Abstract
On the basis of the sequence of the mitochondrial genome in the flowering plant Arabidopsis thaliana, RNA editing events were systematically investigated in the respective RNA population. A total of 456 C to U, but no U to C, conversions were identified exclusively in mRNAs, 441 in ORFs, 8 in introns, and 7 in leader and trailer sequences. No RNA editing was seen in any of the rRNAs or in several tRNAs investigated for potential mismatch corrections. RNA editing affects individual coding regions with frequencies varying between 0 and 18.9% of the codons. The predominance of RNA editing events in the first two codon positions is not related to translational decoding, because it is not correlated with codon usage. As a general effect, RNA editing increases the hydrophobicity of the coded mitochondrial proteins. Concerning the selection of RNA editing sites, little significant nucleotide preference is observed in their vicinity in comparison to unedited C residues. This sequence bias is, per se, not sufficient to specify individual C nucleotides in the total RNA population in Arabidopsis mitochondria.
In plants, RNA editing has been identified as C to U as well as U to C conversions in both mitochondria and chloroplasts (1–4). In chloroplasts between 4 and 25 RNA editing events occur in the total transcribed coding complexity of 110 kilobase (kb), whereas in mitochondria several hundred such changes are estimated to alter the coding text of the RNA population.
Arabidopsis thaliana has evolved into one of the model plant species (5) and is presently the focus of a worldwide genome sequencing project (6). The mitochondrial genome of this plant was the first of the three genomes in the nucleus, the mitochondria and the chloroplasts, respectively, to be fully sequenced (7). This complete higher-plant mitochondrial genome sequence now allows an ordered global analysis of gene expression in Arabidopsis mitochondria. A comprehensive analysis—for practical purposes a cDNA investigation—is required for an overall view of the complexity of RNA editing in this organelle.
To handle the required large numbers of cDNA clones, we have initiated model development of the technology required for the rapidly advancing postgenome-sequencing era. The total Arabidopsis mitochondrial transcript complexity was therefore cloned in a large cDNA library, and the clones were sorted and spotted on high-density filters by using robot technologies (8–10). Sequence and genome-correspondence analysis of the cDNA population has identified functions coded in this genome (11).
In this plant mitochondrial genome with a total length of about 367 kb, only 30 kb are accounted for by known protein coding genes, which specify mostly subunits of respiratory chain complexes (7). Our cDNA sequence analysis covering all of the identified Arabidopsis mitochondrial mRNA and rRNA coding regions reveals the extent of RNA editing modifications in these organelles and their effect on the coded information. The analysis of the total complexity of RNA editing sites may give clues to the experimental investigation and eventual understanding of the specificity determining an individual RNA editing site.
Experimental Procedures
Biological Materials.
Mitochondria were prepared from an A. thaliana var. Columbia cell-suspension culture. Mitochondrial RNA was fractionated by LiCl precipitation, and high molecular-weight RNAs were used to construct a mitochondrial cDNA library primed from random hexamers, as described (10). The fraction of small RNAs was used to amplify and clone tRNA sequences by reverse transcription–PCR.
Isolation of cDNA Clones.
cDNA clones were recovered from a mitochondrial cDNA library, as described (10). Automated technology was used to screen the library rapidly and efficiently (8, 9). To cover the genomic complexity of 367 kb, 50,000 clones were plated and picked into microtiter plates by a picking robot. A spotting robot was then used to produce sets of high-density filters with 25,000 clones each, 2 filters thus representing the entire library. A single hybridization of these filters gives the information necessary to identify individual positive clones in the microtiter plates.
Sequence Analysis.
cDNA clones were analyzed by the chain-termination sequencing method with DNA cycle sequencing by using Cy5-labeled primers. Sequence reactions were analyzed with an ALF express (Pharmacia) automatic sequencer.
Results and Discussion
A total of 65 kb of transcribed sequences was investigated for RNA editing sites in cDNA clones, confirming these 65 kb or 17.8% of the mitochondrial genome of the Arabidopsis mitochondrial genome to be definitely transcribed (Table 1). About half of the analyzed cDNA sequences, 31 kb, are covered by identified protein coding regions, 5.5 kb by rRNAs and tRNAs, 5 kb by unidentified ORFs, 7.5 kb by introns and 16 kb by leader and trailer sequences in the various transcripts.
Table 1.
A total of 65.54 kb of the 367-kb mitochondrial genome of A. thaliana was analyzed in cDNA clones for RNA editing
| Sequence features | Sequence analyzed, kb | cDNA clones analyzed | Sequence not analyzed, kb | Editing sites | Always edited sites* | Editing sites per kb |
|---|---|---|---|---|---|---|
| Identified orfs† | 32.046 | 288 | 0 | 441 | 233 | 13.76 |
| rRNAs and tRNAs | 5.522 | 120‡ | 1.450 | 0 | 0 | 0 |
| Other orfs >100 aa | 4.656§ | 46 | 31.269§ | 0 | 0 | 0 |
| Introns | 7.546¶ | 36 | 17.186∥ | 8 | 7 | 1.06 |
| Leader, trailer | 15.770 | 143 | Unknown | 7 | 1 | 0.44 |
The always edited sites are those found altered in all cDNA clones analyzed, the number of which may vary between 3 and 20 clones covering an individual site.
†The orf110c and orf240a are included here.
‡This represents the ≈40% of the cDNA clones with multiple inserts, one of which usually is an rRNA sequence.
§About 8 kb of these ORFs are not detectably transcribed when analyzed by Northern blots (not shown).
¶The intronic ORF matR is not included here but is included with the identified orfs.
∥This number includes only the cis-splicing introns.
Gene-Specific Distribution of Editing Sites.
The 441 C to U RNA editing modifications detected in ORFs in Arabidopsis mitochondrial transcripts (Table 1) are distributed unevenly between the different genes (Table 2). For example, the transcript of cox1 is not edited at all, whereas the ccb2 mRNA is altered at 39 sites.
Table 2.
RNA editing affects mitochondrial mRNAs at very different rates
| Gene | Number of C/U editing sites in ORFs (N) | Size of the coding region, kb (S) | Editing frequency N/S (no. sites/kb) | ∑N/∑S |
|---|---|---|---|---|
| nad1 | 24 | 0.973 | 24.7 | Complex I ORFs 20.00 |
| nad2 | 31 | 1.495 | 20.7 | |
| nad3 | 12 | 0.359 | 33.4 | |
| nad4 | 32 | 1.484 | 21.6 | |
| nad4L | 9 | 0.302 | 29.8 | |
| nad5 | 27 | 2.005 | 13.5 | |
| nad6 | 10 | 0.617 | 16.2 | |
| nad7 | 28 | 1.180 | 23.7 | |
| nad9 | 7 | 0.572 | 12.2 | |
| Ψsdh4 | 5 | 0.285 | 17.5 | Complex II ORF 17.5 |
| cob | 7 | 1.181 | 5.9 | Complex III ORF 5.9 |
| cox1 | 0 | 1.583 | 0 | Complex IV ORFs 7.3 |
| cox2 | 15 | 0.781 | 19.2 | |
| cox3 | 8 | 0.797 | 10.0 | |
| atp1 | 5 | 1.523 | 3.3 | Complex V ORFs 2.9 |
| atp6-1 | 1 | 1.157 | 0.9 | |
| atp8 (orfB) | 0 | 0.476 | 0 | |
| atp9 | 4 | 0.257 | 15.6 | |
| ccb2 | 39 | 0.620 | 62.9 | Cytochrome c biogenesis ORFs 26.0 |
| ccb3 | 28 | 0.770 | 36.0 | |
| ccb6c | 16 | 1.357 | 11.8 | |
| ccb6n1 | 22 | 1.148 | 19.2 | |
| ccb6n2 | 12 | 0.611 | 19.6 | |
| rpl2 | 1 | 0.923 | 1.1 | Ribosomal protein ORFs 9.3 |
| rpl5 | 10 | 0.557 | 17.9 | |
| rpl16 | 8 | 0.539 | 14.8 | |
| rps3 | 10 | 1.669 | 6.0 | |
| rps4 | 15 | 1.088 | 13.8 | |
| rps7 | 0 | 0.446 | 0 | |
| rps12 | 8 | 0.377 | 21.2 | |
| Ψrps14 | 1 | 0.292 | 3.4 | |
| Ψrps19 | 0 | 0.116 | 0 | |
| mtt2 (orfX) | 24 | 0.860 | 27.9 | Not applicable |
| orf25 | 8 | 0.578 | 13.8 | |
| matR | 9 | 2.018 | 4.4 | |
| orf240a | 1 | 0.720 | 1.4 | |
| orf110c | 4 | 0.330 | 12.1 |
Data for individual mRNA species previously analyzed have been integrated. These include editing sites for nad2 and rps4 (13), nad4L and orf25 (22), nad5 exons d and e (23), cob, rpl5, and Yrps14 (24), orfX (25), and for ccb6n1 and matR (accession nos. X978254 and X98300, respectively). Cytochrome c biogenesis genes (ccb genes) are shown with new gene names; ccb2 is ccb206 (ccmB in Escherichia coli), ccb3 is ccb256 (ccmC in E. coli), ccb6n1 is ccb382, ccb6n2 is ccb203 and ccb6c is ccb452 (ccmF in E. coli, two genes corresponding to the N-terminal part and one gene corresponding to the C-terminal part, respectively).
RNA editing frequencies can be quantified more adequately by comparing the number of sites per kilobase of coding sequence between individual genes (Table 2). The thus obtained editing site densities expressed as sites per kilobase vary from 0 in the cox1 to 62.9 in the ccb2 coding regions. The comparison between gene groups, i.e., genes coding for individual subunits of a protein complex, shows that complex I and cytochrome c biogenesis coding regions are edited at higher frequencies than other gene groups. Transcripts for complexes II, III, IV, V, and the ribosome are less frequently edited, complex V coding regions showing the lowest editing density. Similar frequency distributions have been observed in some plant species but not in others. Although the relatively high level of RNA editing in the ccb coding regions appears to be quite common among flowering plants, a low level of editing in complex V transcripts is predominantly seen in the Brassicaceae. Complex I and V coding regions, although both specifying parts of the respiratory chain, show high divergence in their editing frequencies. These observations suggest that the editing frequency is independent of protein function. Editing level variations are also unrelated to base composition or codon usage. As an example, cox1 and nad2 have similar codon usage and base composition but very different editing frequencies with cox1 altered by 0 but nad2 modified by 20.7 editing sites per kilobase.
RNA Editing Increases the Overall Hydrophobicity of Mitochondrial Proteins.
One of the potential consequences of RNA editing in mRNAs and the corresponding change of the specified amino acid could be a modification of the overall biochemical nature of the affected mitochondrial proteins. The general tendency of the effect of RNA editing in Arabidopsis mitochondria is to increase the proportion of hydrophobic amino acid codons. As an example, the three most frequent amino acid transitions (93 S to L, 80 P to L, and 47 S to F) all result in codons for hydrophobic amino acids. In the overall analysis of RNA editing in Arabidopsis mitochondria, 35% of the modifications are hydrophilic to hydrophobic, and 35% are hydrophobic to hydrophobic codon alterations. Only the 27 P to S codon transitions reverse the tendency by creating codons for hydrophilic amino acids from those for hydrophobic ones. In the 425 modified codons detected, 41.5% specify hydrophobic amino acids before editing and 84.9%, after editing (Fig. 1). Thus RNA editing increases the hydrophobicity of mitochondrial proteins.
Figure 1.
Bar graphs show the overall biochemical nature of amino acids (aa) specified by the 425 edited codons before and after the alteration by RNA editing. RNA editing thus tends to increase the hydrophobicity of the encoded mitochondrial proteins.
RNA Editing in Structural RNAs.
Although mainly found in protein coding regions, RNA editing in plant mitochondria is also occasionally seen in structural RNA sequences, rRNAs and tRNAs, and in untranslated RNAs or in parts of mRNAs such as introns.
Group II Introns.
In Arabidopsis mitochondria, eight editing sites are identified in the 7.5-kb analyzed intron sequences covering about one-third of the total intron complexity (Table 1). The methodological approach taken here to clone and analyze cDNAs does not favor a specific search for cDNA clones covering intron sequences. Nevertheless, numerous clones containing unspliced sequences were identified and analyzed, some of them yielding editing sites within the intron sequences. In unspliced nad1 transcripts, two editing sites are detected in the intron between exons d and e. In nad5 precursor mRNAs, three sites are found in the intron separating exons a and b (Fig. 2A). In nad7 transcripts, one editing site is identified in the intron between exons b and c, and another editing event in this region modifies the first nucleotide of exon c similar to nad4 in wheat mitochondria (ref. 12; Fig. 2B). In nad2, two intron editing sites were already detected (13), one in the a/b intron, the other in the b/c intron.
Figure 2.
Secondary structure model of domains V and VI from two Arabidopsis mitochondrial group II introns. (A) The nad5a/b intron; (B) the nad7b/c intron. Thin black arrows indicate the splice sites. Gray arrows show nucleotides modified by RNA editing. Stars designate nucleotides predicted to be edited to stabilize intron structure but never actually seen altered in unspliced precursor RNAs.
The Arabidopsis mitochondrial intron population is exclusively composed of organellar group II introns (7) with a well-conserved secondary structure, which has been shown to be important for splicing (14, 15). Some of the editing sites affecting group II intron sequences are predicted to improve the quality of the intron folding (Fig. 2) and thus very likely to improve functional splicing. However, as observed in wheat mitochondrial introns (16), not all of the nucleotides predicted to allow better intron folding by a C to U change are actually seen edited (indicated by * in Fig. 2). This suggests that RNA editing, though important, may not in all instances be essential for the splicing activity. On the other hand, only unspliced intron sequences were analyzed here, and editing at such *-sites may actually trigger splicing of these introns and result in a different editing status of spliced introns.
rRNAs.
As in most genetic systems, rRNAs also make up the bulk of the steady-state RNAs in Arabidopsis mitochondria. As a consequence, a large proportion of the cDNA clones composing the randomly primed mitochondrial library used in this analysis (10) cover rRNA sequences. Derived from cloning and library construction artifacts attributed to the blunt-ended cloning procedure, about 40% of the cDNA clones contain more than one insert, one of the fragments generally being a rRNA sequence. Consequently, numerous cloned rRNA fragments were sequenced in this analysis, which in toto cover the entire sequences of the three rRNAs, the 5S, 18S, and 26S rRNAs, several times over. All analyzed complementary rRNA sequences are identical to the genomic sequence and give no indication of any RNA editing in these structural RNAs. No editing in the mitochondrial rRNAs appears to be the rule in higher plants, because in Oenothera mitochondria, where the three rRNAs have also been investigated for editing, two such sites were found only in a single 26S cDNA clone but not in others (17).
tRNAs.
Functionally important RNA editing events have been found in several plant mitochondrial tRNAs in potato and Oenothera (18). To search for editing sites in mitochondrial tRNAs of Arabidopsis, a specific cDNA library screening was performed for a cluster of six tRNA genes, which also includes the hypothetical orf199. Although no clones derived from large precursor transcripts were identified, four different cDNA clones were found covering parts of the spacer sequences between the tRNAs and part of orf199. Some (or all) of the tRNA genes are possibly cotranscribed, and the fragments detected could be maturation products. On the basis of analogy to other plant species, three tRNAs with C⋅A mismatches in stem regions were selected as candidates for editing. These were trnC, trnY, and trnN, the latter of which is edited in Oenothera (19). Sequence analysis of cloned reverse transcription–PCR products revealed no editing modification in these three tRNAs. The remaining tRNAs covering about 1.45 kb were not investigated (Table 1).
Is RNA Editing Connected to Translational Decoding?
To detect any potential connection between RNA editing site selection in protein coding transcripts and their respective codon context, the distribution of the 4% edited codons was analyzed. The investigation of these parameters reveals that of the 37 different C-containing codons, 35 were edited, and the two codon types CAG and AAC were never found altered. The 35 affected codon types are represented with different frequencies among the 425 codons edited in Arabidopsis mitochondrial mRNAs (Table 3). This codon preference for editing may have some functional reason or may simply reflect the overall codon usage in the mitochondrial genes.
Table 3.
The frequency of codons affected by RNA editing displayed against overall codon usage in Arabidopsis mitochondria
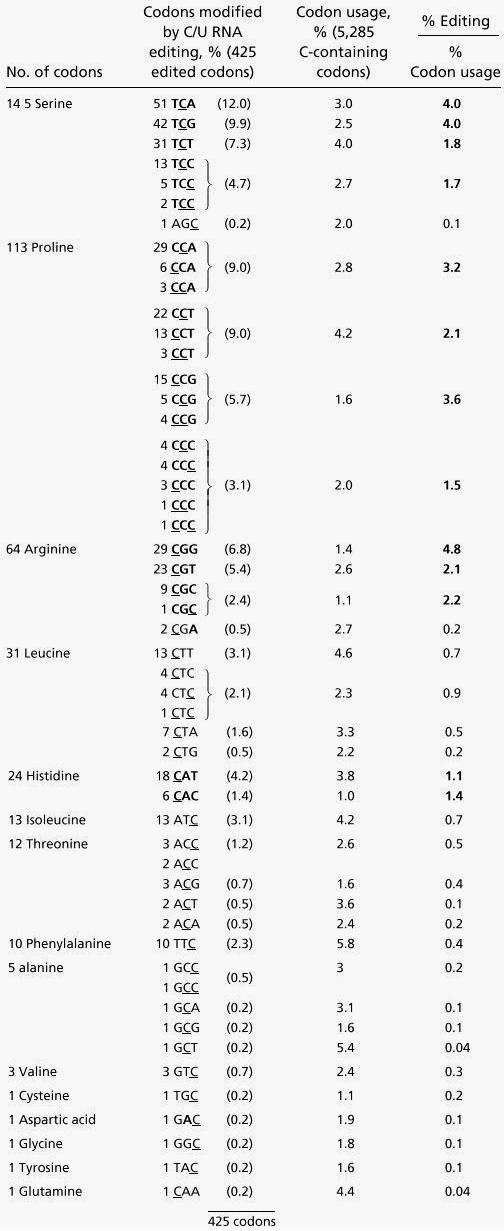 |
Codons are grouped according to the identity of the amino acid encoded and the number of occurrences. Highlighted in bold are the 13 codons edited more frequently than the statistical average. The novel orf110c is included with the identified genes.
To determine the relationship, we calculated the correlation factor between the frequency of a given codon in the population of edited codons and the analogous proportion in the total codon population of all mitochondrial mRNAs (% edited codon/% codon usage column in Table 3). This factor thus indicates whether a codon is proportionally more or less affected by RNA editing than its overall usage in the protein coding regions. A factor of 1 indicates codon editing directly proportional to codon usage, a value between 0 and 1 shows a codon less often edited, and a value over 1 (bold in Table 3) identifies a preferentially edited codon. In the Arabidopsis mitochondrial mRNAs, this factor ranges between 0 and 13.7. The six most frequently edited codons TCA, TCG, CCA, CCT, TCT, and CGG, represent 54% of the edited codons. The presence of a pyrimidine 5′ adjacent to the editing site (see also below) cannot alone be responsible for this codon preference, because the codons TTC, ATC, GCC, and GTC fulfill this criterion but are nevertheless infrequently edited.
The variations of the codon editing correlation factor in Table 3 show that in plant mitochondria, RNA editing is not connected to codon usage. This observation suggests that specific site selection operates independently of translation and translational preferences.
Does RNA Editing Depend on Codon Position?
RNA editing sites are nonrandomly distributed between the three codon positions (Table 4). Although 88.4% of the editing sites are located on the first two positions, only 11.6% of the sites are found at the third position, these reflecting the 12.7% silent editing sites (56 sites). The biased distribution of editing sites within the codons is not explained by the general distribution of cytosines in the Arabidopsis mitochondrial codons, as is apparent from the respective numbers given in Table 4.
Table 4.
Comparison of the RNA editing frequencies at different codon positions with the C composition of codons in Arabidopsis mitochondrial genes shows that the frequency of editing sites does not reflect merely the distribution of Cs
| Codon position | No. of editing sites at each codon position | Distribution of 441 editing sites in ORFs % | Distribution of the 6627 C residues in codons, % |
|---|---|---|---|
| First | 154 | 34.9 | 33 |
| Second | 236 | 53.5 | 36.9 |
| Third | 51 | 11.6 | 30 |
The frequency of edited nucleotides is particularly higher in the second codon position and lower in the third.
According to the universal genetic code and the wobble rules, all C to U editing sites at the third position of a codon are, per se, silent and do not alter the coded amino acid identity. Thus, the only evolutionary pressure exerted to maintain a particular third position editing event besides hypothetical mRNA structural constraints may reside in the preference of the tRNA to attach to the respective codon ending with U rather than with C. Overall codon usage (Table 3) indeed shows that codons ending with U for a given amino acid are nearly systematically preferred to codons ending with any other nucleotide. For example, TCT codons amount to 4.0% of the C-containing codons, whereas TCC contribute 2.7%. Similarly, CCT codons represent 4.2% of the C-containing codons and CCC, only 2.0%. Considering the overall preference for U over C in the third position of the unedited codons, it is possible that this constitutes a “functional” background for these editing sites. However, a real functional investigation, including tests for the relative tRNA-codon affinities, will be required to clarify this possibility.
Alternatively, these third codon position sites are edited because of some chance similarity of their neighboring sequence with the vicinity of a “functional” nonsilent editing site. This similarity could induce an as yet unknown specificity factor such as an antisense “guide” RNA to edit this silent C. Such similarities can indeed be detected between “silent” and “functional” editing vicinities (not shown), but a generally common theme of similarity remains elusive (see also next paragraph).
Nucleotide Identities Around Editing Sites.
In the alignment of sequences surrounding all the RNA editing sites, nucleotide identities are nonrandomly distributed in the vicinity of editing sites, although a strict consensus motif is not readily apparent (Fig. 3). The most prominent example of this nonrandom nucleotide distribution is the identity of the nucleotide immediately 5′ adjacent to the altered C, which is in 93.6% of the editing sites found to be a pyrimidine, including 63.1% uridines and 30.5% cytosines. The next nucleotide upstream at position −2 from the editing site is also most often a pyrimidine (62.8%). On the other, the 3′ side of the edited C, 61.5% of the adjacent nucleotides are purines, including 32.5% guanidines. These observations extend previous comparisons of editing sites in chloroplasts and mitochondria of other higher plant species (20).
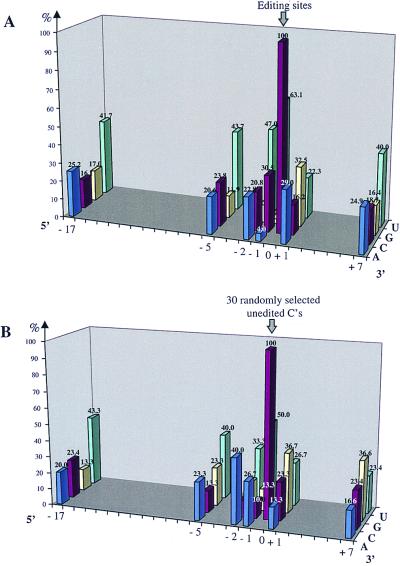
Figure 3.
Distribution of nucleotide identities around editing sites. Histograms show the relatively overrepresented bases around editing sites (position 0) in A, and around 30 randomly selected not edited Cs in B. Meaningful asymmetries could be only those not introduced by the general base composition and the third codon position bias as discussed in the text and as apparent in B. Positions not differing greatly from the overall base composition are not indicated.
Such nucleotide preferences are, however, not confined to the immediate vicinity of the editing sites but are also observed at more distal positions (Fig. 3). Upstream of editing sites, nucleotide biases are observed at positions −17 and −5 with U found in 41.7 and 43.7% of the editing sites, respectively. At the 3′ position +7 downstream of editing sites, 40.0% of the nucleotides are uridines. This preference for U is only partially explained by the elevated usage of U observed in plant mitochondrial coding regions (7), because the overall frequency of 31.4% Us in Arabidopsis mitochondrial mRNAs is much lower than the frequency of U at this nucleotide position. A partial explanation for these at first sight potentially significant nucleotide asymmetries is, however, given by the U preference in the third codon position. Considering that more than half of the editing sites (Table 4) are found in the second codon position, the third codon position U preference will yield a U increase in positions −17 … −5, −2, +1, +4, +7 (Fig. 3). The residual effective codon bias around editing sites is thus restricted to the region −5 to +1.
This nucleotide preference immediately around editing sites, although not sufficient to explain the specificity of editing, may nevertheless have a mechanistic significance (see also next paragraph). For steric reasons, pyrimidines may be preferred by the editosome immediately 5′ of a C to be edited.
Which Specificity Is Required by the Editosome?
The RNA editing activity, termed the editosome to denote the unknown components performing the actual biochemical reaction, needs a specificity discriminator to recognize which cytosine is to be modified to a uridine. This discriminator could act in cis, being a consensus sequence or a conserved fold of the RNA. The specificity factor could also be a trans-acting element, a protein (or part of one), or an RNA molecule (or part of one) guiding the editing activity (20, 21).
Assuming that every single editing site could be specified by a unique sequence, this signature should be at least 10 nt long to identify a unique locus in the 366,924 nt of the mitochondrial genome of Arabidopsis (7) (410 = 1,048,576). Considering only the 31,854 nt constituting the actual protein coding regions, a minimal sequence tag has to be only 8 nt long (48 = 65,536) in order to be unique.
One major characteristic of RNA editing in plant mitochondria is the density and the sheer number of editing sites. In Arabidopsis mitochondria, 8.0% of all of the C-containing codons are edited and 16 of these 5,285 codons are edited twice. In the total number of 6,627 cytosines present in the protein coding regions, almost 6.6% are modified by editing, that is, one of 15 Cs is edited. Accordingly, the specificity required by the editing machinery might not be quite as high as previously suspected. Theoretically, only two strictly conserved nucleotides around a C (42 = 16) could be enough to specify an editing site in Arabidopsis mitochondria. Such always conserved nucleotides are not observed, but their equivalent in an as yet hidden arrangement could still be targeted by only a small number of characteristic factors, possibly even one or very few protein moieties.
Acknowledgments
We thank Dr. Volker Knoop for his advice, Dr. Jörg Kudla and Drs. Kanji Ohyama and Katsuyuki Yamato for their support, and Dr. Géraldine Bonnard for suggesting the nomenclature for the cytochrome c biogenesis (ccb) genes. We are grateful to the anonymous reviewers for constructive suggestions to improve the presentation. This work was supported by a grant from the Bundesministerium für Bildung, Wissenschaft und Technologie.
Abbreviation
- kb
kilobase
Footnotes
References
- 1.Covello P S, Gray M. Nature (London) 1989;341:662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- 2.Gualberto J M, Lamattina L, Bonnard G, Weil J-H, Grienenberger J-M. Nature (London) 1989;341:660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- 3.Hiesel R, Wissinger B, Schuster W, Brennicke A. Science. 1989;246:1632–1643. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- 4.Maier R, Neckermann K, Igloi G, Kössel H. J Mol Biol. 1995;251:614–628. doi: 10.1006/jmbi.1995.0460. [DOI] [PubMed] [Google Scholar]
- 5.Meyerowitz E M, Somerville C R. Arabidopsis. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 6.Blackbourn H, Ingram J. Trends Plant Sci. 1996;1:291–292. [Google Scholar]
- 7.Unseld M, Marienfeld J R, Brandt P, Brennicke A. Nat Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 8.Lennon G G, Lehrach H. Trends Genet. 1991;7:314–317. doi: 10.1016/0168-9525(91)90420-u. [DOI] [PubMed] [Google Scholar]
- 9.Maier E, Bancroft D R, Lehrach H. In: Automation Technologies for Genome Characterization. Beugelsdijk T J, editor. New York: Wiley; 1997. pp. 65–88. [Google Scholar]
- 10.Giegé P, Konthur Z, Walter G, Brennicke A. Plant J. 1998;15:721–726. doi: 10.1046/j.1365-313x.1998.00242.x. [DOI] [PubMed] [Google Scholar]
- 11.Giegé P, Knoop V, Brennicke A. Curr Genet. 1998;34:313–317. doi: 10.1007/s002940050401. [DOI] [PubMed] [Google Scholar]
- 12.Lamattina L, Weil J-H, Grienenberger J-M. FEBS Lett. 1989;258:79–83. doi: 10.1016/0014-5793(89)81620-5. [DOI] [PubMed] [Google Scholar]
- 13.Lippok B, Brennicke A, Unseld M. Biol Chem. 1996;377:251–257. doi: 10.1515/bchm3.1996.377.4.251. [DOI] [PubMed] [Google Scholar]
- 14.Knoop V, Kloska S, Brennicke A. J Mol Biol. 1994;242:389–396. doi: 10.1006/jmbi.1994.1589. [DOI] [PubMed] [Google Scholar]
- 15.Michel F, Ferat J L. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 16.Carillo C, Bonen L. Nucleic Acids Res. 1997;2:403–409. doi: 10.1093/nar/25.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuster W, Ternes R, Knoop V, Hiesel R, Wissinger B, Brennicke A. Curr Genet. 1991;20:397–404. doi: 10.1007/BF00317068. [DOI] [PubMed] [Google Scholar]
- 18.Marechal-Drouard L, Kumar R, Remacle C, Small I. Nucleic Acids Res. 1996;16:3229–3234. doi: 10.1093/nar/24.16.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Binder S, Marchfelder A, Brennicke A. Mol Gen Genet. 1994;244:67–74. doi: 10.1007/BF00280188. [DOI] [PubMed] [Google Scholar]
- 20.Maier R M, Zeltz P, Kössel H, Bonnard G, Gualberto J, Grienenberger J-M. Plant Mol Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 21.Bock R, Hermann M, Fuchs M. RNA. 1997;3:1194–1200. [PMC free article] [PubMed] [Google Scholar]
- 22.Brandt P, Sünkel S, Unseld M, Brennicke A, Knoop V. Mol Gen Genet. 1992;236:33–38. doi: 10.1007/BF00279640. [DOI] [PubMed] [Google Scholar]
- 23.Knoop V, Schuster W, Wissinger B, Brennicke A. EMBO J. 1991;10:3483–3493. doi: 10.1002/j.1460-2075.1991.tb04912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandt P, Unseld M, Eckert-Ossenkopp U, Brennicke A. Curr Genet. 1993;24:330–336. doi: 10.1007/BF00336785. [DOI] [PubMed] [Google Scholar]
- 25.Sünkel S, Brennicke A, Knoop V. Mol Gen Genet. 1994;242:65–72. doi: 10.1007/BF00277349. [DOI] [PubMed] [Google Scholar]