Abstract
One of the major challenges that must be met in developing an HIV-1 vaccine is devising a strategy to generate cellular immunity with sufficient breadth to deal with the extraordinary genetic diversity of the virus. Amino acids in the envelopes of viruses from the same clade can differ by >15%, and those from different clades can differ by >30%. It has been proposed that creating immunogens using centralized HIV-1 gene sequences might provide a practical solution to this problem. Such centralized genes can be generated by employing a number of different strategies: consensus, ancestral, or center of tree sequences. These computer-generated sequences are a shorter genetic distance from any two contemporary virus sequences than those contemporary sequences are from each other. The present study was initiated to evaluate the breadth of cellular immunity generated through immunization of rhesus monkeys with vaccine constructs expressing either an HIV-1 global consensus envelope sequence (CON-S) or a single patient isolate clade B envelope sequence (clade B). We show that vaccine immunogens expressing the single centralized gene CON-S generated cellular immune responses with significantly increased breadth compared with immunogens expressing a wild-type virus gene. In fact, CON-S immunogens elicited cellular immune responses to 3- to 4-fold more discrete epitopes of the envelope proteins from clades A, C, and G than did clade B immunogens. These findings suggest that immunization with centralized genes is a promising vaccine strategy for developing a global vaccine for HIV-1 as well as vaccines for other genetically diverse viruses.
Keywords: consensus gene, cytotoxic T lymphocyte
The creation of a successful HIV-1 vaccine will require the development of a strategy to deal with the extreme genetic diversity of the virus. The amino acid sequences of envelopes of HIV-1 isolates of the same clade can diverge by >15%, and the sequences of isolates of different clades can differ by >30% (1). The problems posed for vaccine development by the genetic diversity of HIV-1 are compounded by the fact that there are multiple clades as well as recombinant forms of the virus circulating in single countries, and population mobility is resulting in increased genetic complexity of the virus in regional epidemics (2, 3). Therefore, it will be important to develop a strategy for generating effective immunity against the diversity of HIV-1 isolates that exist worldwide.
It has recently been proposed that immunogens created by using centralized HIV-1 gene sequences might be a solution for developing a global HIV-1 vaccine (1, 4–7). Such centralized genes can be generated by using a number of different strategies: consensus sequences (aligning available HIV-1 gene sequences and selecting the most commonly used amino acid at each residue of a particular viral protein) (1), the most recent common ancestor sequences (modeling ancestral states of the virus) (1), or center of tree sequences (7, 8). Sequences generated by any of these strategies will reduce the amino acid sequence distance between immunogens and the field virus strains. In fact, a group M consensus sequence is approximately half the genetic distance from any two contemporary clade–disparate virus sequences than those two virus sequences are from each other.
The present study was initiated to evaluate the breadth of cellular immunity generated through immunization of rhesus monkeys with vaccine constructs expressing either a centralized or a single naturally occurring envelope sequence.
Results
Vaccine Trial Design.
We selected previously well characterized HIV-1 genes for use as immunogens in this study: the representative single clade B env HXBc2 with a BaL V3 loop to confer CCR5 coreceptor usage (clade B) (9) and a group M consensus env (CON-S) (1). The envelope gene of CON-S was modified to include a delta CFI mutation so that it corresponded to the B clade gene. Thirty Mamu-A*01-negative Indian-origin rhesus monkeys were distributed into three groups, each consisting of 10 animals. Monkeys received priming immunizations by the intramuscular route at weeks 0, 4, and 8 with 5 mg of plasmid DNA expressing clade B env, CON-S env, or a sham construct. At week 24, monkeys were boosted by intramuscular immunizations with 1011 particles of recombinant E1/E3-deleted adenovirus serotype 5 (rAd) expressing the same gene.
Vaccine-Elicited Immune Responses to the Immunizing Envelopes.
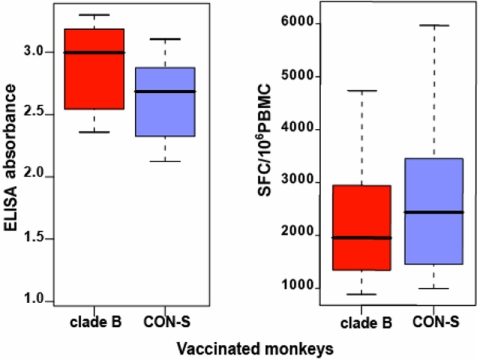
Robust immune responses to the immunizing envelope antigens were detected in both the clade B- and the CON-S-vaccinated monkeys 2 weeks after the rAd boost. The magnitudes of the antibody responses to the immunizing envelopes as measured by ELISA were statistically indistinguishable in the clade B- and CON-S-immunized cohorts of animals (Wilcoxon rank sum, P = 0.19) (Fig. 1Left). IFN-γ ELISpot assays were performed with PBMC from the clade B-immunized monkeys after stimulation by pools of overlapping peptides spanning the entire clade B HXBc2/BaL envelope and with PBMC from the CON-S-immunized monkeys stimulated by peptide pools spanning the group M consensus envelope. IFN-γ ELISpot responses representing the immunizing envelope sequences also were of comparable magnitudes in the two groups of immunized monkeys (P = 0.44) (Fig. 1 Right). Therefore, the differences described below that we observed in the vaccine-elicited immune responses in these groups of animals to heterologous envelopes should reflect the cross-reactivity of the immune responses and not be a consequence of different magnitudes of the vaccine-elicited anti-envelope responses.
Fig. 1.
Humoral and cellular immune responses to the envelope proteins expressed by the vaccine immunogens. (Left) The magnitudes of the antibody responses to the envelope proteins expressed by the vaccine constructs used for immunization as measured by ELISA absorbance at 450 nm. Sera from 10 monkeys vaccinated with immunogens expressing a single clade B envelope were assessed for antibody responses to that clade B envelope protein, and sera from 10 monkeys vaccinated with CON-S envelope immunogens were assessed for anti-CON-S envelope antibody responses. Median and interquartile ranges (IR) of antibody responses for both groups are statistically indistinguishable (Wilcoxon rank sum test, P = 0.19). (Right) PBMC IFN-γ ELISpot responses to peptide pools representing the immunizing envelope sequences. PBMC from monkeys immunized with either the clade B or the CON-S envelope immunogens were assessed in IFN-γ ELISpot assays after exposure to peptide pools spanning the sequences of the envelope proteins used for the immunizations. Median values of SFC per 106 PBMC were of comparable magnitudes for both groups of monkeys (P = 0.44).
Assessment of Vaccine-Elicited Envelope-Specific Cellular Immune Responses.
The breadth of the vaccine-elicited cellular immune responses was then determined by assessing peripheral blood T lymphocyte recognition of 10 different envelope sequences by using a peptide/IFN-γ ELISpot assay. The 10 envelopes chosen as indicator proteins included two clade A, two clade B, four clade C, and two clade G sequences. The criteria for selection were that the sequences were recent and were sampled from diverse geographic locales to represent the contemporary diversity of envelope sequences catalogued in the Los Alamos database (Fig. 2). By assessing T lymphocyte immune responses to each of these selected envelopes, we simulated the exposure of the vaccine-induced cell populations to representative and diverse real virus envelope proteins. PBMCs obtained from each of the monkeys 2 weeks after rAd immunization were evaluated for IFN-γ ELISpot responses to pools of 15 mers overlapping by 11 aa spanning each of the 10 selected envelope sequences.
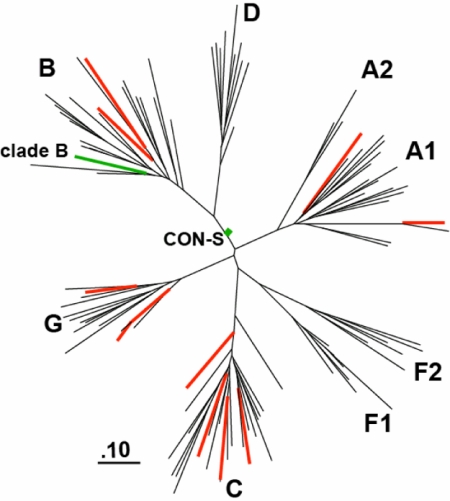
Fig. 2.
Genetic relatedness of immunogens and indicator proteins. A maximum likelihood tree illustrates the genetic distances between the envelope clade B (green) and CON-S (green) immunogen proteins and the indicator envelope proteins (red). The illustrated mean branch lengths (±SD) include CON-S vaccine to circulating forms of virus, 0.34 (0.05); clade B vaccine to other clade B viruses, 0.33 (0.05); and clade B vaccine to non-clade B viruses, 0.62 (0.06). This maximum likelihood tree was built using PHYML (24) branch length summaries obtained with the branch length calculator at the Los Alamos HIV/HCV database.
Clade B Versus Cross-Clade Cellular Immune Responses in Vaccinated Monkeys.
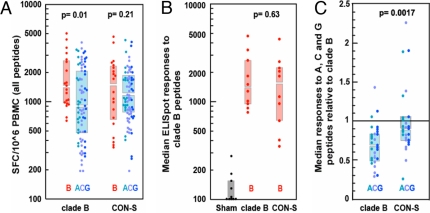
We first evaluated the magnitudes of the IFN-γ ELISpot responses by lymphocytes from the two groups of experimentally vaccinated animals after PBMC exposure to the pooled peptide series representing each of the two clade B (both distinct from the vaccine strain), two clade A, four clade C, and two clade G envelope proteins (Fig. 3A). In the clade-B-vaccinated monkeys, magnitudes of the PBMC responses to clade B peptide pools were significantly higher than their responses to clades A, C, and G peptide series (i.e., within-clade responses were greater than between-clade responses, P = 0.01). In contrast, in CON-S-vaccinated animals there was no statistically significant difference between magnitudes of responses to clade B and other clades' peptide series (P = 0.21). Furthermore, as shown in Fig. 3B, the magnitudes of the PBMC responses to the clade B peptide pools were indistinguishable between the clade B- and CON-S-immunized monkeys (Wilcoxon rank sum test, P = 0.6305). Therefore, the CON-S immunogen elicited as robust a cellular immune response to clade B envelopes as did the clade B immunogen. Thus, the CON-S immunogens elicited within-clade levels of responses.
Fig. 3.
Magnitude of each vaccinated monkey's cellular immune responses to clade B envelope and to each of the envelope peptide pools. (A) IFN-γ ELISpot responses of individual monkeys to the 10 envelope peptide series. Each animal's responses to each peptide series are shown in different colored bullets: clade A (aqua), clade C (blue), and clade G (purple). For both cohorts, responses to clade B peptide series are shown in red bullets in separate box plots to facilitate the comparisons of within-clade and between-clade responses. The boxes indicate the IR, and the gap between the two boxes represents the median. The clade B-immunized monkeys had significantly higher clade B peptide-specific responses than non-clade B peptide pools (Wilcoxon rank sum test, P = 0.01). The CON-S-immunized monkeys had statistically indistinguishable responses to clade B and non-clade B peptides. (B) Magnitudes of responses to clade B peptide series are indistinguishable in the two groups of vaccinated monkeys. The median ELISpot responses to the peptide pools for the two clade B peptide series are shown in red bullets, with the median and IR indicated by the boxes. The responses by the control vaccinees are also shown. (C) Median responses to non-clade B peptides relative to clade B peptides in the two groups of vaccinated monkeys. To simplify the visual representation of the data, the PBMC responses of each monkey to the clade A, C, and G peptide series were normalized by dividing those values by the same animal's median clade B-specific PBMC response. Therefore, if an animal had comparable responses to both clade B and non-clade B peptides, a value near 1 would be achieved. The CON-S-vaccinated monkeys had greater magnitude pooled peptide ELISpot responses than the clade B-vaccinated monkeys to the non-clade B envelope peptide pools (Wilcoxon rank sum test, P < 0.0017). The full dataset was used for the plot and the statistics in A to provide a comprehensive view of the data. The median responses to the peptide sets from each clade were used for each animal for B and C to provide an alternative and more conservative view of the results, thereby providing a framework for a statistical comparison in which each animal is counted only once. Because two A, two B, four C, and two G pooled peptide sets were used per animal, the counts in A are not fully independent.
We were concerned, however, that the apparent benefit associated with using the CON-S immunogen may reflect an inadvertent bias in the data resulting from differences in magnitudes of responses in individual monkeys in the experimental groups. To control for this possibility, we examined the relative magnitudes of the IFN-γ ELISpot responses in the two groups of vaccinated monkeys after normalizing the ELISpot data. The median PBMC responses of each monkey to the clade A, C, and G peptide series were calculated and normalized by dividing those values by the same animal's median clade B-specific PBMC response. The CON-S-immunized monkeys developed significantly larger PBMC responses to the clade A, C, and G envelope peptide series than the clade B-immunized monkeys (Fig. 3C) (Wilcoxon rank sum test, P < 0.0017). The responses elicited by the CON-S immunogens were very comparable across clades. This finding suggests that the vaccine immunogens expressing the single centralized gene CON-S generated consistent cellular immune responses across all four clades, whereas the single clade B envelope gene generated more intraclade B responses (Fig. 3C).
Enumeration of Epitope-Specific T Cell Responses.
To complement these pooled peptide ELISpot studies, we enumerated the epitopes of each of the 10 indicator envelope proteins recognized by PBMCs of each of the experimentally vaccinated monkeys. Epitope enumeration was accomplished by matrix mapping and confirmation with individual peptides. A peptide was scored as positive if it stimulated more than three times the background response and a minimum of 55 spot-forming cells (SFC) per million PBMC. When overlapping/adjacent peptides scored positive, two adjacent peptides were counted as a single epitope and three or four adjacent peptides were counted as two epitopes. The overlapping sets of peptides were created separately for each indicator envelope sequence, and the sequences of those different envelopes often had regions of identity. Therefore, PBMC responses to particular peptides were evaluated more than once for each monkey when peptide sequences from two different envelopes were identical. In those situations, the median of all of the duplicated ELISpot responses to such a peptide was determined for each monkey, and if that value exceeded three times the background, the response was considered positive.
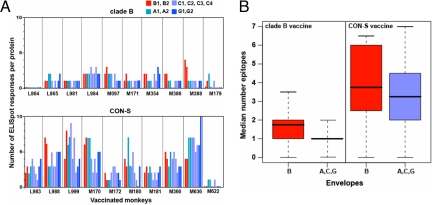
Statistical modeling using Poisson regression showed that the CON-S immunogen elicited a larger number of responses than did the clade B immunogen, with the rate of positive responses larger by a factor of >3.15 (P ≪ 10−6) (Fig. 4A). Within the clade B-vaccinated group of monkeys, significantly more responses were detected to the B clade proteins than to the non-B clade proteins by a factor of 1.55 (P = 0.038), whereas within the CON-S-vaccinated group of monkeys, responses to clade B and to non-clade B proteins were not statistically distinguishable.
Fig. 4.
Epitopes of indicator envelope proteins recognized by T lymphocytes of vaccinated monkeys. PBMCs from each vaccinated monkey were assessed for their recognition of specific epitopes of each of the 10 indicator envelope proteins. This epitope recognition was determined by matrix IFN-γ ELISpot mapping and confirmation with assays employing individual peptides. (A) The number of epitopes recognized of each individual indicator envelope protein by PBMCs of each vaccinated monkey. The epitopes of the indicator envelopes are shown by colored bars: clade B (red), clade A (turquoise), clade C (gray blue), and clade G (sky blue). The CON-S-vaccinated monkeys generated responses to more epitopes than did the clade B-vaccinated monkeys (generalized linear model, P < 0.00002), with the rate of positive responses larger by a factor of >3.15. (B) Median number and IR of epitopes recognized from clade B envelope proteins and non-clade B envelope proteins by PBMCs of each group of vaccinated monkeys. The CON-S-immunized monkeys generated responses to more non-clade B envelope epitopes than did the clade B-immunized monkeys (Wilcoxon rank sum test, P < 0.00047).
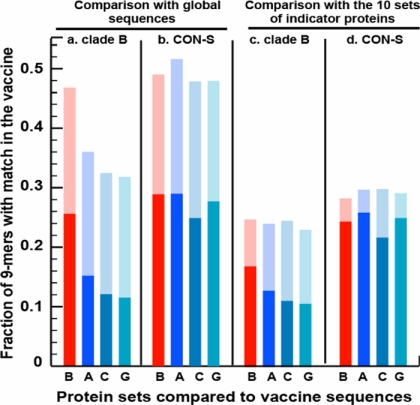
These same data also were analyzed by using an alternative statistical strategy to allow a simple comparison of the number of clade B envelope and non-clade B envelope epitopes recognized by PBMC of the monkeys vaccinated with either the CON-S or the clade B immunogens. The CON-S-immunized monkeys generated responses to more clade A, C, and G envelope epitopes than the clade B-immunized monkeys (Wilcoxon rank sum test, P < 0.00047) (Fig. 4B). The clade B-immunized monkeys generated responses to more clade B envelope epitopes (intraclade responses) than non-clade B envelope epitopes (interclade responses) (paired Wilcoxon test, P = 0.056,), whereas the CON-S-immunized monkeys evidenced no bias toward recognizing epitopes from an envelope of a particular clade of virus (paired Wilcoxon test, P = 0.31) (Fig. 4B). The two groups of immunized monkeys generated responses to comparable numbers of clade B envelope epitopes (Wilcoxon rank sum test, P = 0.083). However, this P value of 0.083, although not significant, suggests a possible trend toward a greater number of responses even to the B clade test proteins in the CON-S-vaccinated animals than in the B clade protein-vaccinated animals. To clarify why this trend toward greater numbers of clade B epitope-specific responses might be observed in the CON-S-vaccinated monkeys, we evaluated the potential T cell epitope coverage [defined as identical matches to all possible 9-aa length fragments, or 9-mer, based on the strategy for comparing potential epitopes (10)] of natural proteins by the B clade and CON-S immunogens. We compared the epitope coverage of the 10 indicator proteins from four clades used for the design of the indicator peptides as well as the coverage of the full set of M group proteins shown in the maximum likelihood tree in Fig. 2 (Fig. 5, dark-shaded bars). As shown in Fig. 5, columns a and b, the CON-S immunogen had better coverage of 9-mer in A, C, and G clade proteins than the clade B immunogen. This result was expected because the CON-S immunogen is the centralized sequence for all group M viruses. However, the CON-S coverage also was better for the clade B proteins.
Fig. 5.
Potential epitope coverage of the two envelope immunogens used in this study. The global sequence set used as a foundation for the phylogenetic tree shown in Fig. 2 made use of a baseline for the comparisons in columns a and b. All proteins in a set were broken down into all possible 9-mer to be considered as potential epitopes. The fraction of perfect matches (dark shades) and the fraction that differed by one amino acid (light shades) from the 9-mer found in the immunogen proteins were calculated. Column a shows the clade B immunogen coverage of a global sampling of envelope proteins. Envelope is highly variable, and only 26% of the 9-mer found in circulating proteins have a perfect match in the clade B immunogen; there is a dramatic reduction in the coverage in other clades, with only 11–15% coverage. Column b shows the CON-S immunogen coverage of a global sampling of envelope proteins. The average coverage of all proteins from the A, B, C, and G clades is comparable with the within-clade coverage shown in column a, with 25–29% of the 9 mers having a perfect match. Columns c and d parallel columns a and b but are restricted to the 10 proteins used for peptide design, and the values are, in general, reduced. This reduction is due to the fact that the database is comprised of proteins sampled over the past 25 years. Older samples tend to be less diverged than the 10 recent samples that were selected to be representative of the contemporary global diversity of sequences. The CON-S immunogen shows better coverage of the global clade B proteins and two clade B indicator proteins than does the clade B immunogen.
This analysis was done by assessing perfect matches between potential epitope sequences in the immunogens and those sequences in either the indicator peptides or the group M envelope proteins catalogued in the Los Alamos database. However, we reasoned that a significant proportion of T cell responses primed by an immunogen to an epitope sequence that differs from a true viral sequence by a single amino acid might still recognize that viral sequence and mediate effective antiviral activity. We therefore repeated this analysis by comparing potential epitope sequences that differed between the immunogen and either the indicator peptides or all group M envelope proteins by a single amino acid (Fig. 5, light-shaded bars). This analysis demonstrated the same degree of potential benefit for using the CON-S immunogen as was seen in the analysis of the perfectly matched epitopes. This finding is compatible with the greater number of clade B peptides that were recognized by PBMC of the monkeys after vaccination with the CON-S than with the clade B immunogens (Fig. 4B). Moreover, this finding was consistent not only for the two clade B proteins selected for creating the indicator peptides (Fig. 5, columns c and d), but in a broad sampling of clade B proteins (Fig. 5, columns a and b).
Discussion
The group M centralized immunogen CON-S was able to stimulate T cell responses that were readily detectable across all clades, whereas T cell responses elicited by the natural clade B immunogen showed significant within-clade specificity. Moreover, the cross-reactive potential of the T cell responses stimulated by the CON-S immunogen within clade B envelope was comparable with that stimulated by the clade B immunogen. Technical issues were considered in interpreting these cellular immune response data. Differences in the production methods and quality control specifications of the rAd vaccine constructs could potentially contribute to differences observed in the immune responses. Arguing against that possibility, however, is the finding that the rAd constructs expressing clade B env and CON-S env boosted antibody and cellular responses to the specific envelope proteins used for immunization that were of comparable magnitudes (Fig. 1). Furthermore, whereas robust immune responses were observed against all four clades of envelopes in the CON-S-vaccinated monkeys, potent intraclade and weak interclade responses were seen in the clade B-vaccinated animals. That this differential recognition of clade B and non-clade B peptides was observed in groups of experimental monkeys that received a single rAd vaccine construct further argues that the increased breadth of envelope reactivity across clades is a consequence of the CON-S gene insert in the rAd vector.
Another strategy for inducing broadly reactive cellular immune responses that has received considerable attention is the use of multiple diverse Env sequences in the same vaccine inoculum (11–14). In depth, systematic studies have not been done to determine whether these immunization strategies elicit T cells that are cross-reactive with diverse clades A, B, and C envelopes that are not included in the vaccine or whether they simply elicit immune responses to vaccine antigens that are additive. Even if this type of approach elicits cross-reactive cellular immune responses to a diversity of heterologous envelopes from multiple viral clades, the use of a CON-S-based immunogen still has the advantage of eliciting broad immunity with a vaccine expressing a single Env protein. Immunogens created with centralized genes and current vaccine technologies are unlikely to induce antibodies capable of neutralizing a diversity of primary patient HIV-1 isolates. This is the case because the extreme genetic diversity of HIV-1 is only one of a number of hurdles standing in the way of the creation of an immunogen that elicits broadly neutralizing antibodies. The current view among investigators is that creating a subunit immunogen with a critical 3D conformation (15), overcoming an immunoregulatory blockade (16), or both will be needed to generate a broadly neutralizing antibody response. However, once the conformational and immunoregulatory issues are clarified, a reasonable strategy for the design of the new vaccines should make use of centralized genes so as to optimize their induction of broad humoral immune responses.
The present data complement a growing body of evidence supporting the utility of centralized genes in the creation of an HIV-1 vaccine. Proteins created from centralized envelope sequences have been shown to have the functional properties of naturally occurring HIV-1 envelopes as well as conserved neutralizing antibody epitopes (4, 5, 17, 18). Because it is not possible to evaluate the breadth of a vaccine-generated cellular immune response in inbred strains of mice, it was necessary to turn to nonhuman primates to assess this critical immune response. The current study in outbred rhesus monkeys makes a compelling argument for a broadening of immunity in individuals vaccinated with centralized gene-based immunogens. Moreover, this validation in primates of the prediction that consensus gene-based immunogens generate broader T cell responses than wild-type gene-based immunogens is a major step forward in HIV-1 vaccine design in that it suggests that future comparisons of T cell immunogen candidates might initially be explored in silico before nonhuman primate and human testing (1). Although the reason for the failure of the Merck HIV T cell vaccine trial is at this point still unresolved, limited breadth of coverage might have been a contributing factor, and we have now demonstrated in a nonhuman primate model that the centralized protein approach offers considerable enhancement of breadth of coverage.
The breadth of cellular immune responses was assessed by using a series of genetically diverse envelope peptide sets. Other strategies have been proposed for evaluating T lymphocyte responses to diverse HIV-1 sequences, including the use of potential T cell epitope peptides and toggled peptides (10). The strategy used in the present study was selected because it simulates the exposure of vaccine-induced memory T cells to genetically diverse real envelopes, allows for a systematic mapping of all recognized epitopes in these proteins, and encompasses a diversity of potential T cell epitopes that is comparable with other design peptide strategies (data not shown).
We have therefore demonstrated that a vaccine based on an M group centralized gene sequence can generate cellular immune responses with considerable breadth. While immune responses elicited by clade B envelope vaccination had little cross-clade reactivity, robust responses were elicited by CON-S envelope vaccination to all clades tested. The CON-S-vaccinated monkeys developed potent cellular immune responses to genetically diverse HIV-1 envelopes as measured by both pooled peptide ELISpot assays and by enumeration of recognized epitopes. These findings suggest that a centralized gene approach may prove useful not only for the creation of an HIV vaccine but also for the creation of vaccines against other genetically diverse pathogens.
Materials and Methods
Experimental Groups and Vaccination Schedule.
Monkeys were housed at Advanced Bioscience Laboratories. The animals were maintained in accordance with National Institutes of Health guidelines. Ten monkeys in each of the following groups were established: (group 1) sham DNA-empty Ad boost, (group 2) DNA-HIV-1 HXBc2/BaL env prime/rAd5-HIV-1 HXBc2/BaL env boost, and (group 3) DNA-CON-S env prime/rAd5-CON-S env boost. The monkeys were immunized intramuscularly with 5-mg per immunization of plasmid DNA at weeks 0, 4, and 8. At week 24, the monkeys were boosted with 1011 particles per immunization of rAd5.
Construction of the Plasmid DNA Vaccines.
The plasmid DNA expressing HXBc2/BaL env gene was constructed as previously described (19). The synthetic gene of the clade B gp160 from HXBc2 was created using the protein sequence of the HXBc2 gp160. The region encoding HIV-1 envelope glycoprotein amino acids 205–361 from HXB2 gp160 was replaced with the corresponding region from the BaL strain of HIV-1, followed by deletion of the fusion and cleavage domains from amino acids 503–536. The interspace between H1 and H2 from amino acid 593 to 620 was also deleted. The gp140ΔCFI version was derived from this sequence by introduction of a termination codon as previously described (9).
The CON-S gp140ΔCFI env gene was constructed as previously described (7). Briefly, the CON-S env gene was generated by converting amino acid sequences of CON-S to nucleotide sequences employing the codon usage of highly expressed human housekeeping genes (20). CON-S gp140ΔCFI was generated by PCR by introducing a stop codon before the membrane-spanning domain as previously described (4, 18). For use as a DNA vaccine, CON-S gp140ΔCFI was subcloned into pCMVR DNA vaccine vector. Endotoxin-free plasmid DNA was produced by Puresyn for the immunization of rhesus monkeys.
Construction of the Recombinant Adenoviruses.
The recombinant adenovirus expressing HXBc2/BaL Env was generated as previously described (17). The rAd5-CON-S vector was produced by homologous recombination of an adaptor plasmid expressing the CON-S Env gene with the relevant cosmid in complementing cells as previously described (21). The plasmids were linearized before transfection of cells using Lipofectamine in T25 flasks. Cells were passaged into T75 flasks after 48 h and maintained until virus cytopathic effect was observed. The vectors were plaque-purified, analyzed for transgene expression, amplified in 24 triple-layer T175 flasks, purified by double CsCl gradient ultracentrifugation, and dialyzed into PBS containing 5% sucrose. Purified rAd vectors were stored at −80°C. Virus particle titers were determined by spectrophotometry. Specific infectivity was assessed by plaque-forming unit assays.
HIV-1 Env Peptide Sets and Design of Peptide Matrices.
Peptides (15-mer) overlapping by 11 aa and spanning the entire HIV-1 Env were used. Ten sets of HIV-1 Env peptides from four different clades were synthesized: two from clade A, two from clade B, four from clade C, and two from clade G. Please refer to the supporting information (SI) Materials and Methods and Table S1 for the criteria for selection of these 10 proteins.
Pooled Peptide and Peptide Matrix IFN-γ ELISpot Assays.
Multiscreen 96-well plates were coated overnight with 5 μg/ml anti-human IFN-γ (B27; BD Pharmingen) in endotoxin-free Dulbecco's PBS (D-PBS) at 100 μl per well. The plates were then washed three times with D-PBS containing 0.25% Tween 20 and blocked for 2 h with RPMI medium 1640 containing 10% FBS. Then peptide pools and 2 × 105 PBMCs were added to each well in 100-μl reaction volumes in triplicate for pooled peptides assays and in duplicate for peptide matrix assays. Each peptide in a pool was present at a 1 μg/ml concentration. After an 18-h incubation at 37°C, the plates were washed nine times with D-PBS/Tween 20 and once with distilled water. The plates were then incubated with 2 μg/ml biotinylated rabbit anti-human IFN-γ (U-Cytech) for 2 h at room temperature, washed six times with D-PBS containing 0.25% Tween-20, and incubated for 2.5 h with a 1:500 dilution of streptavidin-AP (Southern Biotechnology Associates). After five washes with D-PBS/Tween 20 and one wash with D-PBS, the plates were developed with nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt chromogen (Pierce), stopped by washing with tap water, air dried, and read with an ELISpot reader (Cellular Technology). The number of SFC per 106 PBMCs was calculated.
Statistics.
To characterize further the breadth of vaccine-elicited cellular immune responses, detailed statistical modeling was undertaken with the statistical package R (22). Poisson regression models were fit (23) that predicted the expected number of positively scored peptides as a function of the vaccine the animal received and the clade of the envelope epitopes. As is evident from the data, there were significantly variable levels of responses between animals; this was expected because an outbred population of macaques was used to reflect a human vaccine scenario. Items 1 and 2 in the list below provide summaries of some comparisons of interest, where the input data considered were numbers of positive responses, and positivity was defined in terms of the median of the ratio (number of SFC/background) for all identical measurements made on identical peptides.
A Poisson regression with peptide clade (one of clade A, B, C, and G) and vaccine as explanatory variables gave a highly significant enhancement in positive responses for CON-S relative to the B clade vaccine (probability ratio > 3.29, P ≪ 10−6).
Studying only the group that received B-clade vaccine and classifying the peptides as B or not-B (A, C, or G) clade showed a significant enhancement in responses for B clade peptides (probability ratio > 1.55, P < 0.038). In contrast, within the CON-S-vaccinated group, no differences in the B clade and other clade-directed responses were observed.
Further simple comparisons were made between groups by calculating the median number of responses to each of the clade B envelopes in each vaccinated monkey or to the non-clade B (clades A, C and G) envelopes and then comparing the groups of monkeys by using a nonparametric Wilcoxon rank sum statistic (also known as a Mann–Whitney test). When appropriate, a paired test signed rank was performed. All statistics were performed with the statistical computing software R (22).
Supplementary Material
Acknowledgments.
We thank Anjali Nanda, Diana Lynch, Bonnie Ewald, Adam Buzby, and Harikrishnan Balachandran for technical contributions. This work was supported by National Institutes of Health Grant P01-AI61734, National Institute of Allergy and Infectious Diseases Simian Vaccine Evaluation Unit Contract N01-AI60005, National Institutes of Health Contracts N01-AI30033 and N01-AI30034, and a Los Alamos National Laboratory directed research grant for pathogen vaccine design.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803352105/DCSupplemental.
References
- 1.Gaschen B, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 2.Lin HH, et al. Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquired Immune Defic Syndr. 2006;41:399–404. doi: 10.1097/01.qai.0000200663.47838.f1. [DOI] [PubMed] [Google Scholar]
- 3.Arroyo MA, et al. Higher HIV-1 incidence and genetic complexity along main roads in Rakai District, Uganda. J Acquired Immune Defic Syndr. 2006;43:440–445. doi: 10.1097/01.qai.0000243053.80945.f0. [DOI] [PubMed] [Google Scholar]
- 4.Gao F, et al. Antigenicity and immunogenicity of a synthetic human immunodeficiency virus type 1 group m consensus envelope glycoprotein. J Virol. 2005;79:1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doria-Rose NA, et al. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 2005;79:11214–11224. doi: 10.1128/JVI.79.17.11214-11224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rolland M, et al. Reconstruction and function of ancestral center-of-tree human immunodeficiency virus type 1 proteins. J Virol. 2007;81:8507–8514. doi: 10.1128/JVI.02683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao HX, et al. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology. 2006;353:268–282. doi: 10.1016/j.virol.2006.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nickle DC, et al. Coping with viral diversity in HIV vaccine design. PLoS Comput Biol. 2007;3:e75. doi: 10.1371/journal.pcbi.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakrabarti BK, et al. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76:5357–5368. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, et al. Peptide selection for human immunodeficiency virus type 1 CTL-based vaccine evaluation. Vaccine. 2006;24:6893–6904. doi: 10.1016/j.vaccine.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Catanzaro AT, et al. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine. 2007;16:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- 12.Seaman MS, et al. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–2963. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan X, et al. Multi-envelope HIV-1 vaccine devoid of SIV components controls disease in macaques challenged with heterologous pathogenic SHIV. Vaccine. 2005;23:5306–5320. doi: 10.1016/j.vaccine.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, et al. Polyvalent HIV-1 Env vaccine formulations delivered by the DNA priming plus protein boosting approach are effective in generating neutralizing antibodies against primary human immunodeficiency virus type 1 isolates from subtypes A, B, C, D and E. Virology. 2006;350:34–47. doi: 10.1016/j.virol.2006.02.032. [DOI] [PubMed] [Google Scholar]
- 15.Pantophlet R, Burton DR. GP120: Target for neutralizing HIV-1 antibodies. Annu Rev Immunol. 2006;24:739–769. doi: 10.1146/annurev.immunol.24.021605.090557. [DOI] [PubMed] [Google Scholar]
- 16.Haynes BF, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 17.Kothe DL, et al. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology. 2007;360:218–234. doi: 10.1016/j.virol.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kothe DL, et al. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology. 2006;352:438–449. doi: 10.1016/j.virol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 19.Letvin NL, et al. Heterologous envelope immunogens contribute to AIDS vaccine protection in rhesus monkeys. J Virol. 2004;78:7490–7497. doi: 10.1128/JVI.78.14.7490-7497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andre S, et al. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vogels R, et al. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: Efficient human cell infection and bypass of preexisting adenovirus immunity. J Virol. 2003;77:8263–8271. doi: 10.1128/JVI.77.15.8263-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Found for Stat Comput; 2006. [Google Scholar]
- 23.Crawley MJ. Statistical Computing: An Introduction to Data Analysis Using S-Plus. Chichester, UK: Wiley; 2002. [Google Scholar]
- 24.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—A web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–W559. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.