucleotide 784 of this cDNA sequence was a C instead of the reported T, causing amino acid 262 to be annotated as a tryptophan instead of the correct arginine. Our corrected DNA sequence encoded a recognition site for the restriction endonuclease BsmAI. We confirmed the presence of this restriction site by PCR amplification, digestion, and resolution with the cDNA, the BAC F18A8, or genomic DNA from Col-0, phyB-4, and bas1-D phyB-4 as templates (data not shown).
Northern Blot and Reverse Transcription (RT)–PCR Analyses.
For Northern blot analysis, 8-day-old, light-grown (150 μEm−2s−1 of continuous white light) seedlings were used. The protocol was described in ref. 14 with a PCR product of the CYP72B1 cDNA used as a probe. RT-PCR analysis was performed as follows. Wild-type and antisense seedlings were grown in the light for 9 days. For the hypocotyl vs. rosette RT-PCR analysis, seedlings were grown for 5 days in the dark (to induce hypocotyl growth), then for 9 more days in 150 μEm−2s−1 of continuous white light. Total RNA was isolated from frozen tissue (−80°C) with TRIzol Reagent as recommended (Life Technologies). cDNAs from 1 μg of total RNA were synthesized with 500 ng of a 27-mer oligo(dT) and the reverse transcriptase SuperScript (Life Technologies). One-tenth of the cDNA reaction was used for each PCR (see ref. 21 for conditions). Primers spanning the third intron of BAS1 were used (5′-GGTTCAGGACATTGTGGAGG-3′ and 5′-GGATACAACCTTAAAGACTCG-3′). The UBQ10 gene was used as a template control (23). Southern blot analysis of products from PCR runs with varying numbers of cycles was quantified with a PhosphorImager (Molecular Dynamics). The RT-PCR results presented were within the linear range of accuracy.
Recapitulation and Antisense Constructs.
Two constructs were used for recapitulation of the bas1-D mutant phenotype. After restriction of pBAS1K with the endonucleases BamHI and SacI, a 5.7-kb fragment containing the entire CYP72B1 ORF in the context of the four enhancer elements was cloned into the binary vector pPZP212 (24). The second construct was made by cloning a BamHI/KpnI fragment from the T04442 cDNA clone into the binary vector pCHF3, which contains the CaMV 35S full promoter, the RBCS terminator from pea, and confers kanamycin resistance for selection in plants. A construct constitutively expressing BAS1 antisense RNA was made by cloning a BamHI/SacI fragment into the binary vector pCHF1, which contains the CaMV 35S full promoter, the RBCS terminator from pea, and confers gentamicin resistance for selection in plants. In all experiments, seedlings were measured digitally (3). Error bars represent 1 SE. Invisible error bars are smaller than the symbol. To examine the dark-grown phenotype of bas1-D recapitulation lines, T2 seedlings were grown for 6 days in the dark, transferred to growth medium with kanamycin, and imaged with a flat-bed scanner (3). After growth in the light, the hypocotyl lengths of dark-grown, kanamycin-resistant seedlings were determined from the digital image. Tobacco seedlings were treated similarly.
BL Dose Response and Light Conditions.
BL was from CIDtech Research (Mississauga, Ontario, Canada). Light conditions for fluence responses in red and blue light and seedling measurements are described in ref. 3. An E30LED growth chamber (Percival Scientific, Boone, IA) supplied far-red light. Far-red fluences were measured with a portable spectroradiometer (model LI-1800; Li-Cor, Lincoln, NE).
BR Measurements.
Plants were grown on soil (3) in short-day conditions (8 hr of light, 16 hr of dark) for 5 wk before rosettes were harvested in liquid nitrogen. No inflorescence stems were seen at this time. Tissues (100–200 g fresh weight) of phyB-4 and bas1-D phyB-4 were collected. BRs were analyzed according to Fujioka et al. (25). Before feeding experiments, 7-day-old seedlings were transferred to a 200-ml flask containing 30 ml of liquid growth medium and supplemented with 1% sucrose (phyB-4, 50 seedlings; bas1-D phyB-4, 100 seedlings). Five days after transfer, an EtOH solution (50 μl) of 2H6-labeled BL (50 μg) or nonlabeled BL (50 μg) was added. The seedlings were incubated for 1 day at 22°C in the light on a shaker (125 rpm) then extracted with MeOH. The MeOH extract was purified with a cartridge of silica gel (Sep-Pak Vac, 2 g; Waters), which was eluted with 30 ml of chloroform, 3% MeOH in chloroform, and 20% MeOH in chloroform. The last fraction was purified with HPLC on a 150 × 4.6-mm Senshu Pak ODS-1151-D column (Senshu Scientific, Tokyo) with 45% acetonitrile at a flow rate of 1.0 ml/min. The fractions were collected at 1-min intervals (retention time of 1–10 min).
Each fraction was subjected to gas chromatography–mass spectrometry (GC-MS) analysis after derivatization. 26-OHBL was detected at 2–3 min. Authentic OHBL analogs used in this study were chemically synthesized (26). GC-MS analysis was performed on a JEOL Automass JMS-AM 150 mass spectrometer connected to a Hewlett–Packard 5890A-II gas chromatograph. Analysis was conducted under the following conditions: GC column, DB-5 (0.25 mm × 15 m, 0.25-μm film thickness; J & W Scientific, Folsom, CA); injection temperature, 280°C; carrier gas, helium at a flow rate of 1 ml/min; ionization, EI (70 eV); column temperature, 80°C for 1 min, elevated to 320°C at 30°C/min, then maintained at 320°C. The OHBL fraction was treated with pyridine containing methaneboronic acid (20 μg per 10 μl) at 80°C for 30 min and then with 10 μl of N-methyl-N-trimethylsilyltrifluoroacetamide (MSTFA) at 80°C for 30 min.
Tobacco Transformation.
Nicotiana tabacum cv. Xanthi was transformed (27) with the same Agrobacterium strains used for Arabidopsis. T2 seeds were sterilized for 30 min in 10% (vol/vol) bleach with 0.05% Triton X-100, then washed three times with sterile water and plated on 2× growth medium, 0.8% bactoagar, with or without 200 mg/l kanamycin.
Results
bas1-D Is a Suppressor of the phyB-4 Mutation.
The phyB-4 missense allele encodes a pigment capable of nearly normal phototransformation and confers a hypocotyl phenotype intermediate between the wild-type and null phyB alleles (28). The resulting weak signal transduction current makes phyB-4 an ideal target for suppressor analysis, casting a broad net in search of genes involved in light signaling. In a screen of 3000 phyB-4 T1 (primary transformant) seedlings, we identified three dominant mutants whose phenotypes were caused by the amplification or ectopic expression of endogenous genes due to the proximal insertion of the transgene. The bas1-D phyB-4 double mutant had a significantly shorter hypocotyl than phyB-4 (Fig. 1 A–C). T3 seeds from heterozygotes in the T2 generation segregated 470 suppressed kanamycin-resistant and 147 nonsuppressed sensitive plants, indicating that this transgene was located at a single locus. Southern blot analysis confirmed this conclusion (data not shown). All of the kanamycin-resistant plants had the bas1-D phyB-4 phenotype. All kanamycin-sensitive segregants did not, indicating linkage to the transgene.
Figure 1.
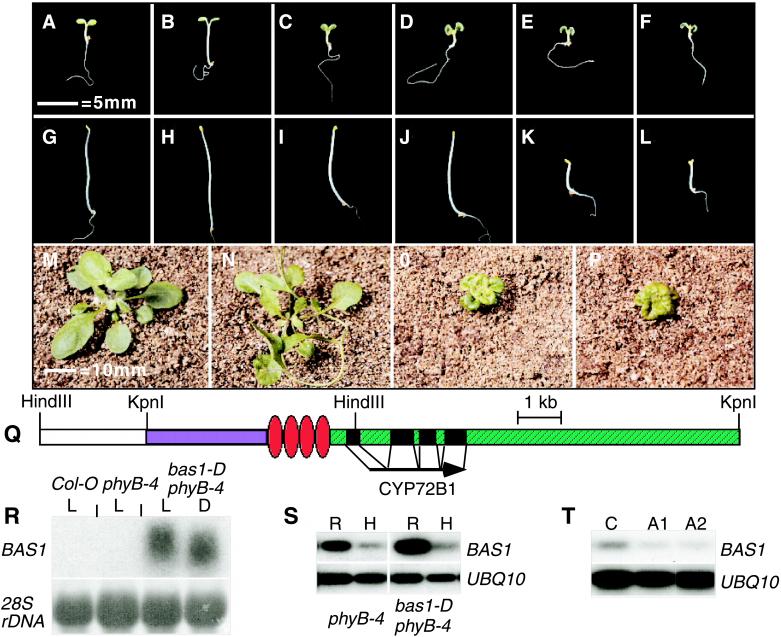
bas1-D suppresses phyB-4 by the enhanced expression of CYP72B1. (A–F) Six-day-old, light-grown seedlings. (G–L) Dark-grown seedlings: Col-0 (A and G), phyB-4 (B and H), bas1-D phyB-4 (C and I), phyB-4 transformed with the bas1-D gene (D and J), phyB-4 transformed with the CaMV35S∷BAS1cDNA (E and K), and det2–1 (F and L). (M–P) Four-week-old plants: Col-0 (M), phyB-4 (N), bas1-D phyB-4 (O), and det2–1 (P). (Q) A diagram of the insertion, the cDNA, and restriction endonuclease sites used for plasmid rescue. White, part of the antibiotic resistance gene. Blue, pBlueScript sequence. Red, the enhancers. Green, genomic DNA. (R) Northern blot analysis of total RNA from light-grown (L) or dark-grown (D) seedlings. (S) RT-PCR analysis of rosette (R) and hypocotyl (H) tissue. (T) RT-PCR analysis of Col-0 (C) and two bas1 antisense lines (A1 and A2).
We cloned genomic DNA adjacent to the right border of the T-DNA by plasmid rescue. blast searches (22) of flanking genomic DNA showed that the site of T-DNA insertion was on chromosome II near the erecta locus. The insertion of the four enhancer elements was 381 nucleotides 5′ to the transcriptional start of the BAS1 gene. Northern blot analysis of total RNA showed that this gene was overexpressed in bas1-D phyB-4 (Fig. 1R). Two other predicted transcripts near the site of T-DNA insertion showed no altered accumulation in the bas1-D phyB-4 mutant compared with either the wild type or phyB-4 (data not shown). The overexpressed transcript encodes a putative cytochrome P450, CYP72B1, and is represented in the Arabidopsis EST database by clone T04442 (29). Sequencing the T04442 clone verified that it encoded a complete cDNA for CYP72B1. phyB-4 mutant seedlings transformed with the mutant gene in context with the enhancer elements or with the cDNA under the control of the CaMV 35S promoter recapitulated the original bas1-D phyB-4 phenotype as light-grown seedlings and adults (Fig. 1 D and E), demonstrating that this gene is responsible for the bas1-D phyB-4 phenotype.
The bas1-D phyB-4 double mutant resembled BR mutants as light-grown seedlings and adults (Fig. 1 C–F, O, and P), suggesting that the suppression of the phyB-4 phenotype is caused by the alteration of BR synthesis or signaling. Unlike plants that lack or are insensitive to BRs, bas1-D phyB-4 seedlings and lines recapitulated with the mutant clone (data not shown) did not have short hypocotyls in the dark (Fig. 1 G–L). In contrast, dark-grown seedlings from four of five independent transformants with the cDNA under the control of the CaMV 35S promoter had short hypocotyls, indicating transcriptional regulation of this gene. Northern blot analysis of the bas1-D transcript in the bas1-D phyB-4 mutant showed no difference between light- and dark-grown seedlings, suggesting that light does not regulate the overall accumulation of bas1-D mRNA (Fig. 1R).
RT-PCR analysis demonstrated differential transcript accumulation between rosettes and hypocotyls of phyB-4 (22 PCR cycles) and the bas1-D phyB-4 (16 PCR cycles) mutants, showing tissue-specific transcriptional regulation for both the wild-type and mutant gene (Fig. 1S). Quantification of RT-PCR products showed a 3-fold higher transcript accumulation in the rosette compared with the hypocotyl in both phyB-4 and bas1-D phyB-4. The expression levels and patterns in the wild type were nearly identical to those in phyB-4 (data not shown), demonstrating that bas1-D suppresses phyB-4 through the amplification of the endogenous BAS1 expression pattern and not by the ectopic expression of this gene. Quantification showed a 50-fold accumulation of BAS1 transcript in bas1-D phyB-4 compared with phyB-4. bas1-D phyB-4 mutants have nearly normal hypocotyls in the dark, arguing for either light regulation of transcript accumulation specifically in the hypocotyl or posttranslational modification of the gene product.
Altered Expression of CYP72B1 Modifies Responses to BL.
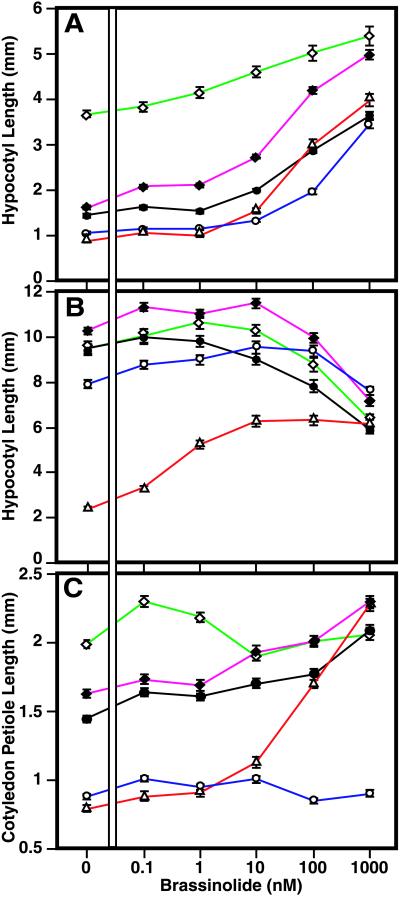
To test the role of the BAS1 gene in wild-type plants, we generated partial loss of function transgenic lines by means of antisense (30). RT-PCR analysis showed that two of these lines had approximately 50% of the wild-type BAS1 transcript accumulation (Fig. 1T). These lines were epistatic to bas1-D phyB-4, further demonstrating that these are bona fide antisense mutants (data not shown). Dose-response experiments showed a hyperresponsivity to BL in the hypocotyls of antisense lines when grown in the light (Fig. 2A), though not when grown in the dark (Fig. 2B). The BR-biosynthesis mutant det2–1 had petioles that are shorter than the wild type in the light, while phyB-4 petioles were longer than the wild type in the light. These phenotypes were rescued by increasing amounts of BL. The bas1-D phyB-4 mutant petioles were always shorter than the wild type for all BL levels tested, indicating that the rosette phenotype of bas1-D phyB-4 is insensitive to BL (Fig. 2C).
Figure 2.
CYP72B1 expression controls BR responses. Hypocotyls of light-grown (A) or dark-grown (B) seedlings were measured after 6 days on varying levels of BL. Cotyledon petioles were measured after 12 days of growth in white light (C) for Col-0 (●, black), phyB-4 (⋄, green), bas1-D phyB-4 (○, blue), BAS1 antisense line A2 from Fig. 1T (♦, purple), and det2–1 (▵, red). Both antisense lines had similar phenotypes.
Altered Accumulation of BRs in bas1-D phyB-4.
To test whether the bas1-D gene product inactivates or degrades BRs, levels of BRs in bas1-D phyB-4 and phyB-4 plants were determined. Studies in Arabidopsis have confirmed the following biosynthetic sequence for both the early and late C6-oxidation pathways for brassinolide: (6-deoxo)teasterone→→(6-deoxo) castasterone→castasterone→brassinolide [Fujioka et al. (25, 31), and unpublished data]. By using gas chromatography-selected ion-monitoring analysis, castasterone and 6-deoxocastasterone were detected in bas1-D phyB-4, but their levels were greatly reduced compared with those in phyB-4. Moreover, BL was not detected in bas1-D phyB-4 (Table 1). Thus, endogenous levels of BRs in bas1-D phyB-4 were greatly diminished, suggesting that this mutation affects BR levels, which may be related to hydroxylation of BR biosynthetic intermediates. There was a concomitant increased accumulation of 6-deoxoteasterone in the bas1-D phyB-4 mutant. This could be caused by up-regulation of biosynthetic enzymes that are feedback-inhibited by the end-product BL, which is not detectable in the bas1-D phyB-4 mutant.
Table 1.
Brassinosteroid (BR) levels (ng/g fresh weight)
| BR | phyB-4 | bas1-D phyB-4 |
|---|---|---|
| 6-deoxoTE | 0.19 | 0.26 |
| 6-deoxoCS | 0.79 | 0.04 |
| CS | 0.13 | 0.02 |
| BL | 0.32 | ND |
6-deoxoTE, 6-deoxoteasterone; 6-deoxoCS, 6-deoxocastasterone; CS, castasterone; BL, brassinolide; ND, not detected.
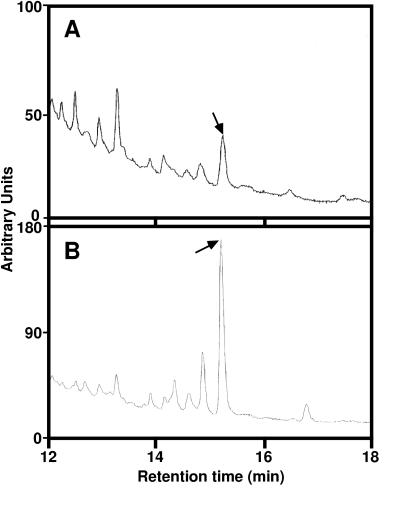
Metabolism of deuterium-labeled and nonlabeled BL was examined by using aseptically grown seedlings. bas1-D phyB-4 and phyB-4 seedlings were fed nonlabeled BL and incubated for 1 day. As possible hydroxylated metabolites of BL, we chemically synthesized 14-OHBL, 20-OHBL, 25-OHBL, 26-OHBL, and 28-OHBL. Relevant fractions from the feeding experiments were analyzed by GC-MS after conversion to methaneboronate-trimethylsilyl derivatives and compared with chemically synthesized OHBL. A prominent peak, comigrating with authentic 26-OHBL, was found in bas1-D phyB-4 (the level was five times higher than in phyB-4 alone). To confirm the identification as 26-OHBL, we performed feeding experiments with 2H6-BL. When 2H6-BL was fed to the seedlings, fragment ions such as m/z 625, 583, and 570 corresponding to m/z 619, 577, and 564 of nonlabeled 26-OHBL methaneboronate-trimethylsilyl derivative were detected. Thus, 2H6-26-OHBL was confirmed to be a metabolite of 2H6-BL. This metabolite was six times more abundant in bas1-D phyB-4 than in phyB-4 (Fig. 3), further demonstrating that BL is converted to 26-OHBL in Arabidopsis seedlings and that the conversion is greater in the bas1-D phyB-4 mutant.
Figure 3.
C-26 hydroxylation is enhanced in bas1-D phyB-4. Before GC-MS analysis of extracted brassinosteroids, 2H6-labeled brassinolide was fed to phyB-4 (A) or bas1-D phyB-4 (B) and compared with authentic 26-hydroxybrassinolide (not shown). Arrows indicate the 2H6-26-hydroxybrassinolide peak.
Fluence Response Analysis.
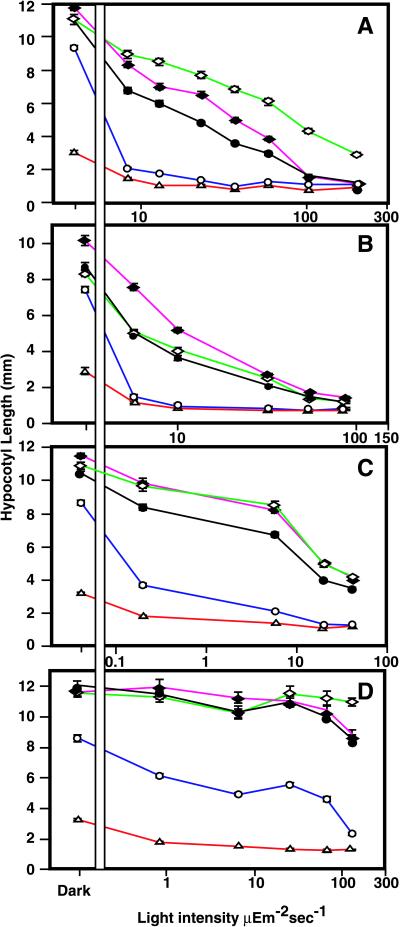
To test the possible role of BAS1 in light signal transduction, we analyzed the response of both overexpressers and underexpressers to varying qualities and quantities of light (Fig. 4). Though the bas1-D phyB-4 mutant was slightly shorter than the wild type in the dark, it was hyperresponsive to continuous white, red, far-red, and blue light. The BAS1 antisense lines had hypocotyls that were slightly longer than the wild type in the dark, showed a reduced responsiveness to white, far-red, and blue light (Fig. 4 A–C), and had a wild-type response to red light (Fig. 4D). As expected, phyB-4 mutants had a reduced response to white and red light when compared with the wild type. It is likely that there is minimal BAS1 activity downstream of red light since the bas1-D phyB-4 mutant was less responsive to red light than to the other light conditions and given that the BAS1 antisense lines responded normally to red light.
Figure 4.
Altered expression of CYP72B1 affects hypocotyl responses to light. Hypocotyls of 6-day-old seedlings were measured after growth in the dark or varying intensities of white light (A), far-red light (B), blue light (C), or red light (D). The lines and symbols are Col-0 (●, black), phyB-4 (⋄, green), bas1-D phyB-4 (○, blue), BAS1 antisense line A2 from Fig. 1T (♦, purple), and det2–1 (▵, red). Both antisense lines had similar phenotypes.
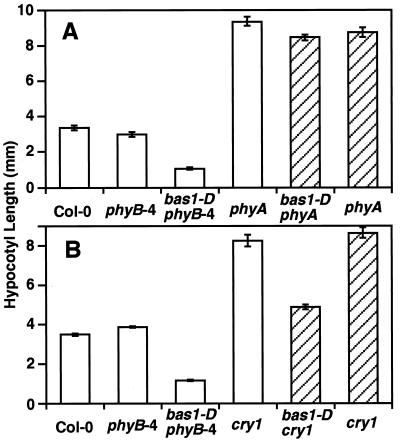
To test which photoreceptors control the activity of BAS1, we made double mutants of bas1-D with null alleles of phyA, phyB, and cry1 (Fig. 5). In continuous far-red light, bas1-D did not suppress a phyA-null mutation (Fig. 5A). In continuous blue light, bas1-D partially suppressed a cry1-null mutation (Fig. 5B). In contrast, bas1-D fully suppressed a phyB-null mutation (data not shown). Taken together with the fluence response analysis (Fig. 4), the bas1-D mutation appears to suppress phyB alleles through the activity of at least phyA and cry1 and can be formally placed as a bypass suppressor of phyB.
Figure 5.
BAS1 acts genetically downstream of phyA and cry1. (A) phyB-4phyA-201 mutants segregating the bas1-D mutation (hatched bars) were compared with the control lines (open bars) in 15 μEm−2s−1 of continuous far-red light for 6 days. (B) phyB-4cry1 mutants segregating the bas1-D mutation (hatched bars) were compared with control lines (open bars) in 20 μEm−2s−1 of continuous blue light for 6 days.
Heterologous Expression.
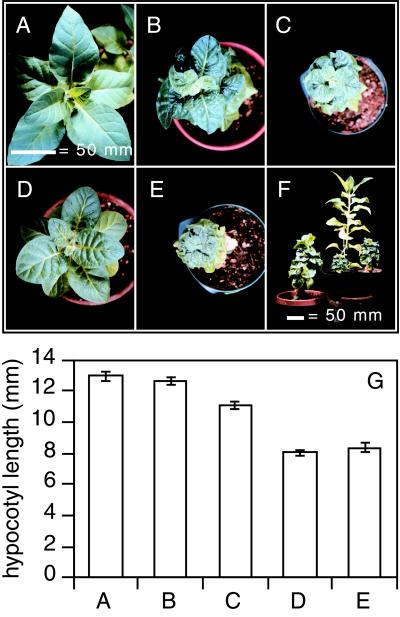
To test BAS1 activity in a heterologous system, we transformed tobacco plants with both recapitulation constructs, which resulted in dwarf plants reminiscent of the original bas1-D phyB-4 mutant in Arabidopsis (Fig. 6). Tobacco plants expressing the bas1-D mutant gene had dark-green, epinastic leaves with short stems and petioles (Fig. 6 B, C, and F) when compared with the wild type (Fig. 6 A and F). Dark-grown seedlings from these plants had hypocotyls similar to the wild type (Fig. 6G), with the more severe of the two (Fig. 6C) having slightly shorter hypocotyls reminiscent of dark-grown bas1-D phyB-4. Tobacco plants expressing the BAS1 cDNA under control of the CaMV 35S promoter were also light-grown dwarfs (Fig. 6 D–F). In the dark, these seedlings had significantly shorter hypocotyls than the wild type (Fig. 6G), with the weaker of the two cDNA expressors (Fig. 6D) having dramatically shorter hypocotyls than the strongest of the two lines expressing the bas1-D mutant gene (Fig. 6C). This shows that BRs can be inactivated by CYP72B1 in tobacco and that there is a similar transcriptional control in this heterologous plant system.
Figure 6.
bas1-D is effective in tobacco. (A–F) Five-month-old, primary tobacco transformants. Seeds from the wild type (A), two typical recapitulation lines using the bas1-D genomic clone (B and C), or two typical recapitulation lines using the CaMV35S∷BAS1 cDNA (D and E) were grown for 9 days in the dark and then measured (G). The letters under the bars in G correspond to Fig. 6 A–E. A side view of these recapitulation lines is shown compared with the wild type (tallest) in F.
Discussion
The identification of bas1-D gives significant insight into two complex signaling processes and how they interact to regulate plant development. The interplay between light and hormone signaling has been studied for years, but mechanisms connecting these pathways are poorly understood. Here, we describe a CYP450 (CYP72B1) that, with enhanced expression, suppresses the long hypocotyl phenotype of the weak photoreceptor mutant phyB-4. It is likely that this enzyme catalyzes inactivation of BRs by means of hydroxylation given that: (i) the bas1-D phyB-4 mutant resembles BR mutants, (ii) there is no detectable BL in the bas1-D phyB-4 mutant, and (iii) the bas1-D phyB-4 mutant converts BL to a C26-hydroxylated form at a greater rate than the wild type. Feeding and dose-response experiments argue that one of the substrates for CYP72B1 is the biosynthetic end product BL. However, measurements of BR biosynthetic precursors show reduced levels of both castasterone and 6-deoxocastasterone in the bas1-D phyB-4 double mutant compared with phyB-4 alone, suggesting that CYP72B1 can also act on BR precursors.
The accumulation of 6-deoxoteasterone in bas1-D phyB-4 is probably caused by increased activity of the steroid hydroxylase, CYP90A1, which catalyzes the C-23 hydroxylation of (6-deoxo)cathasterone to (6-deoxo)teasterone and was originally identified through loss-of-function alleles of cpd, dwf3, and cbb3 (32, 33). The CPD gene is expressed in the cotyledons and young leaves of developing seedlings and is down-regulated by BL (34). Because BL is undetectable in rosettes of bas1-D phyB-4, one would expect higher expression of CPD. Indeed, this is what we observed (data not shown). CYP90B1, a family member with CYP90A1 and originally identified as the dwf4 allele (31), catalyzes the C-22 hydroxylation step prior to CPD activity. This mRNA also has increased accumulation in the bas1-D phyB-4 mutant (data not shown). The accumulation of 6-deoxoteasterone in bas1-D phyB-4 suggests that BAS1 does not efficiently hydroxylate 6-deoxoteasterone, acting downstream of this intermediate in BL biosynthesis and placing BAS1 downstream of both CPD and DWF4.
A putative BR receptor, BRI1, has been identified and shown to have homology with leucine-rich repeat receptor kinases (35). Unlike BR biosynthesis mutants, loss-of-function bri1 alleles are insensitive to BL. It is perplexing that the BRI1 gene seems to be ubiquitously and constitutively expressed throughout Arabidopsis growth and is not regulated by light, a pattern that is similar to expression of the BR biosynthetic gene DET2 (ref. 35, and D. Friedrichsen and J.C., unpublished data). Thus, how do BRs act as hormones if they are synthesized and perceived in the same cell? One way of regulating tissue-specific responses to BL may be by inactivation of the steroid through BAS1-mediated hydroxylation. That BAS1 has tissue-specific transcriptional regulation supports this model. Both gain-of-function and partial loss-of-function bas1 mutations confer altered BR responses in light-grown hypocotyls, suggesting that, in this tissue, the activity of CYP72B1 determines the degree of response to BRs. Though dark-grown bas1-D phyB-4 seedlings are etiolated, they have hypocotyls that are slightly shorter than the wild type. In addition, dark-grown bas1 antisense lines have slightly longer hypocotyls than the wild type, again arguing that it is the activity of CYP72B1 that ultimately controls the hypocotyl response to BRs.
Steroid hormone inactivation by hydroxylation is not an uncommon mechanism. As is the case with BAS1, CYP450-mediated C26-hydroxylation inactivates the insect hormones ecdysteroids, although the precise enzyme has not been identified (36–38). Though ecdysteroids are inactivated in a similar manner to BRs, these insect hormones do not induce BR responses in plants (39). Other insect juvenile hormones can be hydroxylated and presumably catabolized in a similar manner to ecdysone (40). CYP450-dependent C24-hydroxylation of 1,25-dihydroxyvitamin D-3 inactivates this form of vitamin D in both rats (41, 42) and humans (43). In plants, 2β-hydroxylation inactivates gibberellins, targeting them for destruction. There are at least two dioxygenases catalyzing this reaction in pea (44) and at least three in Arabidopsis (45). In both cases, there appears to be some genetic redundancy with overlapping as well as distinct transcriptional patterns in different tissues. A similar genetic redundancy may be present for hydroxylation-mediated inactivation of BRs. Indeed, there is at least one other CYP72 in Arabidopsis (chibi2), which when overexpressed confers a BR-deficient phenotype similar to bas1-D (A. Nagatani, personal communication). C26-hydroxylation is probably not the only way to inactivate BRs. Steroid sulfotransferases have been isolated from both Brassica napus and Arabidopsis and have been shown to inactivate BRs through O-sulfonation (46). Though an in vivo role for these sulfotransferases has yet to be determined, it is clear that there are multiple mechanisms for the control of BRs through their catabolism.
Because the bas1-D mutation is caused by a gain-of-gene-function mutation, we can transfer this genetic information into heterologous plant systems. We have used two constructs to create BR-deficient mutants in tobacco. To date, the only way to study tobacco plants lacking BRs is to grow them on the BR biosynthesis inhibitor brassinazole (47), creating plants that look similar to our weaker transgenic lines. There are multiple advantages to using plants overexpressing BAS1 instead of growth on brassinazole. Depending on the transgene used, we can control the dark-grown phenotype of transgenic lines. The transgenic plants confer a gain-of-function allelic series with some plants being more severe than plants grown on high levels of brassinazole, circumventing the need for brassinazole dose responses in different environmental conditions. Transgenic plants overexpressing CYP72B1 allow us to ask questions that cannot be easily addressed in Arabidopsis. If CYP72B1 can catalyze the C26-hydroxylation of brassinosteroids, can it also catalyze the C26-hydroxylation of ecdysone? This may prevent predators from molting after feeding on these plants. Transgenic bas1-D tobacco plants fed to Manduca sexta may address the involvement of BRs in plant/insect interactions. Because of their larger size, transgenic tobacco lines may also facilitate further biochemical analyses.
Genetic analysis with Arabidopsis photoreceptor-null mutants shows that bas1-D acts downstream of both phyA and cry1. Because the bas1-D mutation suppresses a phyB-null allele, this places it as a bypass suppressor of phyB signaling. It is interesting to note that the long petiole phenotype of light-grown phyB-4 is rescued by increasing levels of BL, suggesting a connection between phyB and BR signaling. Both hypermorphic and hypomorphic mutants of BAS1 expression have altered hypocotyl responses to a variety of light conditions, arguing that the activity of this gene is controlled by multiple light-signaling pathways. Though accumulation of BAS1 mRNA is not regulated by light, there is some control of this activity by photoreceptors. Dark-grown bas1-D phyB-4 seedlings have hypocotyls nearly as long as the wild type, even though there is 50-fold greater accumulation of bas1-D mRNA. Therefore, there must still be some light-dependent mechanism causing hypocotyls of bas1-D phyB-4 mutants to become hyperresponsive to light. This could be caused by light-regulating transcription in the hypocotyl, modifying the gene product, or altering the hypocotyl's sensitivity to BRs. Future genetic studies addressing this light/dark regulation of bas1-D activity in the hypocotyl may elucidate the mechanism of interaction between CYP72B1 and photomorphogenic signal transduction pathways.
Activation-tagging suppressor analysis should be a powerful tool for genetic studies in plants. As with multicopy suppressor studies in yeast (48), it facilitates the genetic identification of genes that are part of a redundant family. Given the large number of redundant genes encoded in the Arabidopsis genome (49), gain-of-function mutations will play an increasingly important role in uncovering the complexities of signal transduction in plants.
Acknowledgments
We thank Dr. Christian Fankhauser for the gift of pCHF1 and pCHF3 and Drs. Detlef Weigel and Igor Kardailsky for the pSKI074 vector. We thank Drs. Karin Schumacher, Christian Fankhauser, and Sioux Christensen for many discussions on activation tagging. We are grateful to Leslie Barden for help with the figures, and Drs. Weigel and Fankhauser for critically reading this manuscript. We thank Bridey Maxwell for naming the bas mutants. This work was supported by National Institutes of Health Grant RO1GM52413 (to J.C.) and a Grant-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Sports and Culture of Japan (10460050) (to S.F.). M.M.N. was supported by National Research Service Award Postdoctoral Fellowship GM17577. J.C. is an Associate Investigator of the Howard Hughes Medical Institute.
Abbreviations
- phy
phytochrome
- cry
cryptochrome
- BR
brassinosteroid
- BL
brassinolide
- BAC
bacterial artificial chromosome
- RT
reverse transcription
- OHBL
hydroxybrassinolide
- GC-MS
gas chromatography–mass spectrometry
- EST
expressed sequence tag
- CaMV
cauliflower mosaic virus
Footnotes
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 27, 1999.
References
- 1.Fankhauser C, Chory J. Annu Rev Cell Dev Biol. 1997;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- 2.Casal J J, Mazzella M A. Plant Physiol. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neff M M, Chory J. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hennig L, Poppe C, Unger S, Schafer E. Planta. 1999;208:257–263. doi: 10.1007/s004250050557. [DOI] [PubMed] [Google Scholar]
- 5.Lasceve G, Leymarie J, Olney M A, Liscum E, Christie J M, Vavasseur A, Briggs W R. Plant Physiol. 1999;120:605–614. doi: 10.1104/pp.120.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chory J. Plant Cell. 1997;9:1225–1234. doi: 10.1105/tpc.9.7.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamiya Y, García-Martínez J L. Curr Opin Plant Biol. 1999;2:398–403. doi: 10.1016/s1369-5266(99)00012-6. [DOI] [PubMed] [Google Scholar]
- 8.Kim B C, Soh M S, Hong S H, Furuya M, Nam H G. Plant J. 1998;15:61–68. doi: 10.1046/j.1365-313x.1998.00179.x. [DOI] [PubMed] [Google Scholar]
- 9.Reed J W, Elumalai R P, Chory J. Genetics. 1998;148:1295–1310. doi: 10.1093/genetics/148.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian Q, Reed J W. Development (Cambridge, UK) 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- 11.Clouse S D, Sasse J M. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 12.Prelich G. Trends Genet. 1999;15:261–266. doi: 10.1016/s0168-9525(99)01749-7. [DOI] [PubMed] [Google Scholar]
- 13.Hoecker U, Tepperman J M, Quail P H. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- 14.Pepper A E, Chory J. Genetics. 1997;145:1125–1137. doi: 10.1093/genetics/145.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walden R, Fritze K, Hayashi H, Miklashevichs E, Harling H, Schell J. Plant Mol Biol. 1994;26:1521–1528. doi: 10.1007/BF00016488. [DOI] [PubMed] [Google Scholar]
- 16.Koornneef M, Rolff E, Spruit C J P. Z Pflanzenphysiol. 1980;100S:147–160. [Google Scholar]
- 17.Reed J W, Nagpal P, Poole D S, Furuya M, Chory J. Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 19.Clough S J, Bent A F. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 20.Valvekens D, van Montagu M, van Lijsebetterns M. Proc Nat Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neff M M, Neff J D, Chory J, Pepper A E. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 23.Callis J, Carpenter T, Sun C W, Vierstra R D. Genetics. 1995;139:921–939. doi: 10.1093/genetics/139.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajdukiewicz P, Svab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 25.Fujioka S, Li J, Choi Y H, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, Sakurai A. Plant Cell. 1997;9:1951–1962. doi: 10.1105/tpc.9.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seto H, Fujioka S, Koshino H, Yoshida S, Tsubuki M, Honda T. Tetrahedron. 1999;55:8341–8352. [Google Scholar]
- 27.Horsch R B, Fry J E, Hoffmann N L, Eichholtz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 28.Elich T D, Chory J. Plant Cell. 1997;9:2271–2280. doi: 10.1105/tpc.9.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baulcombe D C. Plant Mol Biol. 1996;32:79–88. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 31.Choe S, Dilkes B P, Fujioka S, Takatsuto S, Sakurai A, Feldmann K A. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei G P, Nagy F, Schell J, Koncz C. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 33.Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Plant J. 1996;9:701–713. [Google Scholar]
- 34.Mathur J, Molnar G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- 35.Li J, Chory J. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 36.Rees H H. Eur J Entomol. 1995;92:9–39. [Google Scholar]
- 37.Kayser H, Winkler T, Spindler-Barth M. Eur J Biochem. 1997;248:707–716. doi: 10.1111/j.1432-1033.1997.00707.x. [DOI] [PubMed] [Google Scholar]
- 38.Williams D R, Chen J H, Fisher M J, Rees H H. J Biol Chem. 1997;272:8427–8432. doi: 10.1074/jbc.272.13.8427. [DOI] [PubMed] [Google Scholar]
- 39.Clouse S D, Zurek D M, McMorris T C, Baker M E. Plant Physiol. 1992;100:1377–1383. doi: 10.1104/pp.100.3.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sutherland T D, Unnithan G C, Andersen J F, Evans P H, Murataliev M B, Szabo L Z, Mash E A, Bowers W S, Feyereisen R. Proc Natl Acad Sci USA. 1998;95:12884–12889. doi: 10.1073/pnas.95.22.12884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hahn C N, Kerry D M, Omdahl J L, May B K. Nucleic Acids Res. 1994;22:2410–2416. doi: 10.1093/nar/22.12.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar R, Schnoes H K, DeLuca H F. J Biol Chem. 1978;253:3804–3809. [PubMed] [Google Scholar]
- 43.Chen K S, DeLuca H F. Biochim Biophys Acta. 1995;1263:1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- 44.Lester D R, Ross J J, Smith J J, Elliott R C, Reid J B. Plant J. 1999;19:65–73. doi: 10.1046/j.1365-313x.1999.00501.x. [DOI] [PubMed] [Google Scholar]
- 45.Thomas S G, Phillips A L, Hedden P. Proc Natl Acad Sci USA. 1999;96:4698–4703. doi: 10.1073/pnas.96.8.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rouleau M, Marsolais F, Richard M, Nicolle L, Voigt B, Adam G, Varin L. J Biol Chem. 1999;274:20925–20930. doi: 10.1074/jbc.274.30.20925. [DOI] [PubMed] [Google Scholar]
- 47.Asami T, Yoshida S. Trends Plant Sci. 1999;4:348–353. doi: 10.1016/s1360-1385(99)01456-9. [DOI] [PubMed] [Google Scholar]
- 48.Rine J. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- 49.Somerville C, Somerville S. Science. 1999;285:380–383. doi: 10.1126/science.285.5426.380. [DOI] [PubMed] [Google Scholar]