Abstract
Oil-in-water emulsions are potent human adjuvants used for effective pandemic influenza vaccines; however, their mechanism of action is still unknown. By combining microarray and immunofluorescence analysis, we monitored the effects of the adjuvants MF59 oil-in-water emulsion, CpG, and alum in the mouse muscle. MF59 induced a time-dependent change in the expression of 891 genes, whereas CpG and alum regulated 387 and 312 genes, respectively. All adjuvants modulated a common set of 168 genes and promoted antigen-presenting cell recruitment. MF59 was the stronger inducer of cytokines, cytokine receptors, adhesion molecules involved in leukocyte migration, and antigen-presentation genes. In addition, MF59 triggered a more rapid influx of CD11b+ blood cells compared with other adjuvants. The early biomarkers selected by microarray, JunB and Ptx3, were used to identify skeletal muscle as a direct target of MF59. We propose that oil-in-water emulsions are the most efficient human vaccine adjuvants, because they induce an early and strong immunocompetent environment at the injection site by targeting muscle cells.
Keywords: innate immunity, microarray, MF59, alum, CpG, oligonucleotide
Vaccine adjuvants are represented by different classes of compounds, such as microbial products, mineral salts, emulsions, microparticles, and liposomes, which exert their function by diverse and often poorly characterized mechanisms of action (1–3). Based on recent findings, a classification in two major functional groups, Toll-like receptor (TLR)-dependent and -independent, can be made (4, 5). TLR-dependent adjuvants act directly on dendritic cells (DCs), inducing the up-regulation of cytokines, MHC class II, and costimulatory molecules, and promoting DC migration to the T cell area of the lymph node (1, 6). One example of TLR-dependent adjuvant is represented by nonmethylated CpG oligonucleotide (CpG), used as vaccine adjuvant in both preclinical and clinical studies (7, 8). CpG acts through TLR9, expressed by human plasmacytoid DCs and B cells (9).
Among TLR-independent adjuvants, alum has been widely used in human vaccines for >70 years, whereas the squalene-based oil-in-water emulsion MF59 was licensed for human use a decade ago. The molecular mechanism of action and the target cells of alum and MF59 are still unknown. It has been proposed that adsorption to alum increases antigen availability at injection site allowing an efficient uptake by antigen-presenting cells (APCs) (10). Alum could also increase antigen uptake by DCs in vitro, further supporting an antigen delivery function (11). However, several studies suggested that, in addition to antigen delivery, alum might have immunostimulating activities in vivo. Alum i.m. administration resulted in cell recruitment events at the injection site (12, 13). More recently, it has been demonstrated that i.p. injection of alum induced the recruitment of monocytes, which could uptake the vaccine antigen, migrate to the draining lymph nodes, and differentiate into fully competent inflammatory DCs (14).
Similarly to alum, MF59 could promote antigen uptake by dendritic cells in vivo (15). Moreover, it has been shown that, after i.m. injection, MF59 is internalized by APCs that migrate to the lymph node (16). Besides promoting antigen delivery, MF59 might also act as a local pro-inflammatory adjuvant. Indeed, it was observed that MF59: i.m. injection induces the influx of blood mononuclear cells (16).
Several mouse studies reported that MF59 enhances immunogenicity of soluble antigens better than alum and CpG (17–20). Furthermore, recent clinical data have demonstrated that pandemic flu vaccines formulated with oil-in-water emulsions induce superior seroconversion and cross-neutralization compared with nonadjuvanted vaccines or to vaccines formulated with alum (21–23). The adjuvanticity of alum and MF59 is modulated by the addition of CpG (17, 19). In particular, the addition of CpG to MF59 or alum induces a dramatic shift from a Th2 to a Th1 response in BALB/c mice (17, 20).
To better understand the molecular mechanism of action of oil-in-water emulsions and their relative potency when compared with other adjuvants, we initially performed microarray analysis of human peripheral blood mononuclear cells stimulated in vitro. In this assay, MF59 displayed very modest effects on transcription profiles (data not shown). We therefore performed microarray analysis of the whole muscle injected with MF59, alum, CpG, and with a combination of MF59 and CpG. Genes selected by microarray data analysis were used in immunofluorescence experiments to identify MF59 target cells and to monitor cell recruitment events triggered by vaccine adjuvants at the injection site. Finally, the systemic effects of all adjuvants tested were investigated by measuring cytokine concentration in the serum. Here, we show that, whereas all adjuvants tested induced a common set of 168 genes, MF59 induced approximately three times more genes than alum and CpG. MF59 was the strongest inducer of cytokines, cytokine receptors, and genes involved in leukocyte migration and antigen presentation. Immunofluorescence analysis showed that MF59 promoted a more rapid recruitment of CD11b-positive cells. Furthermore, early biomarker expression suggests that muscle fibers are the primary target of MF59.
Results
Differential Modulation of Gene Expression at Injection Site by Human Vaccine Adjuvants.
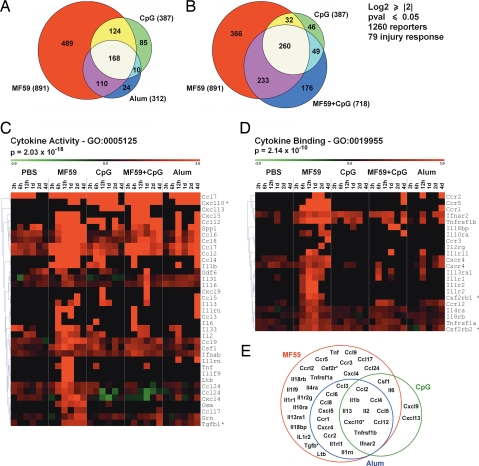
To analyze the local effects on gene expression induced by MF59, alum, CpG, and a combination of MF59 and CpG, mice quadriceps were injected with 50 μl of each adjuvant diluted in PBS and processed at 3, 6, 12, 24, 48, and 96 h for whole-mouse genome microarray analysis, as described in Materials and Methods. The same volume of PBS was used as control. A total of 1,260 genes have been selected with an average log2 ratio ≥ 2 compared with untreated quadriceps and a P value ≤ 0.05 calculated on the three replicates of at least one time point [supporting information (SI) Table S1]. Among these genes, 79 were modulated by all adjuvants and by PBS. The injury produced by the needle and the injection of a relatively large volume of liquid into the muscle might be responsible for regulating this group of transcripts, which included Ccl7, Timp1, Socs3, Mt1, and Mt2. All other genes selected with the threshold criteria described previously were regulated by at least one adjuvant but not by the injection of PBS and therefore were considered adjuvant-responsive genes. MF59 regulated a larger number of genes (891) compared with other adjuvants, and among these, 489 were MF59-selective. Alum regulated 312 genes, and only 24 were alum-selective. CpG modulated 387 genes, of which 85 were selective (Fig. 1A). Interestingly, 168 genes were responsive to all adjuvants; therefore, they were defined as “adjuvant core response genes.” Functional analysis of this group of genes identified three categories significantly enriched: cytokine-cytokine receptor interaction [Kyoto Encyclopedia of Genes and Genomes (KEGG) database; P value = 0.00127], host–pathogen interaction [Gene Ontology (GO): 0030383, P value = 1.07 × 10−18], and defense immunity protein activity (GO: 0003793, P value = 9.58 × 10−4). We also identified 19 genes related to type I IFN response (Table S2).
Fig. 1.
Microarray analysis of transcription profiles induced by vaccine adjuvants in mouse muscle. Genes (1,260) have been selected with an average log2 ratio ≥2 and a P value ≤0.05 in at least one time point. Seventy-nine genes were modulated log2 ratio ≥2 by PBS and were considered injury-response genes, as indicated. (A and B) Venn diagram showing the distribution of genes modulated by MF59, CpG, and alum (A) or by MF59, CpG, and MF59+CpG coadministered (B). In parentheses, the total number of genes modulated by each treatment is indicated. The area of each sector is proportional to the number of genes. (C and D) Cluster analysis of the expression profiles of genes encoding proteins with cytokine activity (C) and cytokine-binding activity (D) after treatment with PBS, MF59, CpG, MF59+CpG, and alum for the indicated times. Database accession number and statistical significance (P value) of each category of genes are indicated. Asterisks indicate genes not identified by the GO database and manually added to the cluster. Each column represents one time point. Each row represents the average kinetic of expression of one gene. Some genes, such as Ccl24, appear more than once in the cluster, because they are represented by multiple unrelated probes in the Agilent 44k Whole-Mouse Genome Array. The expression values are shown as log2 ratio. Color scale ranges from −3 (green, down-regulation) to 3 (red, up-regulation). (E) Venn diagram showing the responsiveness of cytokines and cytokine receptor genes to MF59, CpG, and alum.
As expected, the majority (542) of the genes regulated by coadministered MF59 and CpG were also modulated by MF59 or CpG alone (Fig. 1B). However, some genes (176), including IFN type I Ifnab, Stat6, and Il16, were regulated only when MF59 and CpG were coadministered (Fig. 1B and Table S1). Other IFN pathway genes responsive to both MF59 and CpG, such as Irf1, Irf7, Irf8, Stat1, and Stat2, were further up-regulated in the combination treatment (Fig. S1). We found that CpG regulated the expression profile of a large number of MF59-responsive genes. Indeed, 366 genes modulated by MF59 were not regulated by the combination treatment (Fig. 1B). In particular, CpG inhibited the activation of many inflammatory genes, including Tnf, Il1b, Ltb, Ccr1, Ccr3, and Il1r2 (Fig. 1 C and D).
MF59 Activates Multiple Inflammatory and Host Defense Pathways at Injection Site.
All 1,260 genes selected by microarray have been subjected to functional analysis using the GO, KEGG, and GenMapp databases. The genes belonging to the most significantly enriched categories have been clustered based on the expression profile. All adjuvants regulated the local expression of cytokines and cytokine receptors (Fig. 1 C–E). A group of cytokines (Ccl2, Ccl4, Ccl5, Ccl12, Cxcl10, Il1b, and Il2) was up-regulated at early time points by MF59 and CpG and later also by alum (Fig. 1 C and E). Several other cytokines, such as Tnf, Ccl17, Ccl24, Ltb, and Tgfb1, were specific for MF59; Cxcl9 and Cxcl13 were specific for CpG, whereas we failed to detect cytokines specific for alum (Fig. 1 C and E). MF59 was a more potent inducer of chemokine receptors compared with CpG and alum, triggering the sequential up-regulation of Ccr1 and Cxcr4 (6 h), Ccr5 (12 h), Ccr2 (1 d), and Ccr3 (4 d) (Fig. 1D). In addition, the receptors for Il1, Il2, Il4, and Il10 were induced selectively by MF59. Several transcription factors known to regulate cytokine expression, like Irf1, Irf7, Stat1, and Stat2, were modulated by all adjuvants (Fig. S2). By functional analysis, we identified other significantly enriched gene clusters preferentially activated by MF59, like genes involved in complement activation, prostaglandin synthesis, and Il1 signaling, and genes encoding matrix metalloproteinases (Fig. S2).
MF59 Induces the Recruitment of MHC Class II+ and CD11b+ Cells at Injection Site.
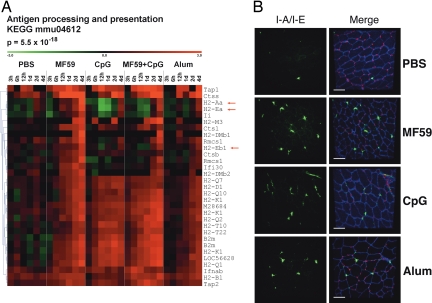
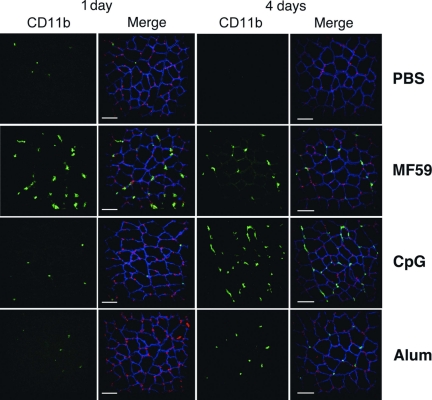
The up-regulation of proinflammatory genes at the injection site suggests that vaccine adjuvants could also drive cell recruitment from the bloodstream into the muscle. This hypothesis was further supported by the significant enrichment of genes involved in leukocyte transendothelial migration (Fig. S2). Another group of genes selected by the functional analysis of microarray data are involved in antigen processing and presentation (Fig. 2A). Within this cluster, MHC class I genes (H2-Q, H2-K, H2-T, H2-D) were up-regulated by all adjuvants, although with different kinetics: MF59 and CpG induced an up-regulation already at 6–12 h after treatment, whereas alum was induced at 1–2 days. MHC class II genes, including H2-Aa, H2-Ea, and H2-Eb1, were up-regulated by all adjuvants at 4 days. MF59 was a more potent inducer of MHC class II transcripts compared with CpG or alum. Interestingly, at earlier time points, CpG down-regulated MHC class II genes both when administered alone and in combination with MF59. Other genes involved in antigen processing and presentation, like cathepsins and B2m, were also up-regulated. The up-regulation of MHC class II genes might result either from activation of resident APCs or from APC recruitment driven by the local expression of chemoattractants and adhesion molecules. To monitor APC recruitment events after adjuvant injection, we performed immunofluorescence analysis of muscle cryosections after i.m. administration of PBS, MF59, CpG, and alum using an anti-MHC class II I-A/I-E antibody. The structure of the muscle was visualized by using an antibody specific for Utrophin, a cytoskeletal protein playing a role in anchoring the cytoskeleton to the plasma membrane and located in the sarcolemma of muscle cells. Very few MHC class II+ cells were observed in the muscle 12 and 24 h after treatment (data not shown). However, at 4 days after injection, all vaccine adjuvants increased the local concentration of MHC class II+ cells in muscle tissue compared with the PBS control (Fig. 2B). Interestingly, the kinetic of MHC class II+ cell recruitment was consistent with the MHC class II gene expression data (Fig. 2A). In contrast with MHC class II genes, another blood cell marker, Itgam/CD11b, was up-regulated at high levels by MF59 already at 12 h (Fig. S2). We monitored the recruitment of CD11b+ cells by immunofluorescence analysis and found that at 1 day after injection, only MF59 induced influx of CD11b+ cells into the muscle (Fig. 3 Left). This finding is consistent with previous data obtained from muscle single-cell suspension, which showed that at 1 day after injection, MF59 induced a influx of mononuclear cells (16). All adjuvants induced the recruitment of CD11b+ cells with similar efficiency 4 days after injection (Fig. 3 Right).
Fig. 2.
MHC class II+ cell recruitment at the injection site by MF59, CpG, and alum. (A) Cluster analysis of genes encoding antigen processing and presentation proteins. The arrows indicate the MHC class II genes. (B) Confocal analysis of muscles collected 4 days after treatment and stained with anti-MHC class II (anti-IA/IE) (green) and anti-Utrophin (anti-UTRN) (blue) antibodies and with the nuclear tracker PI (red). (Left) IA/IE stain; (Right) merge images. (Scale bar, 60 μm.)
Fig. 3.
Analysis of CD11b+ cell recruitment at injection site by MF59, CpG, and alum. Confocal microscopy analysis of muscles collected 1 or 4 days after treatment with PBS, MF59, CpG, or alum and stained with anti-CD11b (green), anti-UTRN (blue), and the nuclear tracker ToPro3 (red). (Left) CD11b+ stain. (Right) Merge images. (Scale bar, 60 μm.)
MF59 Activates the Expression of the Early Biomarkers Ptx3 and JunB in Muscle Fibers.
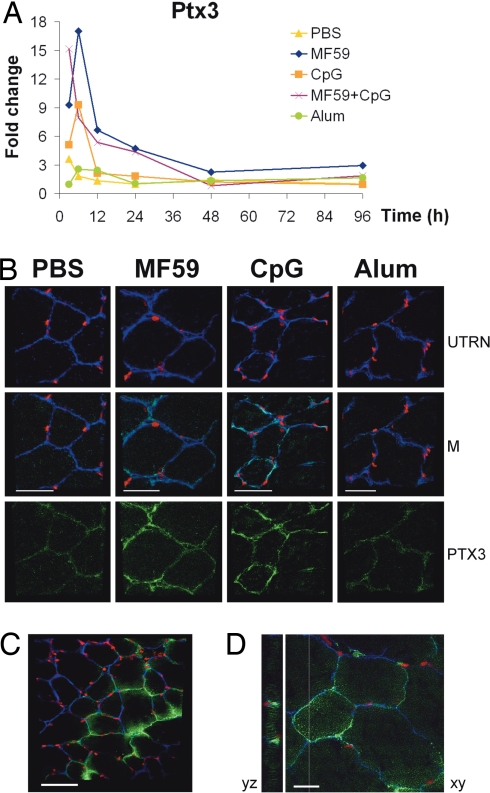
The data described above demonstrate that MF59 acts as a strong immune potentiator at injection site; however, the target cell of MF59 immunostimulating activity is not known. In the attempt to identify MF59 target cell, Pentraxin3 (Ptx3) and JunB, induced by MF59 and CpG 3 h after treatment, were selected as biomarkers for immunofluorescence analysis on muscle cryosections (Fig. 4A and Fig. S3). The long Pentraxin 3 (Ptx3) is a soluble pattern recognition receptor that recognizes pathogens such as Aspergillus fumigatus, facilitating the interaction with mononuclear phagocytes and DCs (24). Consistent with microarray data, immunofluorescence analysis showed an increased expression of PTX3 in muscle fibers at 12 h in both MF59- and CpG-treated mice, whereas there was no significant difference between alum and control (Fig. 4B). Similar results were obtained by using an antibody against JUNB, which detected an up-regulation of the protein in the nuclei of skeletal muscle in response to MF59 and CpG (Fig. S3). The induction of early response proteins JUNB and PTX3 suggests that MF59 activated directly muscle fibers. To study the interaction of MF59 with muscle cells, we injected a 3,3′-dioactadecyloxacarbocyanine (DIO)-labeled form of MF59. At 3 h, MF59 localized inside muscle fibers, further supporting the hypothesis that MF59 directly targets the muscle (Fig. 4 C and D).
Fig. 4.
MF59 targets muscle fibers. (A and B) MF59 and CpG up-regulate PTX3 in muscle fibers. (A) Microarray analysis of Ptx3 expression profile in MF59, CpG, alum, MF59+CpG, or PBS-treated muscles after 3, 6, 12 h, 1, 2, and 4 days. Expression levels are shown in fold change compared with untreated muscles. (B) Confocal microscopy analysis of muscles collected 12 h after treatment with PBS, MF59, CpG, or alum and stained with anti-PTX3 (green), anti-UTRN (blue), and PI (red). M, merge. (Scale bar, 40 μm.) (C and D) MF59 enters muscle fibers at 3 h. Confocal microscopy analysis of muscles collected 3 h after injection of DIO-labeled MF59 (green) and stained with anti-UTRN (blue) and PI (red). (C) ×40 magnification and 60-μm scale bar. (D) ×100 magnification, 20-μm scale bar.
Systemic Response to Vaccine Adjuvants.
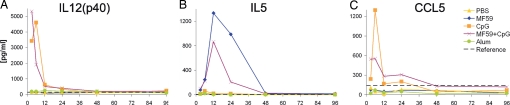
To dissect the local and systemic effects of vaccine adjuvants, we collected the sera of the same mice used for microarray analysis and measured cytokine concentration. CpG was the most potent inducer of a large number of cytokines, including IL12(p40), CCL5, CCL2, and CXCL1, whereas MF59 up-regulated IL5. Alum did not induce any of the tested cytokines (Fig. 5 and data not shown). The systemic expression of IL12(p40) and IL5 is in agreement with the Th1 and Th2 immune responses elicited by CpG and MF59, respectively, in BALB/c mice (17, 20). In addition, Il12p40 and Il5 mRNAs were not up-regulated at the injection site, suggesting that the increase in cytokine levels in the serum derived from the activation of cells of the draining lymph nodes or from circulating blood cells.
Fig. 5.
Systemic expression of cytokines after vaccine adjuvant administration. Cytokine expression profiles were measured in the sera of the same mice subjected to microarray analysis. IL12(p40) (A), IL5 (B), and CCL5 (C) protein expression was measured at the indicated times (expressed in h) after adjuvant administration. The dashed lines indicate the cytokine concentration in untreated mice.
Discussion
Although oil-in-water emulsions are considered the best adjuvants for flu and promising candidates for new human vaccines, their mode of action is still unclear. Here, we show that oil-in-water emulsions, similarly to alum and CpG, activated innate immune reactions at the injection site. The cluster of genes modulated by all adjuvants named “adjuvant core response gene” was characterized by the up-regulation of cytokines, chemokines, and adhesion molecules, suggesting that the establishment of a local immunocompetent environment associated to a nonpathogenic inflammatory process is generally associated to vaccine adjuvanticity. Indeed, we could monitor the recruitment in the muscle of CD11b+ and MHC class II+ blood cells 4 days after administration of all adjuvants. These data are in agreement with previous reports showing that the injection of alum results in local inflammation (12, 13) and with more recent data showing that alum induced monocyte recruitment in the peritoneum (14). Furthermore, two of the adjuvant core response genes identified in mouse muscle, CCL2 and IL1b, were also up-regulated in the peritoneum after alum injection (14). MF59 was a more potent activator of immune related genes than alum and CpG and promoted a more rapid recruitment of CD11b+ blood cells in the muscle. This finding is consistent with previous data obtained from muscle single-cell suspension, which showed that at 1 day after injection, MF59 induced a influx of mononuclear cells (16). The same study demonstrated that MF59-mediated cell recruitment was partially driven by CCR2. Accordingly, in our microarray analysis, MF59 induced Ccr2 at 1–2 days. Moreover, MF59 up-regulated the Ccr2 ligands Ccl2 and Ccl7 at 3 h and Ccl8 at 12 h.
It has been reported that CpG oligonucleotides can modulate the adaptive immune response elicited by MF59 in mice (17, 20). Here, we show that CpG regulated the expression profile of a large number of MF59-responsive genes at the injection site, which may contribute to the modulation of the adaptive response. Moreover, we found that CpG induced stronger systemic responses compared with MF59 and alum, probably reflecting the capability of oligonucleotides to directly activate circulating blood cells such as DCs and B cells.
By using two early biomarkers identified by microarray analysis, JunB and Ptx3, we could identify the skeletal muscle as a target of MF59 immunostimulating activity. Furthermore, we detected labeled MF59 in muscle fibers, supporting a direct activation of the muscle by MF59. However, we never observed a pathological effect of MF59 on the muscle tissue in histological analysis (data not shown). Interestingly, CpG could also activate PTX3 and JUNB expression in muscle cells, suggesting they might respond directly to TLR9 agonists. However, we cannot rule out that early cytokine expression induced by MF59 or CpG in hematopoietic cells contributes to muscle activation. It is well known that skeletal muscle can actively participate in local immune reactions by expressing proinflammatory cytokines, chemokines, adhesion molecules, and TLRs (25). Our data suggest that the skeletal muscle could play an important role in enhancing the efficacy of intramuscularly administered human vaccines. Unlike MF59 and CpG, alum failed to activate muscle fibers, and more work must be performed to identify the target cell responsible for alum-dependent local immunostimulation in the muscle.
We hypothesize that MF59 is a very efficient adjuvant, because it combines antigen delivery function with strong immune-stimulating activity at the injection site. We propose that MF59 induces, in muscle fibers, the production of immune mediators, which in turn activate tissue-resident DCs. MF59 may also promote a sustained antigen-presentation process after vaccination by triggering the recruitment of CD11b+ monocytes, which might differentiate in functional inflammatory DCs expressing high levels of MHC class II, as described for alum (14). Our findings strongly suggest that the mechanism of action of vaccine adjuvants must be addressed in vivo where different cell types cooperate in establishing an integrated immunocompetent environment.
Methods
Mice.
Pathogen-free female BALB/c mice 6–8 weeks of age were obtained from Charles Rivers Laboratories. All animals were housed and treated according to internal animal ethical committee and institutional guidelines. Mice were injected i.m. in both quadriceps with 50 μl per quadricep of PBS alone (control experiment) or supplemented with MF59 (1:1 dilution); 10 μg of CpG; 10 μg of CpG and MF59 diluted 1:1; or 100 μg of Al(OH)3 (Pierce). We choose the amount of adjuvant that gave optimal adjuvanticity in previous studies conducted with various antigens (17–20). Muscles and sera were taken from three mice per group at 3, 6, and 12 h and 1, 2, and 4 days after treatment.
Adjuvants.
MF59 (5% squalene, 0.5% Tween 80, 0.5% Span 85) was prepared in distilled water with a Microfluidizer 110S (MFIC), as described (26, 27). The CpG oligonucleotide sequence used was 5′-TCC ATG ACG TTC CTG ACG TT-3′ with all phosphorothioate backbones (CpG1826). MF59-DIO was prepared by diluting chloroform-resuspended DIO (Invitrogen) in MF59, final concentration 0.25 μg/ml.
Muscle RNA Extraction and Purification.
Whole muscles were homogenized in 7.5 ml of TRIzol (Invitrogen) with an Ultra-Turrax T25 (IKA), and total RNA was extracted from the tissue following the manufacturer's protocol. One hundred micrograms of RNA from each couple of muscles were purified by using the RNeasy RNA purification columns (Qiagen) following the producer's protocol. Residual DNA was removed by an additional on-column DNase digestion step using the Qiagen RNase-free DNase set. RNA quality was assessed by using the automated Experion electrophoresis system (Bio-Rad) coupled with the RNA StdSens kit following the producer's protocol.
RNA Labeling, Microarray Hybridization, and Data Acquisition.
Microarray cDNA probes were prepared from total RNA obtained from treated muscles (test) or from a pool of RNAs extracted from the muscles of 15 naïve mice (reference) using Cy5 and Cy3 dyes, respectively. Twenty-five micrograms of purified total RNA was retrotranscribed at 42°C for 2 h in a 40-μl reaction mix containing 0.625 ng/μl oligo(dT) (Invitrogen), 1 unit/μl of RNAsin (Promega), 10 mM DTT, 2 mM dNTPs-dCTP, and 1 mM dCTP (dNTP mix, Amersham), 2 nmol of Cy3- or Cy5-labeled dCTP (Amersham), and 30 units/μl Super Script II Reverse Transcriptase enzyme (Life Technologies). The RNA was degraded in a 60-μl reaction mix containing 0.3 units/μl RNase One (Promega) and 0.6 units/μl RNaseH (Invitrogen) enzymes for 30 min at 37°C, and then the labeled cDNA was purified by using the QIAquik PCR Purification Kit (Qiagen) following the manufacturer's protocol. The efficiency of incorporation of the Cy5 or Cy3 dyes was measured by Nanodrop analysis. Equal amounts of labeled Cy5 and Cy3 cDNAs were hybridized onto the Agilent 44k Whole Mouse Genome Microarray, detecting >40,000 transcripts, for 17 h at 60°C following Agilent protocol. Images were acquired by using the ScanArray Express microarray scanner (Perkin–Elmer).
Microarray Data Analysis.
Microarray images were first analyzed by using the GenePix 6.0 software (Molecular Devices), and the data were then transferred to the BASE 1.2 database/analysis software (28). For each spot, local background was subtracted, and spot intensities were normalized by the mean fluorescence intensity for each channel. Spots with a signal-to-noise ratio ≤3 in both channels or manually flagged for bad quality were filtered. Four additional hybridizations using the same reference RNA labeled with Cy5 and Cy3 were processed in the same way to determine the dye incorporation bias and to correct the baseline of each spot. The average intensity ratio of each spot from experimental replicates was estimated by geometric mean, and the accuracy and statistical significance of the observed ratios were determined by using Student's t test. Spots with less than two values in the same time point were considered “not found,” and we assigned a log2 ratio of zero. Only genes having t test P values <0.05 and average intensity ratios >4 (log2 ratio ≥2) in at least one time point were selected. Genes were considered responsive to each stimulus if modulated with a fold change of log2 ratio ≥2 compared with untreated muscles. Hierarchical clustering was performed with the TMEV 3.1 software (29) on the log2 ratio transformed dataset applying the Euclidean distance matrix and the average linkage clustering method. Some genes appear more than once in clusters, because they are represented by multiple unrelated probes in the Agilent 44k Whole Mouse Genome Array. Functional analysis of the dataset was performed with GeneSpring GX version 7 software (Agilent Technologies) by using GO, GenMAPP, and KEGG.
Immunofluorescence Experiments.
Cryostat-cut muscle sections (14 μm) were mounted, fixed in PBS and 3% p-formaldehyde for 10 min, and then incubated for another 10 min in blocking and permeabilization solution (PBS, 3% BSA, 1% saponin). The structure of the muscle was visualized by using an antibody specific for Utrophin, a cytoskeletal protein located in the sarcolemma of muscle cells. Tissue sections were incubated for 1 h with the following primary antibodies: goat anti-human Utrophin (Santa Cruz Biotechnology), rabbit anti-mouse PTX3 (Santa Cruz Biotechnology), rabbit anti-mouse JUNB (Santa Cruz Biotechnology), FITC-conjugated rat anti-mouse I-A/I-E (BD PharMingen), and rat anti-mouse CD11b (AbD Serotec). After washing, sections were incubated 30 min with the following secondary antibodies: donkey anti-goat IgG-Alexa Fluor 647 (Molecular Probes), donkey anti-rabbit IgG-Alexa Fluor 488 (Molecular Probes), chicken anti-goat IgG-Alexa Fluor 488 (Molecular Probes), and goat anti-rat IgG-Alexa Fluor 555 (Molecular Probes). Nuclei were stained with propidium iodide (PI) in all experiments, with the exception of CD11b staining in which ToPro3 (Invitrogen) was used. Sections were washed and mounted in Vectashield mounting Medium (Vector Laboratories) and viewed by confocal microscopy (Bio-Rad).
Cytokines Concentration in the Serum.
Cytokine concentrations in the serum have been determined by using the Bio-Plex Cytokine Assay (23-Plex, Bio-Rad) following the producer's protocol. We determined cytokine concentration as an average of three replicates per time point. Three naïve mice were used to detect the background level for each cytokine.
Supplementary Material
Acknowledgments.
We thank M. Tortoli and the Animal Care Facility for mice treatment; B. Baudner (Novartis Vaccines and Diagnostics) for providing DIO-labeled MF59; D. Piccioli for assistance with the Bioplex assay; G. Galli, M. Pizza, U. D'Oro, E. Soldaini, N. Valiante, A. Wack, A. Seubert, and F. Castellino for critical analysis of the data; and A. Covacci and the bioinformatics unit for support. This work was partially supported by Grant MUVAPRED from the European Commission (LSHP-CT-2003-503240). F.M. is the recipient of a Novartis fellowship from the Ph.D. program in Cellular, Molecular and Industrial Biology of the University of Bologna.
Footnotes
Conflict of interest statement: R.R., F.M., E.T., A.M., E.M., F.B., C.I., D.O., and E.D.G. are employees of Novartis Vaccines and Diagnostics.
Data deposition: The complete set of microarray data has been submitted to the ArrayExpress database European Molecular Biology Laboratory–European Bioinformatics Institute, www.ebi.ac.uk/arrayexpress (accession no. E-TABM-506).
This article contains supporting information online at www.pnas.org/cgi/content/full/0804699105/DCSupplemental.
References
- 1.Fraser CK, Diener KR, Brown MP, Hayball JD. Improving vaccines by incorporating immunological coadjuvants. Exp Rev Vaccines. 2007;6:559–578. doi: 10.1586/14760584.6.4.559. [DOI] [PubMed] [Google Scholar]
- 2.Guy B. The perfect mix: Recent progress in adjuvant research. Nat Rev Microbiol. 2007;5:505–517. doi: 10.1038/nrmicro1681. [DOI] [PubMed] [Google Scholar]
- 3.Pashine A, Valiante NM, Ulmer JB. Targeting the innate immune response with improved vaccine adjuvants. Nat Med. 2005;11:S63–68. doi: 10.1038/nm1210. [DOI] [PubMed] [Google Scholar]
- 4.Gavin AL, et al. Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling. Science. 2006;314:1936–1938. doi: 10.1126/science.1135299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemazee D, Gavin A, Hoebe K, Beutler B. Immunology: Toll-like receptors and antibody responses. Nature. 2006;441:E4. doi: 10.1038/nature04875. discussion E4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 7.Boland G, et al. Safety and immunogenicity profile of an experimental hepatitis B vaccine adjuvanted with AS04. Vaccine. 2004;23:316–320. doi: 10.1016/j.vaccine.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 8.Klinman DM. Immunotherapeutic uses of CpG oligodeoxynucleotides. Nat Rev Immunol. 2004;4:249–258. doi: 10.1038/nri1329. [DOI] [PubMed] [Google Scholar]
- 9.Pulendran B, Ahmed R. Translating innate immunity into immunological memory: Implications for vaccine development. Cell. 2006;124:849–863. doi: 10.1016/j.cell.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Hem SL, Hogenesch H. Relationship between physical and chemical properties of aluminum-containing adjuvants and immunopotentiation. Exp Rev Vaccines. 2007;6:685–698. doi: 10.1586/14760584.6.5.685. [DOI] [PubMed] [Google Scholar]
- 11.Morefield GL, et al. Role of aluminum-containing adjuvants in antigen internalization by dendritic cells in vitro. Vaccine. 2005;23:1588–1595. doi: 10.1016/j.vaccine.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 12.Goto N, et al. Local tissue irritating effects and adjuvant activities of calcium phosphate and aluminium hydroxide with different physical properties. Vaccine. 1997;15:1364–1371. doi: 10.1016/s0264-410x(97)00054-6. [DOI] [PubMed] [Google Scholar]
- 13.Goto N, Akama K. Histopathological studies of reactions in mice injected with aluminum-adsorbed tetanus toxoid. Microbiol Immunol. 1982;26:1121–1132. doi: 10.1111/j.1348-0421.1982.tb00261.x. [DOI] [PubMed] [Google Scholar]
- 14.Kool M, et al. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J Exp Med. 2008;205:869–882. doi: 10.1084/jem.20071087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dupuis M, et al. Dendritic cells internalize vaccine adjuvant after intramuscular injection. Cell Immunol. 1998;186:18–27. doi: 10.1006/cimm.1998.1283. [DOI] [PubMed] [Google Scholar]
- 16.Dupuis M, et al. Immunization with the adjuvant MF59 induces macrophage trafficking and apoptosis. Eur J Immunol. 2001;31:2910–2918. doi: 10.1002/1521-4141(2001010)31:10<2910::aid-immu2910>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 17.O'Hagan DT, et al. Synergistic adjuvant activity of immunostimulatory DNA and oil/water emulsions for immunization with HIV p55 gag antigen. Vaccine. 2002;20:3389–3398. doi: 10.1016/s0264-410x(02)00272-4. [DOI] [PubMed] [Google Scholar]
- 18.Singh M, et al. A preliminary evaluation of alternative adjuvants to alum using a range of established and new generation vaccine antigens. Vaccine. 2006;24:1680–1686. doi: 10.1016/j.vaccine.2005.09.046. [DOI] [PubMed] [Google Scholar]
- 19.Vajdy M, et al. Hepatitis C virus polyprotein vaccine formulations capable of inducing broad antibody and cellular immune responses. J Gen Virol. 2006;87:2253–2262. doi: 10.1099/vir.0.81849-0. [DOI] [PubMed] [Google Scholar]
- 20.Wack A, et al. Combination adjuvants for the induction of potent, long-lasting antibody and T-cell responses to influenza vaccine in mice. Vaccine. 2008;26:552–561. doi: 10.1016/j.vaccine.2007.11.054. [DOI] [PubMed] [Google Scholar]
- 21.Bresson JL, et al. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet. 2006;367:1657–1664. doi: 10.1016/S0140-6736(06)68656-X. [DOI] [PubMed] [Google Scholar]
- 22.Leroux-Roels I, et al. Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: a randomised controlled trial. Lancet. 2007;370:580–589. doi: 10.1016/S0140-6736(07)61297-5. [DOI] [PubMed] [Google Scholar]
- 23.Nicholson KG, et al. Safety and antigenicity of nonadjuvanted and MF59-adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: A randomised trial of two potential vaccines against H5N1 influenza. Lancet. 2001;357:1937–1943. doi: 10.1016/S0140-6736(00)05066-2. [DOI] [PubMed] [Google Scholar]
- 24.Garlanda C, et al. Nonredundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 2002;420:182–186. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- 25.Wiendl H, Hohlfeld R, Kieseier BC. Immunobiology of muscle: Advances in understanding an immunological microenvironment. Trends Immunol. 2005;26:373–380. doi: 10.1016/j.it.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Ott G, Barchfeld GL, Van Nest G. Enhancement of humoral response against human influenza vaccine with the simple submicron oil/water emulsion adjuvant MF59. Vaccine. 1995;13:1557–1562. doi: 10.1016/0264-410x(95)00089-j. [DOI] [PubMed] [Google Scholar]
- 27.O'Hagan DT. MF59 is a safe and potent vaccine adjuvant that enhances protection against influenza virus infection. Exp Rev Vaccines. 2007;6:699–710. doi: 10.1586/14760584.6.5.699. [DOI] [PubMed] [Google Scholar]
- 28.Saal LH, et al. BioArray Software Environment (BASE): A platform for comprehensive management and analysis of microarray data. Genome Biol. 2002;3:SOFTWARE0003. doi: 10.1186/gb-2002-3-8-software0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.