Abstract
Accumulation of red phlobaphene pigments in sorghum grain pericarp is under the control of the Y gene. A mutable allele of Y, designated as y-cs (y-candystripe), produces a variegated pericarp phenotype. Using probes from the maize p1 gene that cross-hybridize with the sorghum Y gene, we isolated the y-cs allele containing a large insertion element. Our results show that the Y gene is a member of the MYB-transcription factor family. The insertion element, named Candystripe1 (Cs1), is present in the second intron of the Y gene and shares features of the CACTA superfamily of transposons. Cs1 is 23,018 bp in size and is bordered by 20-bp terminal inverted repeat sequences. It generated a 3-bp target site duplication upon insertion within the Y gene and excised from y-cs, leaving a 2-bp footprint in two cases analyzed. Reinsertion of the excised copy of Cs1 was identified by Southern hybridization in the genome of each of seven red pericarp revertant lines tested. Cs1 is the first active transposable element isolated from sorghum. Our analysis suggests that Cs1-homologous sequences are present in low copy number in sorghum and other grasses, including sudangrass, maize, rice, teosinte, and sugarcane. The low copy number and high transposition frequency of Cs1 imply that this transposon could prove to be an efficient gene isolation tool in sorghum.
Transposable elements as causative agents of variegation and genetic variation were first discovered in maize by Barbara McClintock in the 1940s (1) and since have been found in many organisms. Transposon-induced variegation traits are powerful genetic tools to study gene expression and regulation (2). Although variegation mutants have been described in at least 35 plant species (3), active transposable elements have been isolated from only a minority of these plants. Three well characterized maize transposons—Ac/Ds, En/Spm, and Mu (4–6)—have been used in gene-tagging approaches to isolate a large number of plant genes (3, 7). More recent studies have revealed high levels of microsynteny among grass genomes, suggesting that plants containing small genomes could be used as efficient model systems for gene tagging and isolation (8). Among the cereal grains, sorghum has one of the smallest genomes, and it is closely related to maize (9). Thus, the identification of an active transposable element for gene tagging in sorghum could provide a new route to the isolation of the corresponding maize genes. In addition to serving as a model for other cereals, sorghum is a major source of nutrition in developing countries and ranks fourth in economic importance among grain crops, after maize, wheat, and rice.
In sorghum, the Y gene is required for the production of a red phlobaphene pigment, whereas a mutable allele Yv (Y-variegated; ref. 10), or y-cs (y-candystripe; ref. 11), is associated with variegated pigmentation phenotype in the grain pericarp and other plant tissues. Based on the high frequency of somatic and germinal reversions of y-cs to Y (23%), it has been postulated that the y-cs phenotype may result from the presence of a transposable element in the Y gene (10–12). In these respects, the y-cs allele bears a marked resemblance to the maize P1-vv allele. In maize, the p1 gene regulates the production of a red flavonoid pigment in kernel pericarp and other plant tissues. The P1-vv allele specifies variegated kernel pericarp pigmentation, and this allele contains an Ac transposable element insertion in the P1-rr gene. We have found that the sorghum Y gene encodes a plant MYB domain protein that is closely related to p1 and homologous genes in maize and teosinte (P. Zhang, S.C., and T.P., unpublished data). Here, we show that the y-cs allele contains a large transposon inserted within the second intron of the Y gene. The transposon is named Candystripe1 (Cs1) and is a member of the CACTA family of plant transposable elements. Of several members of this family, the En/Spm element of maize has been best understood at the genetic and molecular levels. It was identified originally as Enhancer (En; ref. 13) and was shown later to be homologous to the Suppressor-Mutator system (Spm; ref. 14) by genetic (15) and molecular tests (16). Besides maize, CACTA elements have been characterized from snapdragon (Tam1; ref. 17), soybean (Tgm1; ref. 18), pea (Pis1; ref. 19), Japanese morning glory (Tpn1; ref. 20), rice (Tnr3; ref. 21), carrot (Tdc1; ref. 22), and petunia (PsI; ref. 23). Similar to these elements, Cs1 has characteristic short terminal inverted repeats (TIR) with conserved 5′-CACTA-3′ ends, and, upon insertion, it seems to generate a 3-bp direct duplication of the target sequence. Another significant commonality between Cs1 and other CACTA elements is the presence of highly structured subterminal regions. Though CACTA elements are known for their large sizes (Tam1; 15.2 kb; ref. 24), Cs1 with its 23,018-bp sequence may be the largest active transposable element described to date.
Materials and Methods
Sorghum Stocks, Plant DNA Isolation, and Southern Analysis.
The original candystripe sorghum was collected from Gedaref, Sudan, by O. Webster, as described previously (11). Inbred candystripe line CS8110419 was kindly provided by J. Bennetzen (Purdue University) and was used to prepare the genomic library from which Y-cs clones were isolated. To study putative transposition events, red revertants were obtained from an agronomically improved candystripe sorghum line developed by the Purdue sorghum genetics program as follows: candystripe line CS8110419 was crossed to white-seeded line Tx2737 and subsequently propagated by self-pollination for 10 generations. In the F10 generation, seven spontaneous full red seed heads (inflorescences) were selected and their progeny seedlings were used for Southern hybridization analysis. Diverse sorghum lines were obtained from the Plant Genetic Resources Conservation Unit, U.S. Department of Agriculture, Agricultural Research Service, University of Georgia. Representative grass species were obtained from Brent Pearce, Iowa State University. Plant genomic DNA was prepared by using the hexadecyl trimethyl ammonium bromide method (25, 26). Restriction enzyme digestions were performed by using enzymes, reagents, and incubation conditions from Promega. Southern blot hybridizations were performed as described previously (27). Southern blots were stripped by washing for 15 min in boiling solution of 0.1% SDS before rehybridization.
Isolation of Genomic λ Clones, Plasmid Subcloning, and DNA Sequencing.
A BamHI size-selected (7.0–10.0 kb) subgenomic library from leaf DNA of candystripe sorghum line CS8110419 was constructed in λ-EMBL3/BamHI vector by following standard methods (28). A total of 160,000 clones were screened with maize P1-rr genomic fragment 8A (29) as probe, and two positive clones were isolated. The clones appeared to be identical based on their restriction digest patterns. The approximately 8.0-kb insert of one of the clones (λCS-16) was restriction-mapped and sequenced. A second library was made from partially Sau3AI-digested leaf DNA of CS8110419 in λ-FIX II/XhoI vector by following the reaction conditions from Stratagene. Genomic fragments were partially filled in to generate ends compatible with the partially filled-in XhoI ends of the vector. From this library, a total of 8 × 105 clones were screened by using the maize P1-rr gene fragment 12 (29). In this screen, three positive clones were identified and the largest clone (λCS-3) was characterized further. DNA fragments were subcloned into pBluescript plasmid vectors by standard methods (28). Plasmid subclones were sequenced with gene- or vector-specific primers by using the Applied Biosystems fluorescent sequencing system at the Iowa State University Nucleic Acids facility.
Sequence Analysis.
DNA sequence data was compiled by using gcg software. Short sequence repeats were identified by the algorithm of Leung et al. (30). Potential splice sites were identified with the splicepredictor program at http://gremlin1.zool.iastate.edu/cgi-bin/sp.cgi (31, 32). Potential genes were identified with the genscan algorithm of Burge and Karlin (33), available at http://CCR-081.mit.edu/GENSCAN.html, and refined with the genegenerator program (34) and the spliced alignment options of splicepredictor (35). None of the gene structure prediction programs had any prior training on sorghum DNA. Results were obtained and analyzed by using both maize and Arabidopsis parameter settings. Database searches were performed with the blast suite of programs available at http://www.ncbi.nlm.nih.gov/BLAST/. Protein percent similarities were obtained from the matching segments in the blast output.
PCR Amplification.
Positions of PCR primers used for excision footprint analysis are indicated in Fig. 2B, and their sequences are: Primer1, 5′-TTGACACTGCGGACGCTGAG-3′ and Primer2, 5′-AAGCTTGAATTCGAGTTCCAGTAGTTCTTGATC-3′. Sorghum genomic sequences containing conserved TIRs of Cs1 were PCR-amplified by using a single primer, 5′-CACTATGTGAAAAAAGCTTA-3′. PCR conditions were the same as described earlier (27). PCR products were cloned in TA cloning vectors from Promega or Novagen and sequenced as described above.
Figure 2.
Molecular characterization of the Y-cs allele. (A) Detection of RFLP. Genomic DNA samples from red revertant (R) and candystripe (C) plants were digested with BamHI and hybridized to the indicated probe fragments. Probe 8A is a genomic DNA fragment of the maize p1 gene containing exons 1 and 2, which encode part of the MYB DNA-binding domain (37). Probes F3, F2, and F1 are restriction fragments of the y-cs clone λCS-16 (B). The approximately 8.0-kb polymorphic band present in candystripe DNA that hybridizes to all the probe fragments is indicated with an asterisk. The 5.0-kb band (arrow) present in red revertant DNA hybridizes to the maize p1-Myb probe 8A and sorghum Y gene intron2 probe F3. Nonpolymorphic bands hybridizing to 8A probe may represent other Myb-homologous genes in sorghum. Molecular sizes are shown in kb. (B) Partial restriction map of the y-cs allele (not to scale). The Y gene exons are represented as black boxes. The Candystripe1 (Cs1) element present in the second intron of the Y gene is indicated by a bold line. Outwardly oriented arrow heads represent 20-bp TIR sequences found at the borders of the Cs1 element. Positions of DNA fragments 8A, 12, F1, F2, F3, and 28E used as hybridization probes are indicated below the y-cs map. Restriction enzyme sites shown are B, BamHI; E, EcoRI; H, HindIII; and X, XhoI. The y-cs map was constructed from overlapping λ clones (Lower). Arrows marked as P-1 and P-2 indicate positions of primer 1 and primer 2, respectively, used for the PCR amplification.
Results
Molecular Basis of Sorghum Candystripe Phenotype.
In sorghum, the candystripe seed phenotype (Fig. 1) has been attributed to y-cs, a mutable allele of the Y gene. Using maize p1 genomic DNA fragments as probes (fragments 8A and 12), we detected restriction fragment length polymorphism (RFLP) between candystripe and red revertant sorghum leaf DNA. Fig. 2A shows that in a BamHI digestion, probe 8A detects a 5.0-kb band in the DNA of a red revertant (lane R), which is replaced by a band of approximately 8.0 kb in the candystripe DNA (lane C). The maize p1 gene fragment 12 hybridizes to the same 5.0-kb band as detected by probe fragment 8A in the red revertant DNA but it detects an approximately 5.5-kb band in candystripe DNA (not shown).
Figure 1.
Sorghum seeds showing “candystripe” pericarp phenotype of the y-cs allele.
To isolate the polymorphic BamHI band, a size-selected λ genomic (candystripe plant DNA) library was probed with the maize p1 gene fragment 8A, and two positive clones were isolated and characterized further. An approximately 8.0-kb BamHI insert from one of the positive clones (λCS-16) was subcloned into a plasmid vector and will be referred to here as pCS16. Three restriction fragments of pCS16, which did not hybridize to the probe fragment 8A, were identified as F1, F2, and F3 (Fig. 2B). These individual fragments were used further as probes on the genomic DNA blot that was hybridized previously to the P1-rr gene fragment 8A. Fragments F1 and F2 hybridized to the 8.0-kb polymorphic band as well as to several other bands in candystripe and red DNA, but did not hybridize to the 5.0-kb band detected by fragment 8A in the red revertant DNA (Fig. 2A). Fragment F3 hybridized to the 8.0-kb polymorphic band in candystripe as well as to the 5.0-kb band in the red revertant plant DNA. These results suggest that the F3 fragment lies in the sorghum Y gene, and fragments F1 and F2 are part of an insertion. These results also indicate that the beginning of the insertion sequence may lie somewhere between fragments F2 and F3. Similar results were obtained with Southern hybridization by using several independent red revertant plants obtained from candystripe sorghum line (data not shown).
Isolation of Candystripe1 (Cs1) Transposable Element.
Analysis of the 7,377-bp insert of pCS16 revealed that it contains two types of sequences. The first 3,400 bp showed an overall nucleotide similarity of 82% with the maize P1-rr gene sequence. The remaining approximately 3,900 bp of pCS16 did not detect any similarity to P1-rr or to any other sequences in the public databases. This latter region of pCS16 comprises restriction fragments F1 and F2, which were shown by DNA gel blot analysis to be part of an insertion (Fig. 2A). However, because no landmarks of a transposable element were identified in this sequence, we further isolated the 3′ end of the sorghum y-cs sequence. A λ genomic library prepared from a partial Sau3A digest of candystripe leaf DNA was screened with a probe containing the 3′ end of intron 2 and the 5′ region of exon 3 of the maize p1 gene (fragment 12). Three positive clones were isolated; the largest clone (λCS-3), whose 5′ end overlapped with the pCS16 sequence, was subcloned and sequenced. Comparison of the composite sequence of the y-cs allele with the maize P1-rr gene sequence allowed us to identify a 23-kb insertion in the second intron of the Y gene (Fig. 2B). The insertion in y-cs also was confirmed by comparing sequences of y-cs and Y red revertant in the region flanking the insertion site (not shown). The insertion sequence has the structural features of a transposable element, and, in reference to the “candystripe” phenotype, we have named it Candystripe1 (Cs1).
Transposition of Cs1.
To analyze the transposition of Cs1, we characterized seven putative germinal excision events obtained as spontaneous full red head plants from a candystripe population. Southern blots of genomic DNA from the red revertants and their corresponding candystripe siblings were hybridized with probe fragments of Cs1. Results from three pairs of red and candystripe DNA digests probed with Cs1 fragment 28E are presented in Fig. 3. The approximately 8.0-kb BamHI band identified previously (Fig. 2) as the y-cs polymorphic band is present in all the lanes carrying DNA from candystripe plants (lanes C). Red revertant DNA samples (lanes R) show new bands hybridizing with the probe. These results thus demonstrate that the full red head plants (Y/Y or Y/y-cs) are derived from an excision of the Cs1 element from the y-cs allele. Furthermore, PCR amplification of DNA from two independent red revertant plants by using flanking primers 1 and 2 (Fig. 2B) shows that the complete 23,018-bp Cs1 element excised from the Y gene and left behind a 2-bp footprint. The presence of a new Cs1-hybridizing band in red revertant plants shows that the Cs1 transposon can reinsert elsewhere in the genome after it excises from the Y gene.
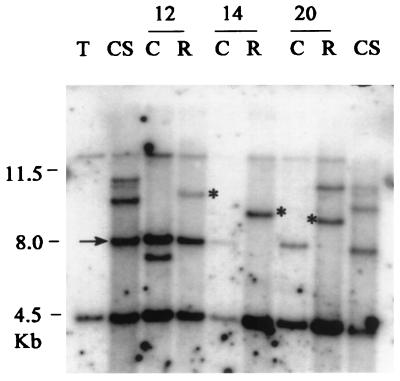
Figure 3.
Transposition of Candystripe1 element. Southern blot analysis was done on pairs of candystripe and red revertant sibling plants (Materials and Methods). Results obtained from three pairs (nos. 12, 14, and 20) are presented. Genomic DNA from parental lines Tx2737 (white pericarp, lane T) and CS8110419 (candystripe, lane CS) and three pairs of candystripe (C) and red revertant (R) siblings was digested with BamHI, gel-fractionated, and blotted. The blot was hybridized with Cs1 probe 28E, a 1.1-kb subfragment of the F1 fragment (Fig. 2B). The 8.0-kb polymorphic BamHI band present in the parental candystripe line (CS) and sibling candystripe plants (C) is shown by an arrow. This band is absent from homozygous (genotype YY) red revertants 14-R and 20-R. Transposition of Cs1 to a new genomic site is demonstrated by the presence of a new Cs1-hybridizing band, indicated by an asterisk in these red revertants. Note that red revertant 12-R shows the presence of the 8.0-kb y-cs band as well as a second band in the genome; progeny analysis confirmed that this revertant is of a heterozygous Y/y-cs genotype.
Cs1 Copy Number and Homologs in Sorghum Lines and Other Grasses.
Copy number of the Cs1 element was estimated from hybridization of different fragments of Cs1 to gel blots carrying genomic DNA isolated from sorghum and other grasses. Cs1-homologous sequences are detected at low copy number (<10) in candystripe line CS8110419 and seven other diverse sorghum lines, as well as sudangrass, rice, and sugar cane. A higher copy number of Cs1-homologous bands were observed in teosinte and maize, whereas no hybridizing bands were detected in tripsacum, wheat, and oats under these stringent hybridization conditions (Fig. 4). In a BamHI restriction enzyme digestion, an approximately 4.5-kb hybridizing band is common in most of the sorghum lines and some of the grass species. To determine whether deletion derivatives of Cs1 exist in the sorghum genome, PCR amplifications were performed on DNA from candystripe or red revertant plants by using a single forward primer synthesized from the 20-bp TIR sequence of Cs1. Four amplification products ranging in size from 0.7 to 1.4 kb were detected. The 0.7-kb fragment was sequenced and found to contain sequences homologous to the 5′ 220 bp and the 3′ 505 bp of Cs1; the internal 22,293 bp corresponding to the central region of Cs1 are deleted.
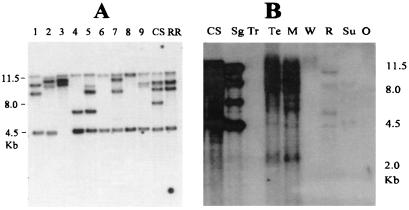
Figure 4.
Homologs of Candystripe1 in sorghum races and grasses. Cs1-homologous sequences in diverse sorghum lines (A) and grass species (B) were identified through Southern blot analysis. Gel blots prepared from BamHI-digested leaf DNA were hybridized to the Candystripe1 probe fragment 28E (Fig. 2B). Plant introduction (PI) numbers and country of collection of diverse sorghum races used in A are: 1, Bicolor (PI147837, Ethiopia); 2, Caudatum (PI267459, India); 3, Caudatum (PI276837, Ethiopia); 4, Caudatum (PI570719, Sudan); 5, Durra (PI534132, Ethiopia); 6, Durra (PI248317, India); 7, Guinea (PI534070, Nigeria); 8, Guinea-Caudatum (PI533871, Nigeria); 9, Kafir-Caudatum (PI152621, Sudan). Sorghum breeding lines used are CS, CS8110419 (candystripe); and RR, red revertant. Grass species included in B are Sg, Sudangrass; Tr, Tripsacum; Te, Teosinte; M, Maize; W, Wheat; R, Rice; Su, Sugarcane; and O, Oats.
Structure and Sequence Analysis of Candystripe1.
Salient features of the 23,018-bp Cs1 sequence are presented in Fig. 5. Similar to other members of the CACTA family, Cs1 ends have a short TIR sequence, 5′-CACTATGTGAAAAAAGCTTA-3′, and these termini are flanked by a 3-bp target site duplication. Subterminal regions, 250 bp interior to the TIR, contain multiple copies of direct and indirect repeats. A 12-bp conserved (80%) sequence motif (5′-TTATTACAGACG-3′) is repeated eight and six times, respectively, in the 5′ and 3′ subterminal regions. Sequences similar to the subterminal repeat motif also are present at seven sites in the central region of the Cs1 transposon. Interspersed within the Cs1 element are other tandem repeats as well as several copies of HCSRs (high-copy short interspersed repeats). Several of these repeat sequences have high similarity (up to 95%) to a Sb1 Tourist element of sorghum as well as to other MITEs (miniature inverted repeat transposable elements; ref. 36).
Figure 5.
Structure of Candystripe1 transposon. Line diagram showing the structural features of the Cs1 element. Arrow heads at the end represent the 20-bp TIR sequences. Subterminal ends are enlarged to show the arrangement of the 12-bp repeat motifs. Predicted genes 1, 2, 3, and 4 are indicated with exons as black boxes and introns as diagonal lines joining the exons. Other sequence elements are indicated.
Coding capacity of the Cs1 element was analyzed by database comparisons and by gene prediction algorithms. The Cs1 sequence contains 129 ORFs coding for peptides ranging from 30 to 418 aa. The genscan gene-prediction algorithm predicts four potential genes in the Cs1 sequence (Fig. 5). Predicted genes 1 and 3 did not show any similarity to sequences in the public databases. Gene 2 is predicted from the complementary strand (from 6,386 to 2,245) and contains eight exons, producing a deduced protein sequence of 1,001 aa. The predicted gene 2 product has weak similarity (24%) to the TNP1 protein of Tam1 (accession no. X57297), TNPA of En-1 (accession no. AAA66268), and to other hypothetical proteins from sorghum (accession no. AAD27567) and Arabidopsis (accession nos. AC005356, AAD28689, and AC005897). The genscan-predicted gene 4 contains 11 exons and its deduced protein is 876 aa long. Refined splice site analysis with splicepredictor strongly supports a shorter gene 4 product (462 aa) involving only the last five exons. This predicted gene 4 protein shows significant similarity to a predicted sorghum protein (accession no. AAD27562: 53% over a 198-aa segment) and to two other hypothetical proteins of Arabidopsis (accession no. AAC14510: 46% over 180 aa; accession no. AAD25557: 38% over 314 aa). None of the Cs1 ORFs show any similarity to the putative transposase proteins TNP2 and TNPD encoded by autonomous CACTA elements Tam1 and En1, respectively.
Discussion
In maize, the p1 gene encodes a Myb-homologous transcription factor that regulates the biosynthesis of the red phlobaphene pigment in kernel pericarp (37). The striking similarities in pericarp pigmentation phenotypes controlled by the maize p1 and sorghum Y genes suggested the possibility that these genes are orthologous. In a previous study, a probe derived from the 3′ untranscribed region of the maize P1-rr locus (P1-rr-15) was mapped to linkage group “a” of the sorghum genome corresponding to the maize chromosome 1S region containing the p1 gene (38). However, the P1-rr-15 probe did not detect any RFLP between candystripe and red pericarp plant DNA (11). Moreover, no linkage was detected between the Y-controlled seed pigmentation phenotype and the sorghum locus homologous to P1-rr-15 (11). Here, we followed a generalized approach based on the premise that a p1 homolog should contain a conserved Myb-homologous DNA-binding domain (37). Using DNA fragments encoding the maize P-Myb domain as probes, we detected RFLPs between candystripe and red pericarp plant DNA. We subsequently isolated the y-cs allele and identified a transposable element (Candystripe1) inserted within the second intron of the Y gene. Excision of the Cs1 element was correlated with reversion of the Y gene, thus establishing the causative role of the Cs1 element in y-cs variegation. It remains to be determined what is the nature of the sorghum locus occupying the syntenic position with the maize p1 locus.
Sequence analysis of Candystripe1 reveals several features that are conserved among transposable elements of the En/Spm family. The Cs1 element is bordered by a 20-bp perfect TIR sequence that contains conserved 5′-CACTA-3′ ends. Most other CACTA elements have 13-bp TIR sequences, with the exception of Tpn1 from Japanese morning glory, which has the longest TIRs of 28 bp (20). Transposable elements of the CACTA family are of relatively large sizes (En/Spm, 8.2 kb; Tam1, 15.1 kb); the 23,018-bp Cs1 element is the largest known member of this family. Like other CACTA elements, Cs1 appears to generate a 3-bp duplication of the target sequence upon insertion. Additionally, the Cs1 element has a subterminal repetitive region containing a 12-bp repeat motif reiterated eight times at the 5′ end and six times at the 3′ end. The En/Spm element contains a similar subterminal structure in which a 12-bp motif is repeated 9 times at the 5′ end and 15 times at the 3′ end. Some of these subterminal repeats, along with the TIR sequence, have been shown to act as cis-determinants for the excision of the En/Spm element (5). The autonomous CACTA elements En-1 and Tam1 encode two trans-factors that bind to the terminal and subterminal elements and mediate transposon excision (6). In the case of En/Spm, the TNPA protein binds to a tail-to-tail dimer of the 12-bp subterminal repeats (39, 40). The binding of TNPA protein to the subterminal repeats also has been shown to mediate the suppression of alleles carrying defective En/Spm elements (41). A hypothetical model for the formation of a transposition complex (transpososome) of the En/Spm element has been proposed (5). In this model, the binding of the TNPA protein to the subterminal repeats promotes the synapsis of the two ends of the transposon leading to the formation of a stem-loop structure. A second En-1-encoded putative transposase protein, TNPD, is then proposed to bind to the TIR sequences and cut at the ends of the element (42). The similar subterminal repeat structures of the Cs1 and En/Spm elements may indicate that the mechanism of Cs1 excision could be similar to that proposed for En/Spm.
From the sequence analyses of Cs1, we found that the deduced protein product of Cs1-predicted gene 2 shows a weak similarity (24% over a stretch of 150 aa) to the TNP1 protein of the Tam1 element of Antirrhinum. Earlier studies detected no similarity between TNP1 of Tam1 and TNPA of En/Spm. However, these two proteins were proposed to be functional analogs because both proteins have been shown to bind to the subterminal repeats of their respective elements (5). In the case of TNPA, a DNA-binding domain has been localized between residues 122 and 427 (40). A multiple sequence alignment shows that a 50-aa region is fairly conserved among TNPA, TNP1, Cs1-gene 2 protein, and another hypothetical sorghum protein (Fig. 6). Interestingly, the conserved region of TNPA (position 293–342) falls within the region containing the DNA-binding domain (40).
Figure 6.
Multiple sequence alignment of a Cs1 predicted protein. Sequence alignment of 50 aa regions of proteins encoded by gene 2 of Cs1 (position 344–393), TNP1 of Tam1 (position 191–240; accession no. S23817), SOR-BAC, a hypothetical protein from sorghum (position 215–264; accession no. AAD27567), and TNPA protein of En-1 (position 293–342; accession no. AAA66268). Conserved residues are highlighted in black, and similar residues are shown in gray.
Previous genetic studies concluded that a transposon at the y-cs locus should be autonomous or tightly linked with an autonomous element, based on the observation that control of y-cs variegation does not segregate independently of the y-cs locus (11). However, none of the ORFs of Cs1 shows similarity to the putative transposase proteins TNPD, TNP2, and PTTB of CACTA elements En-1, Tam1, and Ps1, respectively. The absence of any detectable sequence similarity with putative CACTA transposase proteins might suggest that the Cs1 element is nonautonomous, in which case mobility of Cs1 would depend on the production of transposase functions by an autonomous member of the Cs1 family. Alternatively, Cs1 may encode a highly diverged transposase protein with unrecognizable primary sequence similarity to other CACTA transposase proteins. Interestingly, Arabidopsis genome sequences contain several segments (7–9 kb) flanked by CACTACAAGAAAACA inverted repeats with significant but partial similarity to both TNPD/TNP2 and TNPA/TNP1 or PTTA (not shown). Similar to Cs1, it is unclear whether these sequences represent autonomous elements with substantially diverged but functional transposase and suppressor proteins or whether they are evolutionary traces of previously active elements.
Interpretation of the Cs1 sequence on the basis of database comparisons presents a challenge. On the one hand, sequence similarities to known proteins are, for the most part, statistically weak. On the other hand, there are stretches with similarity to proteins encoded by known CACTA elements. These similarities are revealed in database searches as the relatively best-scoring “hits.” In a typical database search, for example, with the blast programs, similarities are evaluated relative to benchmark expectations for random letter sequences to distinguish biologically significant similarities from chance events (43, 44). In view of the great expansion of sequence databases and the concomitant increased probability of picking up chance events in searches, much effort has been devoted to improving sensitivity and specificity of search algorithms in the “twilight zone” of highly diverged but homologous sequences (45, 46). Our experience with Cs1 suggests a different approach to database searches in similar contexts. Rather than testing for significant deviation from expectations based on random sequence models, one might want to test more specific biological hypotheses. For Cs1, the hypothesis would be that this sequence encodes proteins related to the known transposase and suppressor proteins of CACTA-family transposons. That among more than 398,000 nonredundant protein sequences currently in GenBank, TNP1, and related sequences score in the top 20 sequences in a blast database search then can be interpreted as strong evidence for the hypothesis.
The predicted product of Cs1 gene 4 shows strong similarity to several hypothetical proteins from Sorghum and Arabidopsis. The function of these proteins is unknown, but they appear to be different members of a gene family. It is possible that the gene 4 sequence present in Cs1 has been transduced in a manner similar to that shown recently in the case of the Tpn1 element of Japanese morning glory (47) and other transposable elements (exon shuffling model; reviewed in ref. 6).
In summary, sequence analysis by using homology searches and gene prediction algorithms has identified four putative genes within the 23,018-kb Cs1 sequence. These predictions provide a theoretical framework for the design of experimental approaches required to determine the actual transcripts and proteins encoded by the Cs1 element.
The application of plant transposons to gene tagging was first demonstrated by the cloning of the bronze gene in maize by using the Ac element (48). Since then, a growing number of plant genes have been cloned or identified by using Ac-Ds, En/Spm, or Mu. Until recently, transposon tagging in plants has been restricted to species with active and well characterized transposon systems, such as maize and Antirrhinum (49). Transposition of maize Ac/Ds and En/Spm elements in other species such as Arabidopsis, tomato, and rice has expanded the use of tagging systems in other readily transformed species (50). However, for plant species such as sorghum, which are not amenable to routine plant transformation methods, an endogenous active transposable element provides the only means for gene tagging. Our characterization of Cs1 shows that it has certain characteristics advantageous for gene tagging, including low copy number and high transposition frequency. Another interesting feature of Cs1 is that its transposition frequency appears to be sensitive to environmental conditions; it has been observed that somatic and germinal mutability is higher in field-grown than greenhouse-grown plants (ref. 11; S.C. and T.P., unpublished data). Transposition of the Antirrhinum Tam3 element is sensitive to temperature, and this provides a means to control the frequency of Tam3 transposition (51). Similarly, it may be possible to modulate the frequency of Cs1 transposition to optimize gene-tagging experiments.
Acknowledgments
We thank Drs. J. Bennetzen and C. Carvalho for providing genetic stocks, Dr. Brent Pearce for providing grass species, and Terry Olson for expert technical assistance. This work was supported by a Carver Trust Grant from Iowa State University and by U.S. Department of Agriculture–National Research Initiative Grant 9701354. This is Journal Paper No. J-18552 of the Iowa Agriculture and Home Economics Experiment Station (Ames, IA), Project No. 3297, and is supported by Hatch Act and State of Iowa funds.
Abbreviations
- TIR
terminal inverted repeat
- RFLP
restriction fraction length polymorphism
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF206660).
References
- 1.McClintock B. Carnegie Inst Wash Yearbook. 1948;47:155–169. [Google Scholar]
- 2.Kidwell M G, Lisch D. Proc Natl Acad Sci USA. 1997;94:7704–7711. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevers P, Shepherd N A, Saedler H. Adv Bot Res. 1986;12:102–203. [Google Scholar]
- 4.Bennetzen J L. In: Transposable Elements. Saedler H, Gierl A, editors. Berlin: Springer; 1996. pp. 195–229. [Google Scholar]
- 5.Gierl A. In: Transposable Elements. Saedler H, Gierl A, editors. Berlin: Springer; 1996. pp. 145–159. [Google Scholar]
- 6.Kunze R, Saedler H, Lonning W-E. Adv Bot Res. 1997;27:331–470. [Google Scholar]
- 7.Osborne B I, Baker B. Curr Opin Cell Biol. 1995;7:406–413. doi: 10.1016/0955-0674(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 8.Chen M, SanMiguel P, De Oliveira A C, Woo S-S, Zhang H, Wing R A, Bennetzen J L. Proc Natl Acad Sci USA. 1997;94:3431–3435. doi: 10.1073/pnas.94.7.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennetzen J L, Freeling M. Trends Genet. 1993;9:259–261. doi: 10.1016/0168-9525(93)90001-x. [DOI] [PubMed] [Google Scholar]
- 10.McWhirter K S. Abstr Genet. 1973;74:170. (abstr.). [Google Scholar]
- 11.Zanta C A, Yang X, Axtell J D, Bennetzen J L. J Hered. 1994;85:23–29. [Google Scholar]
- 12.Hu G, Kofoid K D, Liang G H. Hereditas. 1991;115:163–167. [Google Scholar]
- 13.Peterson P A. Genetics. 1953;38:682–683. [Google Scholar]
- 14.McClintock B. Carnegie Inst Wash Yearbook. 1954;53:254–260. [Google Scholar]
- 15.Peterson P A. Am Nat. 1965;99:391–398. [Google Scholar]
- 16.Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. EMBO J. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonas U, Sommer H, Harrison B J, Saedler H. Mol Gen Genet. 1984;194:138–143. [Google Scholar]
- 18.Vodkin L O, Rhodes P R, Goldberg R B. Cell. 1983;34:1023–1031. doi: 10.1016/0092-8674(83)90560-3. [DOI] [PubMed] [Google Scholar]
- 19.Shirsat A H. Mol Gen Genet. 1988;212:129–133. doi: 10.1007/BF00322455. [DOI] [PubMed] [Google Scholar]
- 20.Inagaki Y, Hisatomi Y, Suzuki T, Kasahara K, Iida S. Plant Cell. 1994;6:375–383. doi: 10.1105/tpc.6.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Motohashi R, Ohtsubo E, Ohtsubo H. Mol Gen Genet. 1996;250:148–152. doi: 10.1007/BF02174173. [DOI] [PubMed] [Google Scholar]
- 22.Ozeki Y, Davies E, Takeda J. Mol Gen Genet. 1997;254:407–416. doi: 10.1007/s004380050433. [DOI] [PubMed] [Google Scholar]
- 23.Snowden K C, Napoli C A. Plant J. 1998;14:43–54. doi: 10.1046/j.1365-313x.1998.00098.x. [DOI] [PubMed] [Google Scholar]
- 24.Nacken W F, Piotrowiak R, Saedler H, Sommer H. Mol Gen Genet. 1991;228:201–208. doi: 10.1007/BF00282466. [DOI] [PubMed] [Google Scholar]
- 25.Saghai-Maroof M A, Soliman K M, Jorgensen R A, Allard R W. Proc Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise R P, Schnable P S. Theor Appl Genet. 1994;88:785–795. doi: 10.1007/BF01253987. [DOI] [PubMed] [Google Scholar]
- 27.Chopra S, Athma P, Peterson T. Plant Cell. 1996;8:1149–1158. doi: 10.1105/tpc.8.7.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Lechelt C, Peterson T, Laird A, Chen J, Dellaporta S, Dennis E, Peacock W J, Starlinger P. Mol Gen Genet. 1989;219:225–234. doi: 10.1007/BF00261181. [DOI] [PubMed] [Google Scholar]
- 30.Leung M-Y, Blaisdell B E, Burge C, Karlin S. J Mol Biol. 1991;221:1367–1378. doi: 10.1016/0022-2836(91)90938-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleffe J, Hermann K, Vahrson W, Wittig B, Brendel V. Nucleic Acids Res. 1996;24:4718–4728. doi: 10.1093/nar/24.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brendel V, Kleffe J. Nucleic Acids Res. 1998;26:4748–4757. doi: 10.1093/nar/26.20.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burge C, Karlin S. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 34.Kleffe J, Hermann K, Vahrson W, Wittig B, Brendel V. Bioinformatics. 1998;14:232–243. doi: 10.1093/bioinformatics/14.3.232. [DOI] [PubMed] [Google Scholar]
- 35.Usuka, J., Zhu, W. & Brendel, V. (1999) Bioinformatics, in press. [DOI] [PubMed]
- 36.Wessler S, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 37.Grotewold E, Athma P, Peterson T. Proc Natl Acad Sci USA. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hulbert S H, Richter T E, Axtell J D, Bennetzen J L. Proc Natl Acad Sci USA. 1990;87:4251–4255. doi: 10.1073/pnas.87.11.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gierl A, Lutticke S, Saedler H. EMBO J. 1988;7:4045–4053. doi: 10.1002/j.1460-2075.1988.tb03298.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trentmann S M, Saedler H, Gierl A. Mol Gen Genet. 1993;238:201–208. doi: 10.1007/BF00279548. [DOI] [PubMed] [Google Scholar]
- 41.Grant S R, Gierl A, Saedler H. EMBO J. 1990;9:2029–2035. doi: 10.1002/j.1460-2075.1990.tb07369.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frey M, Reinecke J, Grant S, Saedler H, Gierl A. EMBO J. 1990;9:4037–4044. doi: 10.1002/j.1460-2075.1990.tb07625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doolittle R F. Science. 1981;214:149–159. doi: 10.1126/science.7280687. [DOI] [PubMed] [Google Scholar]
- 44.Karlin S, Brendel V. Science. 1993;257:39–49. doi: 10.1126/science.1621093. [DOI] [PubMed] [Google Scholar]
- 45.Altschul S F, Boguski M S, Gish W, Wootton J C. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Madden T L. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi S, Inagaki Y, Satoh H, Hoshino A, Iida S. Mol Gen Genet. 1999;261:447–451. doi: 10.1007/s004380050987. [DOI] [PubMed] [Google Scholar]
- 48.Fedoroff N V, Furtek D B, Nelson O E. Proc Natl Acad Sci USA. 1984;81:3825–3829. doi: 10.1073/pnas.81.12.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walbot V. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:49–82. [Google Scholar]
- 50.Jones J D G, Carland F M, Maliga P, Dooner H K. Science. 1989;244:204–207. doi: 10.1126/science.244.4901.204. [DOI] [PubMed] [Google Scholar]
- 51.Harrison B J, Fincham J R S. Heredity. 1964;19:237–258. [Google Scholar]