Abstract
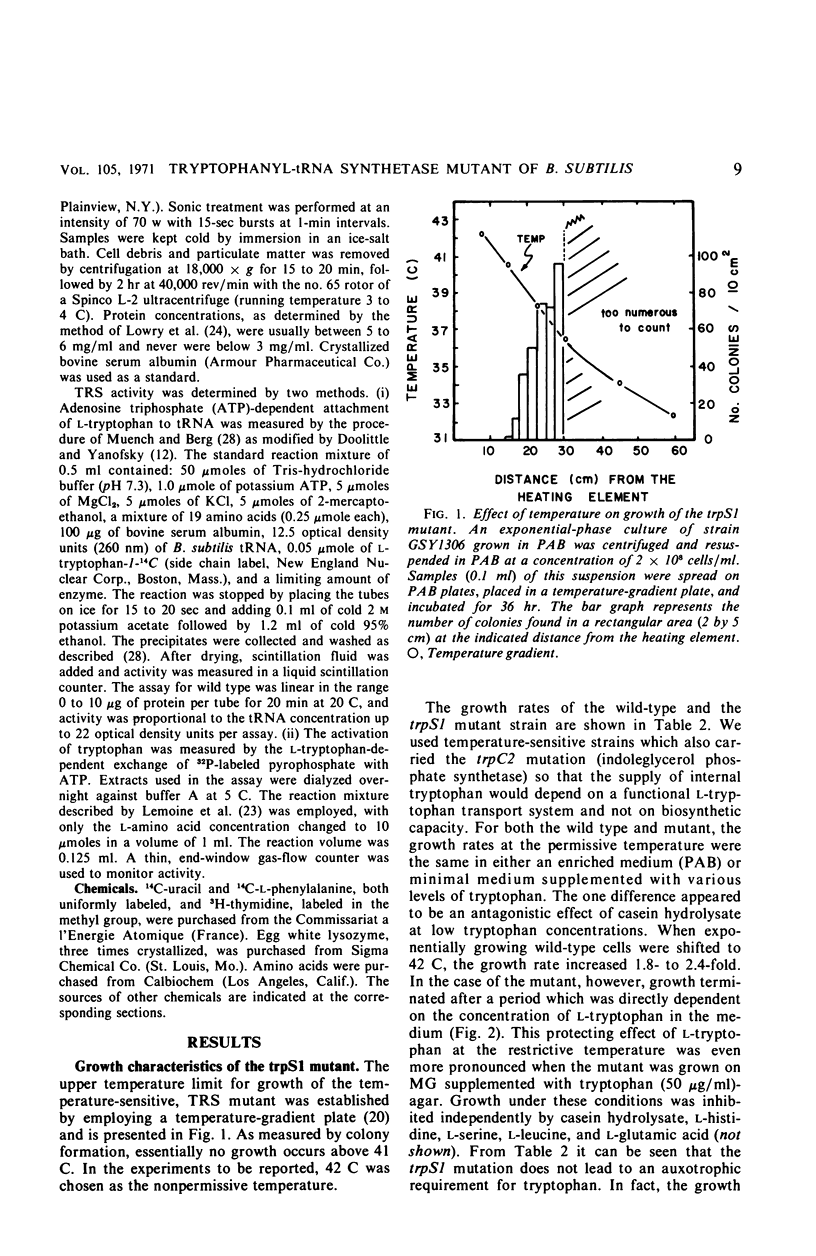
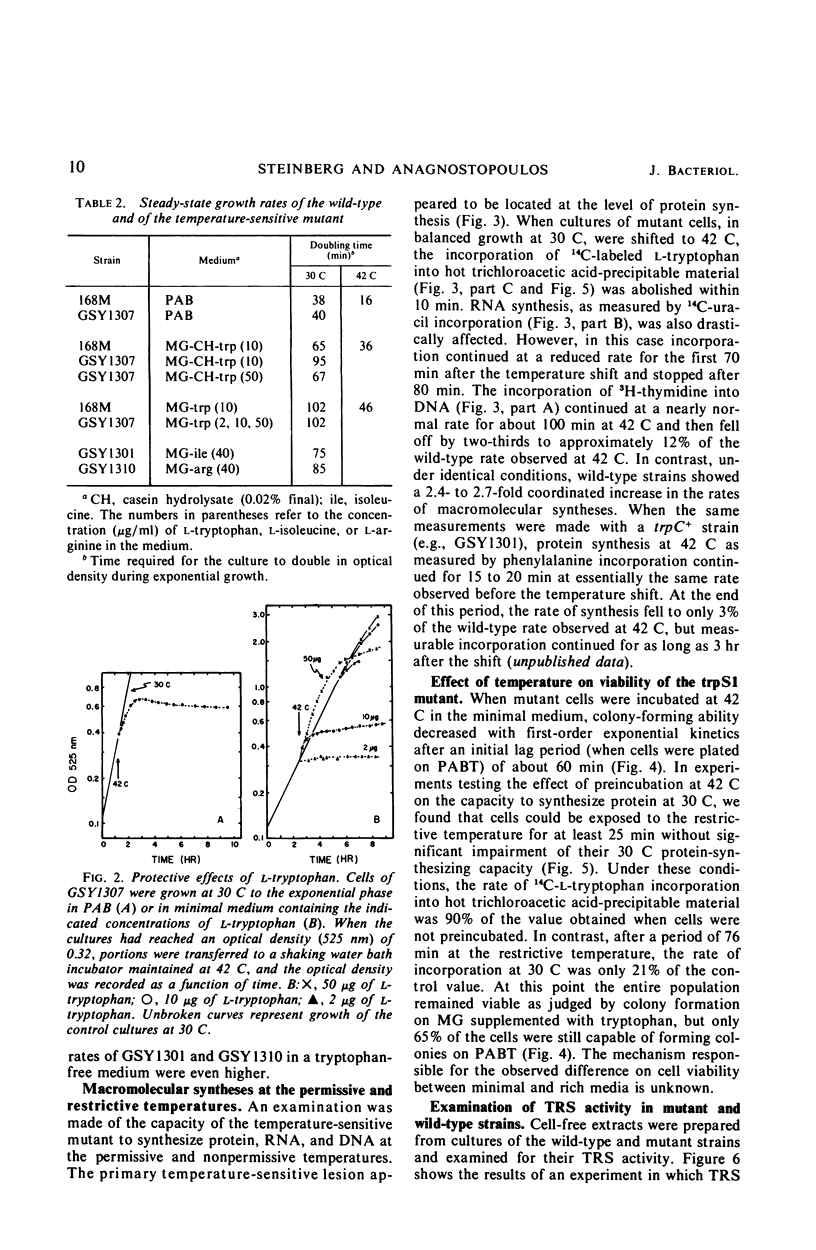
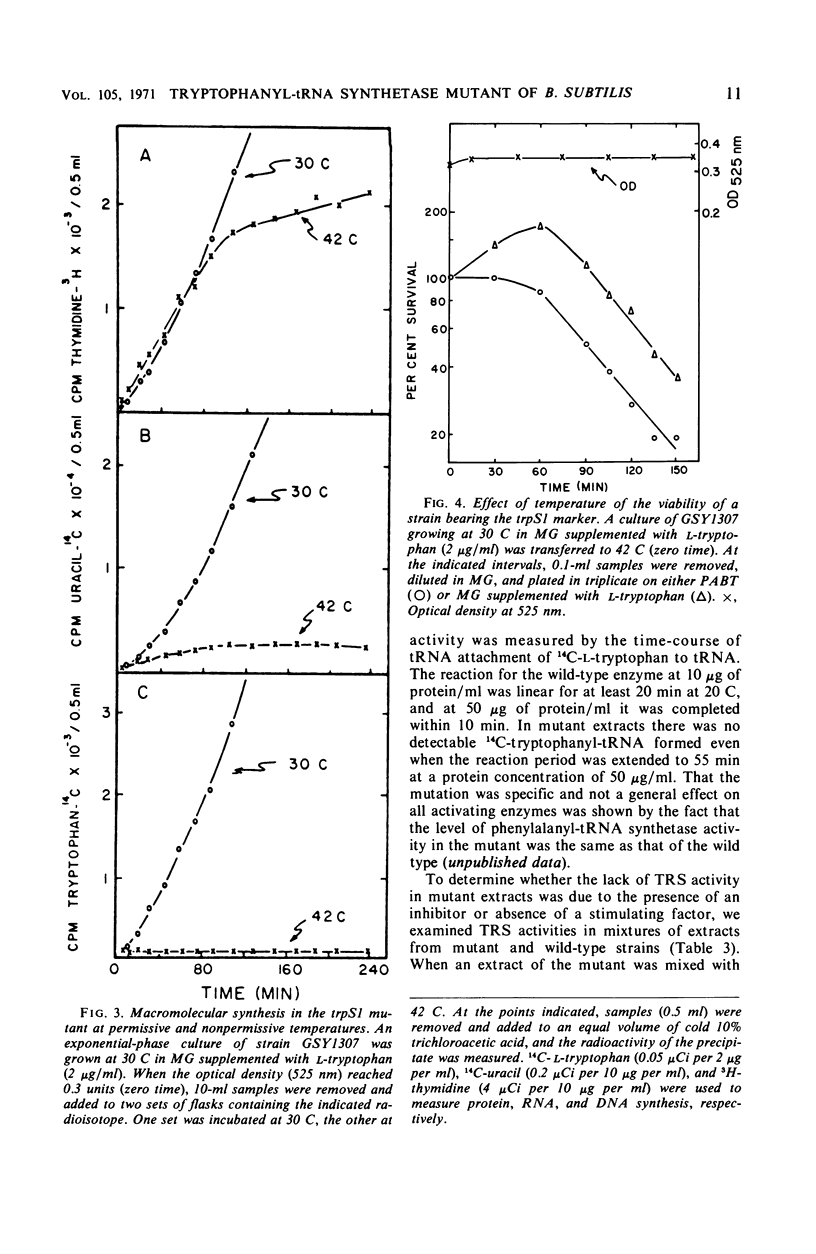
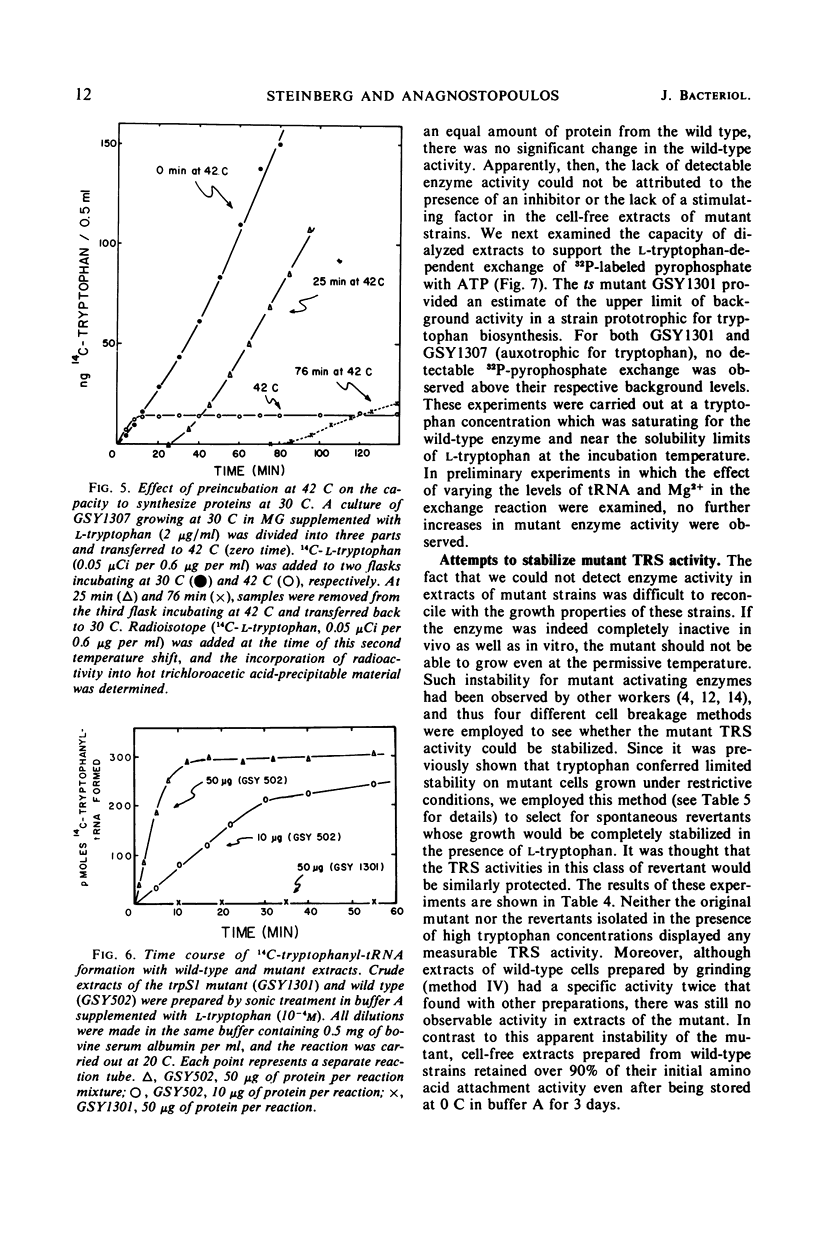
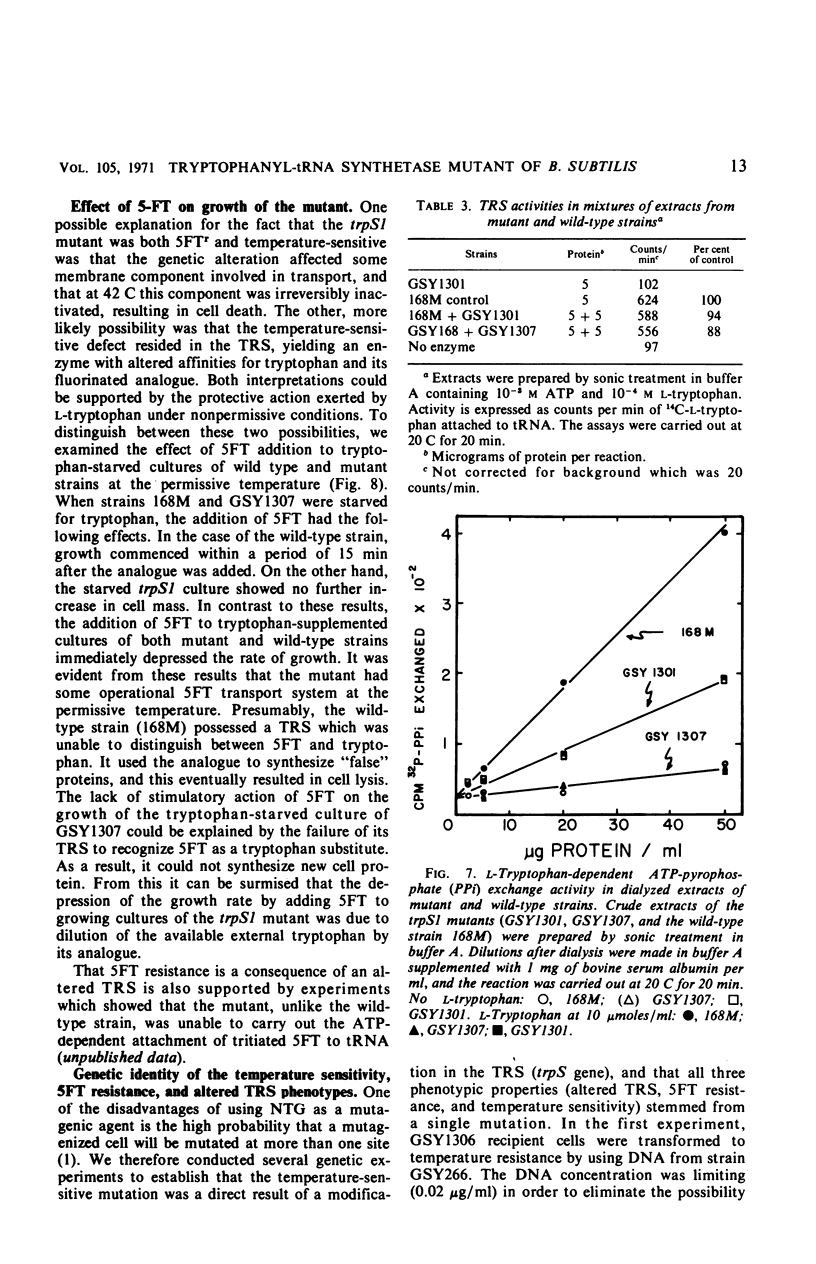
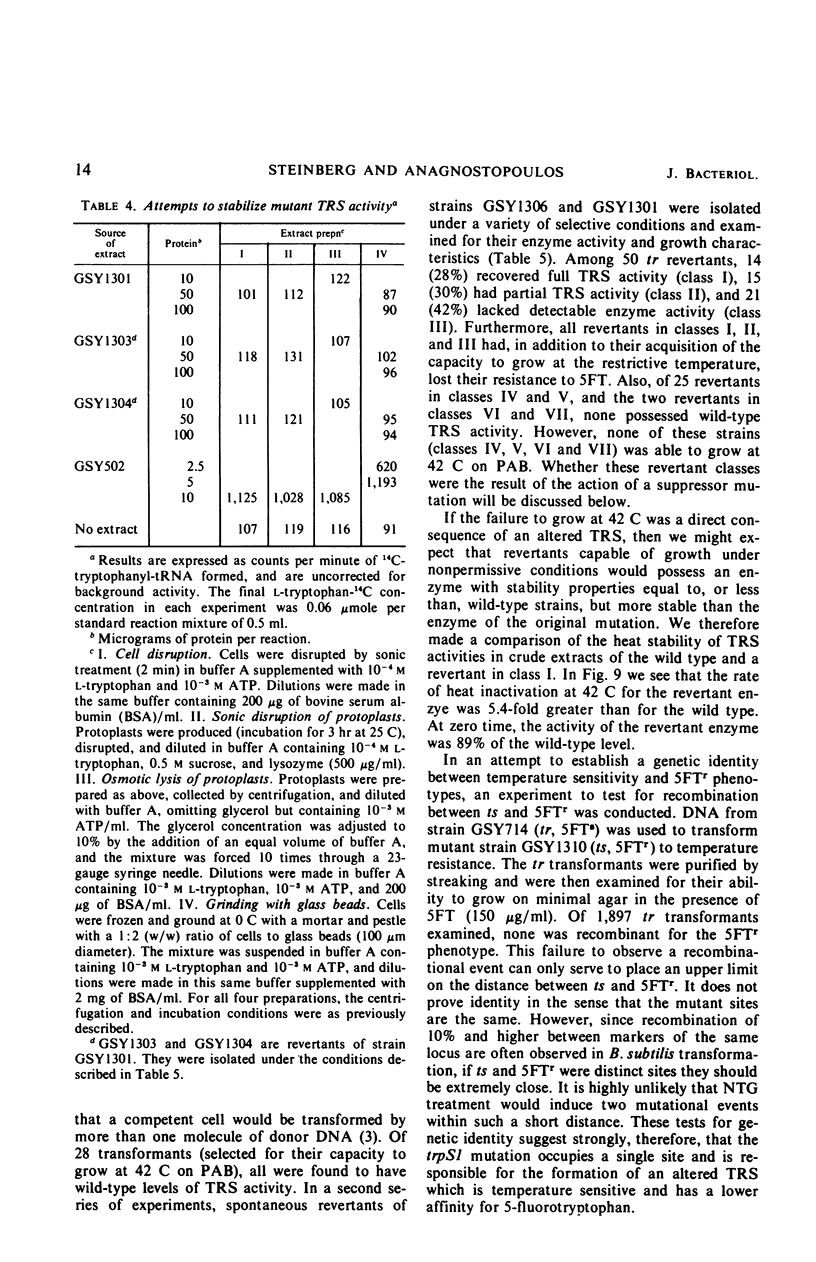
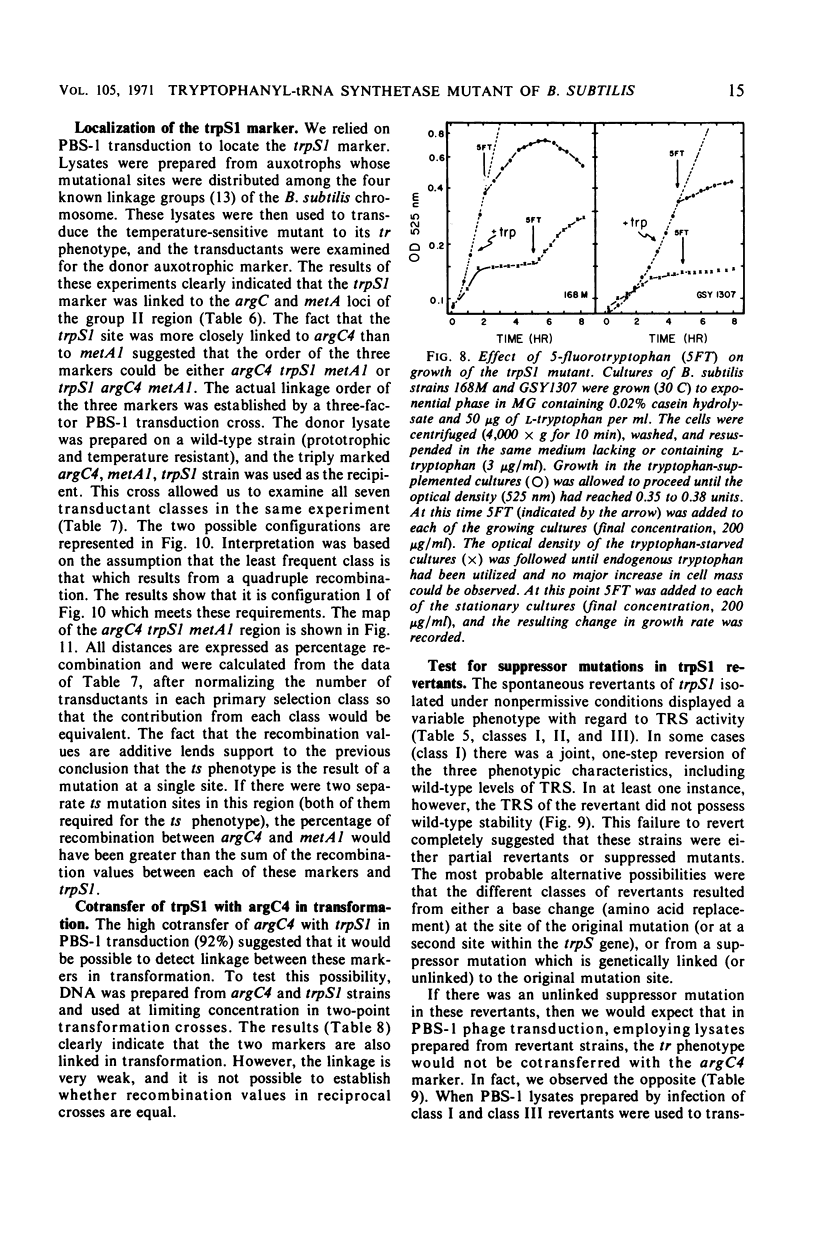
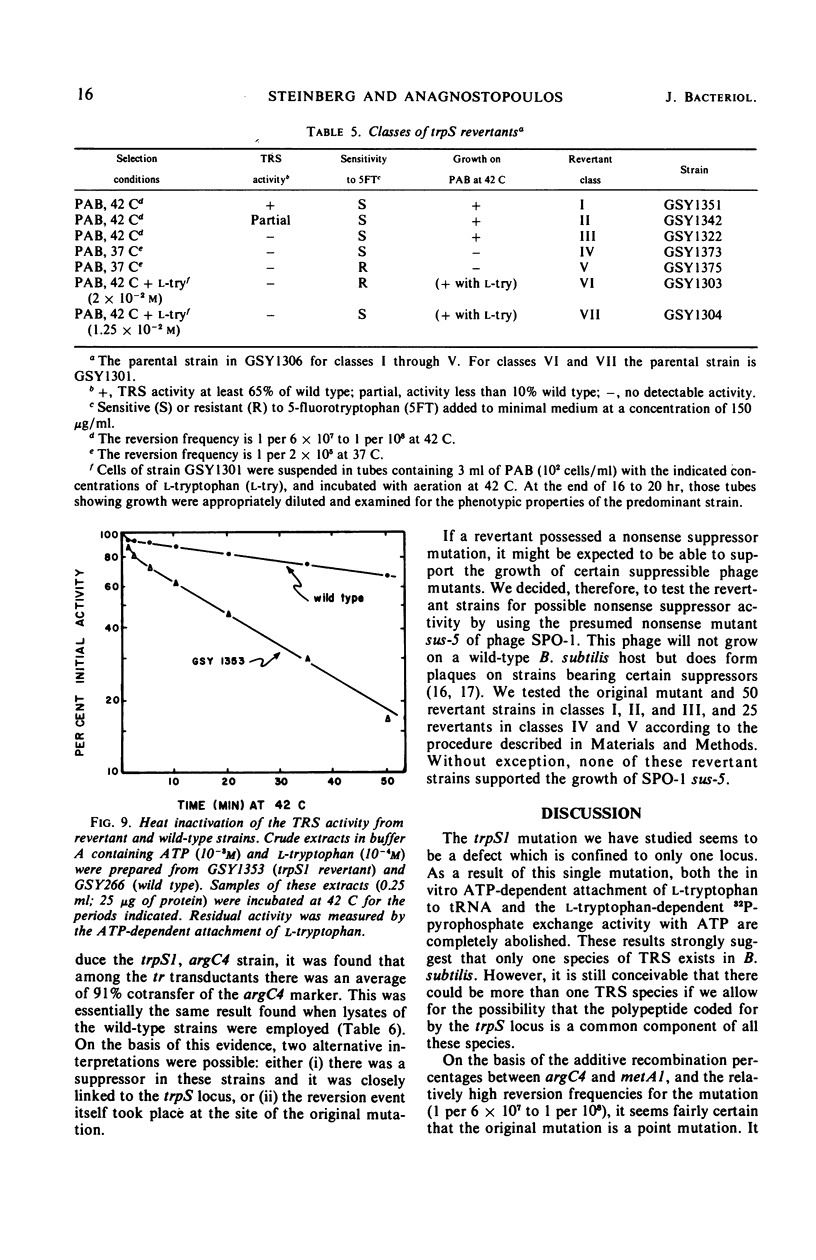
A temperature-sensitive, 5-fluorotryptophan (5FT)-resistant mutant of Bacillus subtilis was isolated which forms an altered tryptophanyl transfer ribonucleic acid synthetase [l-tryptophan: sRNA ligase (AMP), EC 6.1.1.2]. The mutant grows well at 30 C but not at 42 C. At the latter temperature, protein and ribonucleic acid (RNA) synthesis are abolished while deoxyribonucleic acid (DNA) synthesis proceeds for a considerable time. Tryptophanyl-transfer RNA (tRNA) synthetase activity is not detectable in the extracts of the mutant grown at 30 C whether this activity is measured by the attachment of l-tryptophan to tRNA or the l-tryptophan-dependent exchange of 32P-pyrophosphate with adenosine triphosphate. Mixing experiments with extracts from the wild type and the mutant have ruled out the presence of an inhibitor or the absence of an activator as possible causes. Attempts to retrieve enzyme activity in vitro by various means (different conditions for cell disruption, addition of l-tryptophan, and adenosine triphosphate to the extraction buffer containing glycerol) were unsuccessful. The mutation in the locus of the tryptophanyl tRNA synthetase (trpS) was mapped on the bacterial chromosome by transformation and transduction. It is located between argC and metA. All temperature-resistant transformants recover wild-type levels of tryptophanyl tRNA synthetase activity and sensitivity to 5FT. Spontaneous revertants to temperature resistance are 5FT sensitive, but their levels of tryptophanyl tRNA synthetase activity and the thermolability of this enzyme in cell-free extracts varies. These revertants do not support the growth of a presumed nonsense mutant of phase SPO-1. Transduction experiments with phage PBS-1 indicated that reversion must be the result of an event at the site of the original mutation or at a site extremely close to it.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Crawford I. P. Le groupe des gènes régissant la biosynthèse du tryptophane chez Bacillus subtilis. C R Acad Sci Hebd Seances Acad Sci D. 1967 Jul 3;265(1):93–96. [PubMed] [Google Scholar]
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BURTON K. A study of the conditions and mechanism of the diphenylamine reaction for the colorimetric estimation of deoxyribonucleic acid. Biochem J. 1956 Feb;62(2):315–323. doi: 10.1042/bj0620315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott K. F., Wilson G. A. Development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1967 Sep;94(3):562–570. doi: 10.1128/jb.94.3.562-570.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A., Faiman L. E., Neidhardt F. C. Biochemical and genetic characterization of a mutant of Escherichia coli with a temperature-sensitive valyl ribonucleic acid synthetase. J Bacteriol. 1966 Oct;92(4):1076–1082. doi: 10.1128/jb.92.4.1076-1082.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck A., Neidhardt F. C. Genetic mapping of phenylalanyl-sRNA synthetase in Escherichia coli. Science. 1967 Jul 7;157(3784):78–79. doi: 10.1126/science.157.3784.78. [DOI] [PubMed] [Google Scholar]
- Böck A. Relation between subunit structure and temperature-sensitivity of mutant phenylalanyl RNA synthetases of Escherichia coli. Eur J Biochem. 1968 Apr;4(3):395–400. doi: 10.1111/j.1432-1033.1968.tb00225.x. [DOI] [PubMed] [Google Scholar]
- Chrispeels M. J., Boyd R. F., Williams L. S., Neidhardt F. C. Modification of valyl tRNA synthetase by bacteriophage in Escherichia coli. J Mol Biol. 1968 Feb 14;31(3):463–475. doi: 10.1016/0022-2836(68)90421-x. [DOI] [PubMed] [Google Scholar]
- Demerec M., Adelberg E. A., Clark A. J., Hartman P. E. A proposal for a uniform nomenclature in bacterial genetics. Genetics. 1966 Jul;54(1):61–76. doi: 10.1093/genetics/54.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F., Yanofsky C. Mutants of Escherichia coli with an altered tryptophanyl-transfer ribonucleic acid synthetase. J Bacteriol. 1968 Apr;95(4):1283–1294. doi: 10.1128/jb.95.4.1283-1294.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Goldthwaite C., Smith I., Marmur J. Genetic mapping in Bacillus subtilis. J Mol Biol. 1967 Jul 14;27(1):163–185. doi: 10.1016/0022-2836(67)90358-0. [DOI] [PubMed] [Google Scholar]
- EIDLIC L., NEIDHARDT F. C. PROTEIN AND NUCLEIC ACID SYNTHESIS IN TWO MUTANTS OF ESCHERICHIA COLI WITH TEMPERATURE-SENSITIVE AMINOACYL RIBONUCLEIC ACID SYNTHETASES. J Bacteriol. 1965 Mar;89:706–711. doi: 10.1128/jb.89.3.706-711.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FANGMAN W. L., NEIDHARDT F. C. DEMONSTRATION OF AN ALTERED AMINOACYL RIBONUCLEIC ACID SYNTHETASE IN A MUTANT OF ESCHERICHIA COLI. J Biol Chem. 1964 Jun;239:1839–1843. [PubMed] [Google Scholar]
- Georgopoulos C. P. Suppressor system in Bacillus subtilis 168. J Bacteriol. 1969 Mar;97(3):1397–1402. doi: 10.1128/jb.97.3.1397-1402.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O., Anagnostopoulos C., Crawford I. P. Enzymes of the tryptophan operon of Bacillus subtilis. Biochem Biophys Res Commun. 1969 Jun 27;35(6):838–844. doi: 10.1016/0006-291x(69)90700-1. [DOI] [PubMed] [Google Scholar]
- Kaneko I., Doi R. H. Alteration of valyl-sRNA during sporulation of bacillus subtilis. Proc Natl Acad Sci U S A. 1966 Mar;55(3):564–571. doi: 10.1073/pnas.55.3.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LANDMAN O. E., BAUSUM H. T., MATNEY T. S. Temperaturegradient plates for growth of microorganisms. J Bacteriol. 1962 Mar;83:463–469. doi: 10.1128/jb.83.3.463-469.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lazar M., Yaniv M., Gros F. Sur les propriétés d'une alanyl-t-RNA synthétase modifiée dans une souche d'Escherichia coli à croissance thermosensible. C R Acad Sci Hebd Seances Acad Sci D. 1968 Jan 29;266(5):531–534. [PubMed] [Google Scholar]
- Lee M. L., Muench K. H. Prolyl transfer ribonucleic acid synthetase of Escherichia coli. I. Purification and evidence for subunits. J Biol Chem. 1969 Jan 25;244(2):223–230. [PubMed] [Google Scholar]
- Lemoine F., Waller J. P., van Rapenbusch R. Studies on methionyl transfer RNA synthetase. 1. Purification and some properties of methionyl transfer RNA synthetase from Escherichia coli K-12. Eur J Biochem. 1968 Apr 3;4(2):213–221. doi: 10.1111/j.1432-1033.1968.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Malcolm N. L. Subunit structure and function of Micrococcus cryophilus glutamyl transfer RNA synthetase. Biochim Biophys Acta. 1969 Oct 22;190(2):347–357. doi: 10.1016/0005-2787(69)90085-9. [DOI] [PubMed] [Google Scholar]
- McLaughlin C. S., Magee P. T., Hartwell L. H. Role of isoleucyl-transfer ribonucleic acid synthetase in ribonucleic acid synthesis and enzyme repression in yeast. J Bacteriol. 1969 Nov;100(2):579–584. doi: 10.1128/jb.100.2.579-584.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F. C. Roles of amino acid activating enzymes in cellular physiology. Bacteriol Rev. 1966 Dec;30(4):701–719. doi: 10.1128/br.30.4.701-719.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo S., Yanagida T. Isolation of a suppressor mutant in Bacillus subtilis. J Bacteriol. 1968 Mar;95(3):1187–1188. doi: 10.1128/jb.95.3.1187-1188.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Ames B. N. Histidine regulatory mutants in Salmonella typhimurium II. Histidine regulatory mutants having altered histidyl-tRNA synthetase. J Mol Biol. 1966 Dec 28;22(2):325–333. doi: 10.1016/0022-2836(66)90135-5. [DOI] [PubMed] [Google Scholar]
- SARIN P. S., ZAMECNIK P. C. ON THE STABILITY OF AMINOACYL-S-RNA TO NUCLEOPHILIC CATALYSIS. Biochim Biophys Acta. 1964 Dec 16;91:653–655. doi: 10.1016/0926-6550(64)90018-0. [DOI] [PubMed] [Google Scholar]
- Smith I., Dubnau D., Morrell P., Marmur J. Chromosomal location of DNA base sequences complementary to transfer RNA and to 5 s, 16 s and 23 s ribosomal RNA in Bacillus subtilis. J Mol Biol. 1968 Apr 14;33(1):123–140. doi: 10.1016/0022-2836(68)90285-4. [DOI] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehler B. L., Hendley D. D., Hirsch G. P. Evidence of a codon restriction hypothesis of cellular differentiation: multiplicity of mammalian leucyl-sRNA-specific synthetases and tissue-specific deficiency in an alanyl-sRNA synthetase. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1751–1758. doi: 10.1073/pnas.57.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M., Gros F. Studies on valyl-tRNA synthetase and tRNA from Escherichia coli. 3. Valyl-tRNA synthetases from thermosensitive mutants of Escherichia coli. J Mol Biol. 1969 Aug 28;44(1):31–45. doi: 10.1016/0022-2836(69)90403-3. [DOI] [PubMed] [Google Scholar]
- Young F. E., Smith C., Reilly B. E. Chromosomal location of genes regulating resistance to bacteriophage in Bacillus subtilis. J Bacteriol. 1969 Jun;98(3):1087–1097. doi: 10.1128/jb.98.3.1087-1097.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]



