Abstract
Population diversity data have recently provided profound, albeit inferential, insights into meiotic recombination across the human genome, revealing a landscape dominated by thousands of cross-over hotspots. However, very few of these putative hotspots have been directly analyzed for cross-over activity. We now describe a search for very active hotspots, by using extreme breakdown of marker association as a guide for high-resolution sperm cross-over analysis. This strategy has led to the isolation of the most active cross-over hotspots yet described. Their morphology, sequence attributes, and cross-over processes are very similar to those seen at less active hotspots, but their activity in sperm is poorly predicted from population diversity information. Several of these hotspots showed evidence for biased gene conversion accompanying cross-over, in some cases associated with variation between men in cross-over activity and with two hotspots showing complete presence/absence polymorphism in different men. Hotspot polymorphism is very common at less active hotspots but curiously was not seen at any of the most active hotspots. This contrasts with the prediction that extreme hotspots should be the most vulnerable to attenuation by meiotic drive in favor of mutations that suppress recombination and should therefore show rapid rate evolution and thus variation in activity between men. Finally, these very intense hotspots provide a valuable resource for dissecting meiotic recombination processes and pathways in humans.
Keywords: conversion, hotspot, meiosis, polymorphism, recombination
The recombinational exchange of DNA between homologous chromosomes at meiosis is vital to ensure correct chromosome segregation and also plays a major role in increasing haplotype diversity within populations. In humans, the combination of low average cross-over frequency [≈1% recombination frequency (RF) per Mb of DNA] and small numbers of informative meioses in pedigree studies has limited the resolution of current linkage maps to the megabase level (1, 2). Much higher resolution profiles of recombination can instead be obtained indirectly through examining patterns of marker association [linkage disequilibrium (LD)], established through population dynamic processes and eroded by recombination, or directly through labor-intensive screening of millions of sperm for recombinant DNA molecules within short DNA intervals (typically <10 kb).
Population LD (3, 4) and sperm DNA (5–14) analyses have firmly established that most cross-over events in humans cluster into narrow hotspots spaced, on average, 50 kb apart. Recently, the International HapMap Project (15, 16) has mapped the LD landscape genome-wide at the kilobase level. These data allowed inference of the global recombination landscape at high resolution (4) by using coalescent analyses whereby observed haplotypes are explained through in silico reconstruction with variable historical recombination rates. These analyses have identified ≈33,000 putative cross-over hotspots (LD hotspots) throughout the genome (4, 16, 17) and have provided insights into hotspot distribution and historical cross-over activity, as well as identifying DNA sequence motifs associated with hotspots (16, 18).
In contrast, few human recombination hotspots have been directly characterized in sperm, and it is still unclear whether LD landscapes can accurately predict and locate genuine hotspots or correctly estimate their historical activity. To date, sperm surveys have only covered a total of 0.6 Mb of human DNA, identifying seven hotspots in a 216-kb region of the major histocompatibility complex (6, 10, 11), eight in a 206-kb region on chromosome 1 (7–9, 13), one in the β-globin gene cluster (5), and one in the SHOX gene located in the Xp/Yp pseudoautosomal region PAR1 (12). A near-contiguous 103-kb segment of chromosome 21 has also been screened for cross-overs in sperm (14). These surveys have shown a good, if not perfect, concordance between the location of LD hotspots and sperm hotspots. They have revealed additional phenomena that could not have been detected from population data, including variation in hotspot activity between men (7–9, 14) and complete on/off polymorphism despite no changes in local DNA sequence (13). Meiotic drive in favor of a cross-over-suppressing variant within the hotspot has been detected at two loci (8, 19), suggesting a mechanism for hotspot extinction (8, 20). Conversely, active sperm hotspots have been observed within regions of strong LD, consistent with these hotspots being young (9). These findings suggest that cross-over hotspots might be transient features of the genome, turning over rapidly in evolutionary time; this possibility is consistent with the markedly divergent LD landscapes of humans and chimpanzees (21–23).
The autosomal cross-over hotspots analyzed to date show sperm RFs ranging from 0.0005% (6) to 0.14% (5). These hotspots were identified in regions that were not unusually active in meiotic recombination as judged from linkage maps. It is therefore likely that the most active hotspots have yet to be characterized. Indeed, an intense hotspot with 1.1% RF in sperm has been found in mice despite extremely limited surveys of the mouse genome (24). We now expand the current repertoire of human cross-over hotspots by targeting sperm cross-over assays to short intervals showing the most extreme LD breakdown in HapMap genotypes. This panel of “superhotspots” will provide a valuable resource for population geneticists to explore the relationship between recombination and DNA diversity. It will also aid further studies into recombination through the analysis of frequencies and distributions of cross-overs and gene conversions, helping to elucidate factors contributing to the regulation and evolutionary turnover of human cross-over hotspots.
Results and Discussion
Selecting Strong LD Hotspots from Genotype Data.
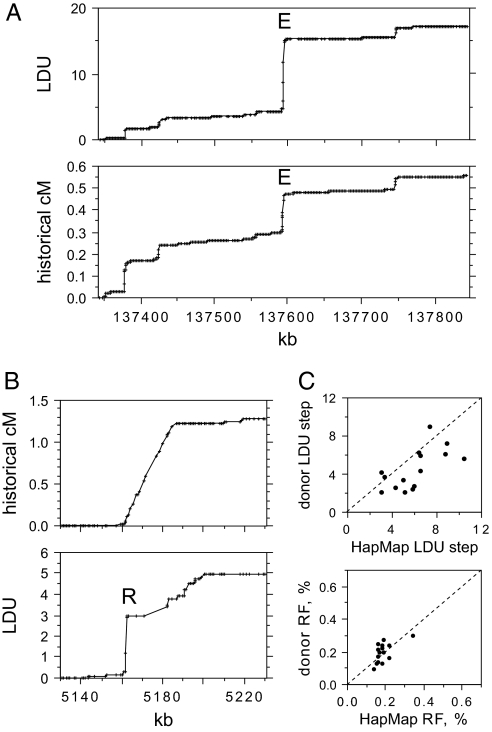
Strong recombination hotspots should create intervals of very substantial or complete breakdown of marker association. Such intervals can be identified by LD mapping (25), which provides a profile of the rate of LD breakdown along a chromosome in linkage disequilibrium units (LDUs). LD maps show a good correlation at the megabase level with linkage maps (26), and also with historical cross-over rates estimated by coalescent analysis of genotype data (17). We therefore constructed LD maps of all of the human autosomes by using Phase II HapMap genotype data (16) on 2.4 million single-nucleotide polymorphisms (SNPs) from 60 individuals from a Utah population with northern and western European ancestry (CEU). We scanned these maps for intervals of 5 kb containing a strong (>3) LDU step and good SNP density (>5 genotyped SNPs) and compared these intervals with coalescent analysis data available from HapMap (Fig. 1A). Fifteen strong LD hotspots (A–Q), each characterized by a major LDU step and substantial historical activity, were selected for cross-over analysis in sperm. We also analyzed the most active LD hotspot reported by HapMap (16) (historical RF estimated at 1.2% by coalescent analysis). This LD hotspot is, however, unusually broad (25 kb) (Fig. 1B), and the LD map appeared to resolve it into a moderately strong LD hotspot 2.5 kb wide (R; chosen for further analysis) followed apparently by a succession of other weaker LD hotspots.
Fig. 1.
Identifying strong candidate recombination hotspots from genotype data. (A) Metric LD map of a 500-kb region of chromosome 8 determined from Phase II HapMap genotypes of 60 unrelated CEU individuals compared with the historical recombination map estimated by coalescent analysis of the same data and shown below (data in cM taken from HapMap). The strong LD hotspot E is marked. (B) Similar analysis of a 100-kb interval on chromosome 20 containing the strongest LD hotspot reported by HapMap (16). (C) LDU step sizes and historical recombination activity at the 15 strong LD hotspots A–Q selected for sperm cross-over analysis, estimated from Phase II HapMap data and from genotypes of 94 north European semen donors. HapMap and donor LDU steps were both estimated across the same 15-kb interval genotyped in donors. LD hotspot R was omitted because of uncertainties in historical activity (B) and lack of markers preventing HapMap LDU step estimation in the 15-kb interval.
Each of the 16 targets was regenotyped in a panel of 94 north European semen donors by using HapMap SNPs plus additional markers in dbSNP. The LDU step at each target was confirmed, although 12 showed a drop in LDU step size, in some cases substantial (Fig. 1C). This drop is significant (paired t test, P = 0.005) and is caused in part by increased marker density closing “holes” (intervals of free association between adjacent markers) in the HapMap LD map (26) and reducing step size. Ascertainment bias probably also contributes to this reduction. Extreme LDU steps will tend to arise at those hotspots where haplotypes sampled in CEU HapMap individuals happen by chance to overinflate LDU step size; genotyping additional individuals will remove this bias and thus reduce step size. In contrast to LDU, historical cross-over rate estimates from coalescent analysis were not significantly different between CEU individuals and north European semen donors (P > 0.05) (Fig. 1C) but showed little variation between hotspots, with most showing a historical RF of ≈0.2%.
Sperm Cross-Over Profiles at Strong LD Hotspots.
Sperm DNA was assayed for cross-over molecules across each of the 16 LD hotspots, with each hotspot analyzed in three different informative men to test for cross-over frequency variation (6). Reciprocal cross-overs (aB and Ab cross-overs in a man heterozygous for haplotypes AB and ab) were analyzed separately in each man to verify cross-over frequency estimates, which proved to be very reproducible (see Materials and Methods) and to test for reciprocal cross-over asymmetry (19). Excluding a few instances of unusually low cross-over frequencies, ≈230 cross-overs were typically analyzed per hotspot per man. This survey yielded in total 11,200 cross-overs, all of which were mapped to locate cross-over breakpoints.
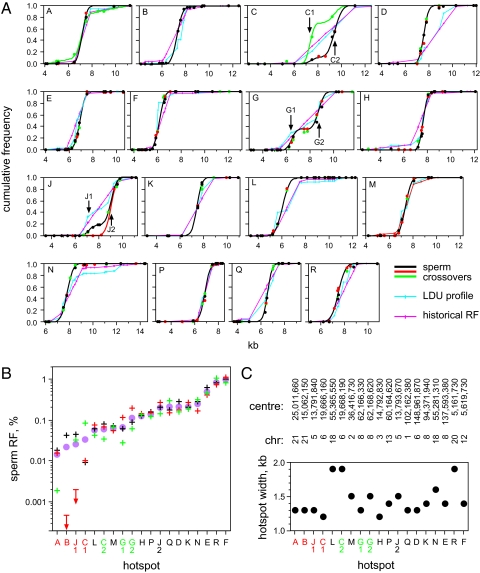
Each test interval showed a highly nonrandom distribution of cross-overs (Fig. 2A). In most cases (13 of 16), these distributions were consistent with a single cross-over hotspot. Targets C, G, and J each showed a broader distribution that, in men with sufficient marker density, could be seen to resolve into two hotspots in each case separated by 2 kb. This survey therefore yielded a total of 19 sperm-verified cross-over hotspots.
Fig. 2.
Cross-over hotspots identified by sperm typing. (A) Cumulative frequency of sperm cross-overs across each test interval (A–R) established for three informative men (black, red, green circles) per target. Each man was assayed for reciprocal cross-overs, and the results were combined. Only two men were available for typing at target B; one showed a very low cross-over frequency and is not shown. Best-fit cumulative normal distributions (6) are shown in black or in different colors for men showing unusual distributions. These distributions are compared with LDU profiles and historical recombination profiles determined from genotypes of 94 semen donors and normalized to 1 for each test interval. Double hotspots are arrowed. (B) Sperm RF per hotspot for each man, color coded as in A, plus mean RF (circle). Hotspots are ranked by mean activity. Hotspots labeled in red show major variation in activity between men; those marked in green showed weaker variation with a 2- to 4-fold difference in activity between the most and least active man. (C) Hotspot widths within which 95% of cross-overs occur, estimated from best-fit normal distributions, together with chromosomal coordinates of hotspot centers in the National Center for Biotechnology Information 36 assembly of the human genome.
Some Strong LD Hotspots Are Very Active in Sperm Cross-Over.
Despite the similar estimates of LDU step size and historical activity at these hotspots, they showed a wide range of sperm cross-over frequencies, varying from 0.015% to 1.0% (median 0.13%) (Fig. 2B). These activities are considerably higher than at previously characterized human autosomal hotspots (0.0005–0.14%, median 0.011%) (5–11, 13), with eight of the hotspots being the most active yet described. This elevated activity at the present hotspots is significant (Mann–Whitney test, P = 0.0005), indicating that our strategy of identifying very active hotspots from intervals of extreme LD breakdown was successful. Although these hotspots are very active, their flanking DNA is as suppressed for recombination as for previously reported hotspots [only 55 cross-overs mapped outside hotspots, giving a mean RF of 0.16% per Mb, consistent with previous estimates (6, 8, 13)].
LD Profiles Are an Imperfect Surrogate for Cross-Over Profiles.
Sperm cross-over profiles can, with sufficient SNP marker density, locate hotspot centers with considerable accuracy, within ±50 bp or less (6). Comparison of sperm profiles with LD maps and historical rate profiles (Fig. 2A) showed an erratic relationship between DNA diversity and cross-over. In some cases (e.g., hotspot P), sperm and diversity profiles were very closely congruent. In other cases (e.g., hotspot D), there were significant discrepancies with the hotspot center mislocated by up to 1 kb. Coalescent analysis by using LDhat (17) did not have the resolution to resolve any of the double hotspots, whereas LD mapping only succeeded in detecting a doublet at hotspot J. Thus, diversity analyses would interpret at least two of these regions (C, G) as a single broad hotspot centered within a region between sperm hotspots that is in fact recombinationally suppressed. Finally, LD mapping identified a second putative hotspot at 12 kb in target N (LDU step = 0.74). However, only three cross-overs were seen in this region in 240,000 molecules screened, indicating that this hotspot, if it exists in the men tested, is extremely weak (RF 0.001%).
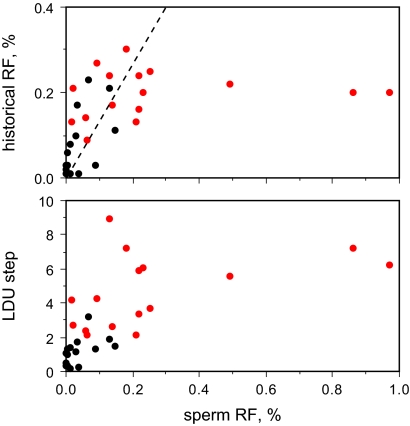
The quantitative relationship between sperm cross-over frequency and both LDU step size and historical rates estimated by coalescent analysis was far from perfect (Fig. 3). Both population estimates appeared to plateau above 0.2% RF in sperm, presumably because of free association arising through saturation of LD breakdown. Below 0.2%, the overall trend for sperm cross-over frequencies followed that predicted from historical rates. There was, however, substantial scatter, with individual hotspots showing anywhere from 30-fold less sperm activity, compared with historical rates, to a 5-fold excess. It is unclear whether these discrepancies reflect imperfections in historical rate estimation by coalescent analysis or instead arise through individual- and sex-specific differences in cross-over activity at specific hotspots (10) together with rapid evolutionary turnover of hotspots (8, 9, 13, 19, 21–23). In any event, neither LD mapping nor historical rate estimation is a good predictor of current sperm cross-over activity.
Fig. 3.
Relationship between DNA diversity and sperm cross-over activity. Historical cross-over activity and LDU step size were estimated from semen donor genotype data for each of the 16 strong LD hotspot regions (in red), with double hotspots combined, and for 15 previously characterized sperm hotspots genotyped in the same panel of semen donors (in black) (5–13). The dotted line shows the expected relationship between sperm and historical activity, corrected for the greater mean cross-over frequency seen in female meiosis (27).
Properties of Sperm Cross-Over Hotspots.
All hotspots showed fairly uniform widths of 1.2–1.9 kb (mean 1.45 kb; Fig. 2C), very similar to the widths of previously characterized but less active hotspots (5–7, 9–13). Thirteen were intergenic, whereas the remaining six were in introns, with hotspots L and N located in the same gene (CCBE1) just 84 kb apart. Variants of the motif CCTCCCTNNCCAC, reported as being associated with LD hotspots (16), were found within 70 bp of the center of hotspots C2, D, F, K, L, M, and P (37% of hotspots), in two cases (D, K) mapping exactly at the center defined by sperm typing. Although each of these associations was significant (P < 0.05), such motifs were not predictive of hotspot location, with examples of equally, if not better, matched motifs being present in the recombinationally inert regions flanking all of these hotspots. In summary, the properties of these very active hotspots in terms of width, genomic location, and sequence motifs are very similar to those of previously characterized but less active hotspots (5–13).
Complex Cross-Overs.
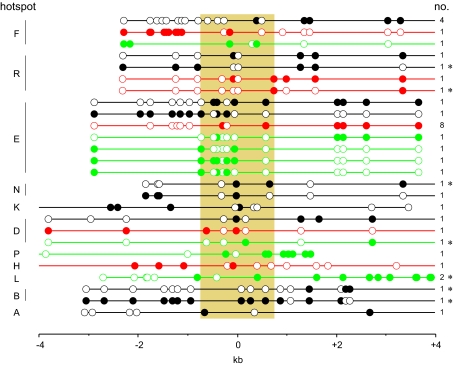
As with previously characterized sperm cross-overs (5–13), the great majority of exchanges were simple, with each cross-over mapping to a single interval between heterozygous SNPs. However, 37 of the 11,200 mapped cross-overs (0.33%) were more complex, showing haplotype switching near the site of exchange presumably arising through patchy repair of heteroduplex DNA generated during recombination (Fig. 4). Most of these events were located within the hotspot, but 22% of them (8 of 37) extended beyond the hotspot, reminiscent of the complex cross-overs seen in mismatch repair-deficient mice (28). Given that different hotspots were analyzed in different men, it was not possible to test whether these complex events were preferentially restricted to specific men nor whether they might associate with impaired mismatch repair functions. These complex events were, however, not randomly distributed across hotspots (P < 0.001), with the most active hotspots E, F, and R showing the highest proportion of complex cross-overs (1.1%, compared with 0.15% at the remaining hotspots). The reason for this correlation is unclear, but it may in part be the result of the higher mean number of markers within these very active hotspots (3.6 per hotspot per man vs. 2.2 in the remaining hotspots) allowing short conversion patches to be more readily detected.
Fig. 4.
Complex cross-over molecules recovered from sperm DNA. SNPs are indicated by circles, filled for one progenitor haplotype or empty for the other. Each hotspot is indicated on the left, the man of origin of each cross-over is color coded as in Fig. 2A, and the number of cross-overs of each class shown on the right. Cross-overs are aligned with respect to the hotspot centers, with the mean hotspot width of 1.45 kb indicated by shading. Cross-overs marked with an asterisk show haplotype switching extending outside the hotspot.
Polymorphism of Hotspot Activity.
Four hotspots (A, B, C1, and J1) showed major (>10-fold) variation in cross-over frequency between the men tested (Fig. 2B). Three other hotspots (C2, G1, and G2) showed minor (2- to 4-fold) interindividual variation in activity. These variations in RF are significant (P < 0.001), but we cannot exclude the possibility that such minor variation might arise from subtle shifts in DNA quality and/or PCR efficiency affecting cross-over recovery. Hotspots B and J1 showed the most extreme variation, with one man in each case showing no cross-overs mapping to the hotspot region, indicating substantial or complete suppression of cross-over activity (suppression >90-fold and >30-fold at B and J1, respectively; P = 0.95). Only one example of presence/absence polymorphism of a hotspot has been previously reported (13).
Curiously, variation in cross-over activity was restricted to the less active hotspots showing <0.1% RF (Fig. 2B). This restriction is significant (Mann–Whitney test, P = 0.0047 if marginally polymorphic hotspots are scored as negative or P = 0.018 if positive). It therefore appears that hotspot polymorphism associates preferentially with weaker hotspots. Data on the current hotspots plus previously characterized hotspots (5–13) indicate that these polymorphisms are common, with ≈55% of weaker hotspots (12 of 22) showing evidence of cross-over frequency variation between men.
It is puzzling that strong hotspots show less variation in activity between individuals. Intense cross-over activity should accelerate the spread and fixation of recombination-suppressing mutations within the hotspot through preferential overtransmission of such variants to recombinant progeny (8, 19). This predicts that the most intense hotspots should be the most ephemeral in human populations and thus the most likely to show activity variation between individuals (20). These hotspots showed no unusual features, and we therefore have no explanation for this apparent paradox; but any model of how hotspots arise and persist in the face of meiotic drive in favor of recombination suppressors will need to take this phenomenon into account.
Activity Polymorphism and Biased Gene Conversion.
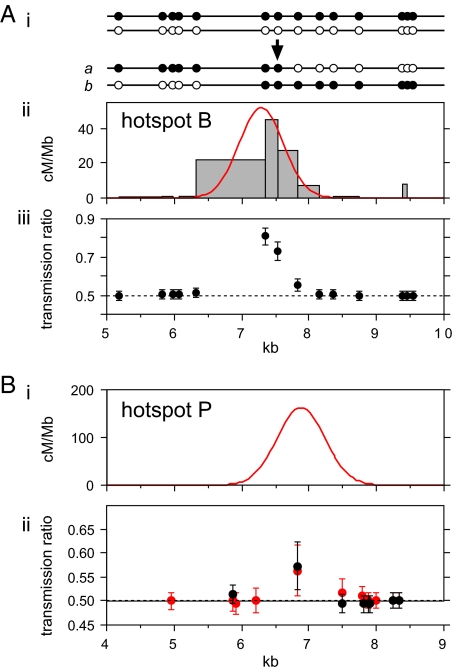
If the two haplotypes in a man differ in their frequency of recombination initiation, then disparities between reciprocal cross-over distributions will arise, with systematic overtransmission of markers from the suppressed haplotype into cross-over progeny (19). Two hotspots (B and J1) each showed very strong disparity in at least one man, with markers close to the hotspot center showing strongly distorted transmission ratios (83:17) into cross-over progeny (Fig. 5A). Similar levels of segregation distortion have been seen at other hotspots (8, 19). Hotspots B and J1 were also the two hotspots that showed presence/absence polymorphism between men (Fig. 2B). This association is significant (P = 0.006) and provides further evidence that strong transmission distortion is the result of men with the hotspot carrying both active and suppressed haplotypes (with inactive men homozygous for suppressed haplotypes) and is not solely caused by strong mismatch repair biases acting on heteroduplex DNA generated during recombination (19).
Fig. 5.
Biased transmission of hotspot markers to cross-overs. (A) Strong transmission distortion in hotspot B. (i) The two haplotypes in the man assayed, plus typical cross-overs in orientation a and b. (ii) Cross-over activity in each marker interval, with the best-fit normal distribution for a plus b cross-overs indicated in red. (iii) Frequency of alleles from the black haplotype in reciprocal cross-overs, normalized to equal numbers of a plus b cross-overs and with 95% confidence intervals indicated. (B) Weak transmission distortion in hotspot P. (i) Best-fit normal distribution of a plus b cross-overs (Fig. 2A). (ii) Transmission frequencies in combined a plus b cross-overs in two men, color coded as in Fig. 2A.
A second, more subtle, type of transmission distortion was also observed. Twenty-three cross-over assays at hotspots excluding B and J1 had markers sufficiently close to the hotspot center (<100 bp) to allow asymmetry to be tested. Of these assays, five showed significant asymmetry (P < 0.05 in each case; P = 0.0002 for the entire dataset) but with very minor biases in transmission ratios (56:44) (Fig. 5B). These biases were seen in one or two men at hotspots D, E, P, and Q and preferentially affected markers at the hotspot center. Such biases could arise through subtle disparities between haplotypes in recombination initiation rates; a difference of just 20% would be sufficient to create these biases but would be impossible to detect on the basis of the very minor (1.2-fold) predicted variation in sperm cross-over frequency between men. Alternatively, they could arise through subtle mismatch repair biases leading to biased gene conversion in cross-overs. Although the effect is subtle, the population consequences could be very significant, particularly at the most active hotspots; if meiotic drive affects a specific variant in the hotspot, then the population level of transmission distortion (up to 50.015:49.985 for the very active hotspot E) is strong enough essentially to guarantee eventual fixation of the overtransmitted variant (8, 19). Even more subtle biased gene conversion might occur during crossing-over, but much larger surveys of cross-over molecules would be needed to detect it.
Summary.
This survey has more than doubled the number of human meiotic recombination hotspots characterized by direct sperm analysis. Features established from studies of less active hotspots, such as narrow (1–2 kb) width, symmetric and quasinormal distribution of cross-over exchange points, generally simple exchange events and hotspot clustering, fully apply to these highly active hotspots. Comparisons of cross-over profiles with LD patterns show that population diversity data are excellent predictors of hotspots and can generally locate them with reasonable accuracy but are poor at predicting hotspot intensity. This in turn implies that the present survey has not detected all of the most intense hotspots in the human genome. It is likely that a full survey will only be possible through information gained, not from population level DNA diversity data, but from very high-resolution genome-wide mapping of cross-overs detected by extensive family analyses. However, current surveys lack the numbers of meioses and marker density required to locate reliably even the most active hotspots at the kilobase level (2).
The hotspots in this survey establish that variation between men in hotspot activity is common, particularly at less active hotspots, and provide examples of on/off polymorphisms suitable for in-depth analysis to determine the causes of hotspot appearance and disappearance (9, 13, 19, 21–23). They have further revealed extreme and also more subtle biased gene conversion processes operating during crossing-over, both of which could substantially influence the population behavior of markers within hotspots. These very active hotspots will provide a valuable resource for characterizing nonexchange gene conversion events within cross-over hotspots (29), and also for beginning to explore recombination initiation and the processing of recombination intermediates in human meiosis.
Materials and Methods
DNA Samples.
Blood and semen samples were collected with approval from the Leicestershire Health Authority Research Ethics Committee and with informed consent. DNAs were extracted and manipulated under conditions designed to minimize the risk of contamination as described in ref. 7. Routine genotyping, optimization of allele-specific primers, and linkage phasing were performed on genomic DNAs that had been whole-genome-amplified by using a GenomiPhi HY DNA amplification kit (GE Healthcare Bio-Sciences).
LD Mapping and Coalescent Analysis.
LDMAP (http://cedar.genetics.soton.ac.uk/pub/PROGRAMS/LDMAP) was used to create genome-wide LD maps in nonoverlapping 500-SNP sections by using 2,382,648 SNPs from 60 unrelated CEU individuals from the nonredundant filtered Phase II HapMap dataset (release 20) (http://www.hapmap.org). LD hotspots A–D were selected from LD maps constructed from early releases of Phase II HapMap data for chromosomes 6 and 21. Hotspots E–Q were selected from Phase II HapMap LD maps covering all autosomes combined with Phase I coalescent analysis information on historical cross-over activity available through HapMap. Strong LD hotspots were identified on the basis of a major (>3) LDU step and high historical activity within a 5-kb window, then ranked by the product of LDU step and historical rate. Hotspots E–Q were chosen from the top 19, with others rejected because of low SNP marker density flanking the LD hotspot. LD hotspots A–D were ranked >500th, 87th, 25th, and 10th, respectively, on this list. More recent data available from Phase II HapMap (16) showed little difference between Phase I and II historical rates at these hotspots. Coalescent analysis was carried out on targets regenotyped in semen donors by using LDhat (17) (http://www.stats.ox.ac.uk/∼mcvean/LDhat/) with 3,000,000 iterations, sampling every 2,000th iteration and discarding the initial 10% of iterations as the burn-in period. Historical RFs were estimated assuming an effective population size of 10,000.
Regenotyping Semen Donors.
A 15-kb interval spanning each LD hotspot was amplified by long PCR as three overlapping amplicons and regenotyped on a panel of 94 unrelated semen donors of north European origin by allele-specific oligonucleotide (ASO) hybridization as described in ref. 10. All Phase II HapMap SNPs were typed, plus some additional SNPs in dbSNP, yielding on average 26 HapMap SNPs plus 12 additional informative SNPs per target. Target sequence information and donor genotypes are available at www.le.ac.uk/genetics/ajj/superhotspots.
Sperm Cross-Over Assays.
For each LD hotspot, three semen donors were chosen for analysis with suitable SNP heterozygosities flanking the hotspot that allowed cross-over molecules to be recovered by repulsion-phase allele-specific long PCR (10), plus sufficient internal markers to allow cross-over breakpoints to be mapped. In total, 34 men were assayed for cross-overs, at one to three LD hotspots per man. Assay intervals were 5.5–9.6 kb long depending on marker location. Allele-specific primers were optimized by PCR-amplifying DNA from men homozygous for the correct or incorrect allele at varying annealing temperatures. SNP markers across the LD hotspot were phased in each man by allele-specific PCR amplification between selector SNPs upstream and downstream of the hotspot.
Cross-over DNA molecules were selectively amplified from sperm DNA by nested repulsion-phase allele-specific long PCR across the hotspot by using methods described in ref. 10. PCR products were analyzed by agarose-gel electrophoresis and staining with ethidium bromide, allowing each PCR to be scored as positive or negative for a cross-over molecule. Cross-over breakpoints were mapped by reamplifying PCR products using PCR primers located internally to the allele-specific primer sites and typing these PCR products by ASO hybridization (10). Primer sequences and cross-over assay conditions for each hotspot are available at www.le.ac.uk/genetics/ajj/superhotspots.
The cross-over frequency at each hotspot was initially estimated by a pilot assay on pools of sperm DNA containing 50–1,600 amplifiable molecules of each haplotype per PCR. Assay specificity was verified by parallel analysis of blood DNA; no mitotic exchanges were seen at any of the hotspots. The full-scale (96-well) assay used the pilot cross-over frequency estimate to amplify pools of sperm DNA of various size containing 0.7–1.8 cross-over molecules per PCR, yielding typically 110 cross-overs per plate. Poisson correction of the numbers of cross-overs of each type, identified by mapping exchange points, to take into account DNA pools containing more than one cross-over molecule is described in ref. 10. A single-molecule PCR efficiency of 50% (one amplifiable DNA molecule of each haplotype per 12 pg of sperm DNA) was assumed throughout, based on extensive previous data on single-molecule long PCR (6, 10).
Reciprocal a and b cross-over assays using independent sets of allele-specific primers (Fig. 5) gave very similar RFs with a median difference of 1.2-fold (range 1.0–1.7 over 42 different reciprocal assays). After Bonferroni correction, none of the differences was significant (P > 0.05), establishing that RF estimates were robust. Reproducibility was also tested by reassaying three men for cross-overs using different sperm DNA preparations; again, indistinguishable RF estimates were obtained (1.1- to 1.3-fold differences between assays).
Screening Cross-Over Hotspots for Sequence Motifs.
The sequence of each hotspot interval assayed for cross-overs was progressively scanned for motifs that showed 0, 1, 2, 3 … mismatches with the sequence CCTCCCTNNCCAC reported to be associated with LD hotspots (16), and at each level of mismatch the distance between the sperm cross-over hotspot center and the nearest motif was measured. The probability that any of the motifs detected at each level would map by chance within this distance of the hotspot center was then estimated, and a significant association was declared at P < 0.05. These motifs are shown on sequences available at www.le.ac.uk/genetics/ajj/superhotspots.
Acknowledgments.
We thank Jane Blower (Leicester Royal Infirmary) and volunteers for providing blood and semen samples; Rita Neumann, Joan Sutherland and Julie Turner for technical assistance; and colleagues for helpful discussions. This work was supported by the Medical Research Council, Wellcome Trust Grant 081227/Z/06/Z, the Royal Society, and the Louis-Jeantet Foundation (A.J.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Kong A, et al. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 2.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 3.Chakravarti A, et al. Nonuniform recombination within the human β-globin gene cluster. Am J Hum Genet. 1984;36:1239–1258. [PMC free article] [PubMed] [Google Scholar]
- 4.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- 5.Holloway K, Lawson VE, Jeffreys AJ. Allelic recombination and de novo deletions in sperm in the human β-globin gene region. Hum Mol Genet. 2006;15:1099–1111. doi: 10.1093/hmg/ddl025. [DOI] [PubMed] [Google Scholar]
- 6.Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 7.Jeffreys AJ, Murray J, Neumann R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell. 1998;2:267–273. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 8.Jeffreys AJ, Neumann R. Factors influencing recombination frequency and distribution in a human meiotic crossover hotspot. Hum Mol Genet. 2005;14:2277–2287. doi: 10.1093/hmg/ddi232. [DOI] [PubMed] [Google Scholar]
- 9.Jeffreys AJ, Neumann R, Panayi M, Myers S, Donnelly P. Human recombination hot spots hidden in regions of strong marker association. Nat Genet. 2005;37:601–606. doi: 10.1038/ng1565. [DOI] [PubMed] [Google Scholar]
- 10.Jeffreys AJ, Ritchie A, Neumann R. High-resolution analysis of haplotype diversity and meiotic crossover in the human TAP2 recombination hotspot. Hum Mol Genet. 2000;9:725–733. doi: 10.1093/hmg/9.5.725. [DOI] [PubMed] [Google Scholar]
- 11.Kauppi L, Stumpf MP, Jeffreys AJ. Localized breakdown in linkage disequilibrium does not always predict sperm crossover hot spots in the human MHC class II region. Genomics. 2005;86:13–24. doi: 10.1016/j.ygeno.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 12.May CA, Shone AC, Kalaydjieva L, Sajantila A, Jeffreys AJ. Crossover clustering and rapid decay of linkage disequilibrium in the Xp/Yp pseudoautosomal gene SHOX. Nat Genet. 2002;31:272–275. doi: 10.1038/ng918. [DOI] [PubMed] [Google Scholar]
- 13.Neumann R, Jeffreys AJ. Polymorphism in the activity of human crossover hotspots independent of local DNA sequence variation. Hum Mol Genet. 2006;15:1401–1411. doi: 10.1093/hmg/ddl063. [DOI] [PubMed] [Google Scholar]
- 14.Tiemann-Boege I, Calabrese P, Cochran DM, Sokol R, Arnheim N. High-resolution recombination patterns in a region of human chromosome 21 measured by sperm typing. PLoS Genet. 2006;2:e70. doi: 10.1371/journal.pgen.0020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McVean GA, et al. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 18.Myers S, et al. The distribution and causes of meiotic recombination in the human genome. Biochem Soc Trans. 2006;34:526–530. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- 19.Jeffreys AJ, Neumann R. Reciprocal crossover asymmetry and meiotic drive in a human recombination hot spot. Nat Genet. 2002;31:267–271. doi: 10.1038/ng910. [DOI] [PubMed] [Google Scholar]
- 20.Coop G, Myers SR. Live hot, die young: Transmission distortion in recombination hotspots. PLoS Genet. 2007;3:e35. doi: 10.1371/journal.pgen.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ptak SE, et al. Fine-scale recombination patterns differ between chimpanzees and humans. Nat Genet. 2005;37:429–434. doi: 10.1038/ng1529. [DOI] [PubMed] [Google Scholar]
- 22.Ptak SE, et al. Absence of the TAP2 human recombination hotspot in chimpanzees. PLoS Biol. 2004;2:e155. doi: 10.1371/journal.pbio.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winckler W, et al. Comparison of fine-scale recombination rates in humans and chimpanzees. Science. 2005;308:107–111. doi: 10.1126/science.1105322. [DOI] [PubMed] [Google Scholar]
- 24.Guillon H, de Massy B. An initiation site for meiotic crossing-over and gene conversion in the mouse. Nat Genet. 2002;32:296–299. doi: 10.1038/ng990. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis N, et al. The first linkage disequilibrium (LD) maps: Delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci USA. 2002;99:2228–2233. doi: 10.1073/pnas.042680999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tapper W, et al. A map of the human genome in linkage disequilibrium units. Proc Natl Acad Sci USA. 2005;102:11835–11839. doi: 10.1073/pnas.0505262102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matise TC, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20:563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 29.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36:151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]