Abstract
Bachground
Activating BRAF mutations are present in approximately 50% of melanomas. Although different downstream target genes of the most common mutant V600E have been identified, the contribution of activating BRAF mutations to malignant transformation needs further clarification.
Methods
Microarray gene analysis was performed for human melanoma cell lines harboring BRAFV600E mutations in comparison to cell lines without this mutation.
Results
This analysis revealed a more than two fold down-regulation of 43 and an increase of 39 gene products. BAALC (Brain and acute Leukaemia, cytoplasmatic) was most prominently regulated, since it was up-regulated in mutated cell lines by a mean of 11.45. Real time PCR analyses with RNA from melanoma cell lines (n = 30) confirmed the BRAF-activation dependent up-regulation of BAALC.
Conclusion
BAALC, which has been associated with cell dedifferentiation and migration, may function as a downstream effector of activating BRAF mutations during melanomagenesis.
Background
Activating mutations of the protooncogene BRAF have been observed in approximately fifty percent of malignant melanomas [1]. The V600E mutant accounting for over ninety percent of these mutations, obviates the requirement for segment phosphorylation of the T599 and S602 residues which is essential for a regular activation of BRAF [2,3]. Thus, the BRAFV600E mutation leads to a continuous stimulation of the MAP kinase cascade which results in a variety of cellular changes such as proliferation and dedifferentiation [4]. However, the role of activating BRAF mutations with respect to course and stage of melanoma is still not defined. On the one hand, comparable incidence of activating BRAF mutations in invasive cutaneous melanomas and benign melanocytic nevi indicate that BRAF mutations alone are insufficient to cause malignant transformation [2,3]. In addition, low frequency of BRAF mutations in radial growth phase melanomas, i.e. the early phase of melanoma progression, suggests a correlation with progression rather than initiation [5]. On the other hand, in patients with metastatic melanoma the presence of BRAF mutations is associated with a significantly poorer prognosis [6]. Moreover, introduction of BRAFV600E in melanocytes rendered them tumorigenic in nude mice [7], while another group demonstrated that BRAFV600E expression in human melanocytes induced cell senescence leading to cell cycle arrest. Hence, additional mutations have to occur to overcome this cell cycle arrest and make the cells tumorigenic [8]. Various genes have been identified as possible targets of the RAS/RAF/MAP kinase pathway. In this regard, microarray gene expression profiling allows measuring the expression of a large number of genes at the same time and thus providing a method for predicting the impact of oncogenes on the expression of possible down stream target genes. For example, microarray analyses of transfected murine embryonic fibroblasts with oncogene expressing adenoviruses helped to identify complex genetic alterations caused by genes such as HRAS, MYC and the E2F family [9]. Similarly, using standard cDNA microarray chips, gene expression signatures were reported for malignant melanoma cell lines harboring mutations in the BRAF gene when compared to wild type cell lines [10-12]. In this study, we used customized microarrays to identify additional target genes of the constitutively active MAPK pathway. The BRAFV600E dependent expression of a newly identified potential effector gene, BAALC (brain and acute leukaemia, cytoplasmatic) was confirmed by real time PCR analyses in complimentary experiments in melanoma cell lines.
Materials and methods
Melanoma cell lines and cell culture
16 human malignant melanoma cell lines harbouring BRAFV600E mutations as well as 9 melanoma cell lines with activating RAS mutations and 5 human malignant melanoma cell lines without mutations in these genomic sections were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (Table 1). Prior to RNA isolation the presence or absence of the V600E mutation was confirmed by direct sequencing PCR amplicons of BRAF exon 15.
Table 1.
BRAF and RAS mutational status of the analyzed cell lines.
| Wild type BRAF/RAS1 | BRAF mutated2 | RAS mutated3 |
| MV3 | FM88 (V600K) | MaMel79 (Q61K)5 |
| M19 | IF-6 (V600E) | MaMel91 (Q61K) |
| MaMel24 | Mel2a (V600E) | MaMel28 (Q61R) |
| MaMel71 | FM55 (V600E) | MaMel26a (Q61R) |
| MaMel15 | FM82 (V600E) | M26 (Q61R) |
| MaMel19 (V600E) | FM79 (Q61L) | |
| MaMel83 (V600E) | BLM (Q61R) | |
| MaMel86a (V600E) | MaMel5 (Q61R) | |
| MaMel85 (V600E) | MaMel27 (G12D) | |
| MaMel13 (V600E) | ||
| MaMel63a (V600E) | ||
| MaMel6 (V600E) | ||
| MaMel7 (V600E)5 | ||
| MaMel4 (V600E) | ||
| MaMel92 (V600E) | ||
| MaMel80a (V600E)5 |
1 Respective cell lines possess neither a BRAF nor a RAS mutation
2 Listed cell lines are mutated for BRAF. The mutation is given in brackets.
3 Listed cell lines are mutated for RAS. The mutation is given in brackets.
4 the MaMel cell lines are short term biopsy-derived cell lines demonstrating strong concordance with the mutational status of the corresponding tissues [24].
5 Cell lines are hemi- or homozygote for the respective mutation.
Gene expression analysis
Three melanoma cell lines (Mel2A, IF-6, FM88) with BRAFV600E mutation and three without this mutation (M26, MV3, M19) were examined by cDNA microarray analyses as described elsewhere [13]. In brief, total cellular RNA was isolated with the RNAeasy kit (QIAGEN, Hilden, Germany) and a subsequent DNase digestion was included. Hybridization probes were generated by indirect labeling with Cy3 and Cy5 dyes, using the CyScribe cDNA Post Labeling Kit RPN5660 (Amersham Biosciences Europe, Freiburg, Germany). All procedures were performed according to the manufacturer's instructions with 60 μg of RNA. Each experiment was performed as sandwich hybridization, i.e. instead of a cover slip a second microarray slide was used. The 4.6 K cDNA chips were generated by the group of M. Krause [http://www.imt.uni-marburg.de (Research, Microarray Unit)] and contained the GF200 set of Research Genetics cDNAs. Fluorescence labeled cDNA was spotted in duplicates. A flip color experiment was included. Samples were hybridized to microarrays for 16 h at 55°C. Chips were scanned with a GMS 418 fluorescent scanner (MWG-Biotech), and the images were analyzed with IMAGENE 3.0 software.
Quantitative RT-PCR analyses for BAALC
Relative expression of BAALC was determined by real time PCR analyses in Sybr green technology using the comparative ΔΔCT method. Total RNA was isolated from approximately 3 × 106 cells of human melanoma cell lines or from 25 six μm thick kryosections of melanoma tissue samples. Samples of total RNA were subjected to reverse transcription. Primers for BAALC were designed with Primer Express software (Applied Biosystems, Weiterstadt, Germany) and read as following: sense 5'-AGC-CGC-CGC-CAG-AGC-CGA-CAG-3'; antisense 5'-GG-GAT-CCA-GTG-CCG-TGA-AGG-3'. For the evaluation of BAALC expression the thermal cycling conditions comprised an initial denaturation step at 95°C for 10 min, then 43 cycles of three-step PCR including 94°C for 30 sec, 60°C for 30 sec and 72°C for 40 sec. GAPDH (Applied Biosystems) served as endogenous control. The relative expression level of BAALC, normalized to GAPDH and relative to the randomly selected human melanoma cell line FM79 (BRAF wt/wt) was calculated as with ΔΔCT = (CT BAALC, sample - CT GAPDH, sample) - (CT BAALC, FM79 - CT GAPDH, FM79). CT is defined as the cycle when the threshold level of fluorescence is reached.
Statistical analysis
In microarray analyses, the threshold value for up or down regulation of gene expression was defined as a more than two fold change compared to the mean value. Data of real time PCR studies were expressed as box-and-whiskers plot. Differences between the values were evaluated by Mann-Whitney test. p < 0.05 was considered to be significant.
Results and Discussion
Activating mutations of RAS and BRAF result in a constant activation of the MAP kinase pathway and eventually contribute to proliferation and dedifferentiation of cancer cells [2,14]. Although numerous target genes of the RAS/BRAF MAP-kinase pathway have been identified so far, the mechanism of action by which activated BRAF contributes to the malignant transformation of melanoma cells needs to be further elucidated. Therefore, we investigated gene expression of human melanoma cell lines harboring activating BRAF mutations (Mel2A, IF-6, FM88) by cDNA microarray analyses and compared their gene expression patterns to melanoma cell lines devoid of mutations in this genomic section (M26, MV3, M19). To this end, In BRAFV600E mutated cell lines a more than two fold decrease of 43 and increase of 39 gene products was detected (table 2). Due to the limited number of cell lines used in the microarray experiments each candidate revealed by this analysis should be confirmed in analysis of larger numbers of samples. Only such confirmation assays ascertain the respective genes to be effected by activated BRAF. Nevertheless, the most interesting gene found by these analyses was BAALC (brain and acute leukaemia, cytoplasmatic), since (i) it was highest up-regulated in all three cell lines harboring BRAF mutations by an average of 11.25 fold and (ii) it was not yet described by other groups analyzing the effect of BRAF mutation on gene expression in melanoma [10-12,15]. The latter, i.e. finding a prominent regulation of gene expression in one study but not in others, seems at first curious. However, it has been noticed that even when the same microarray chips are used, different results are obtained. The use of different expression pattern detection algorithms and lab-dependent differences were identified as source for such inconsistency. Lab-dependent differences include selection and treatment of samples to mRNA isolation, cDNA probe generation, chip hybridization conditions, chip lot and even the use of different chip scanners [16,17]. These findings sustain the notion that confirmation assays are mandatory in order to confirm different regulated expression for genes detected by microarray analysis.
Table 2.
Changes of gene expression in V600E mutated melanoma cell lines compared to wild type melanoma cell lines as revealed by microarray gene expression profiling
| Increase of gene expression | Decrease of gene expression | ||||
| symbol | gene name | fold change | symbol | gene name | fold change |
| SLC2A10 | solute carrier family 2 (facilitated glucose transporter), member 10 | 3,64 | SELK | selenoprotein K | 2,14 |
| ARG99 | ARG99 protein | 2,25 | LPHN2 | latrophilin 2 | 2,11 |
| SOD3 | superoxide dismutase 3, extracellular | 3,71 | ADRB2 | adrenergic, beta-2-, receptor, surface | 2,81 |
| MAN1C1 | mannosidase, alpha, class 1C, member 1 | 2,49 | LOC51159 | colon carcinoma related protein | 2,56 |
| DHX29 | DEAH (Asp-Glu-Ala-His) box polypeptide 29 | 2,09 | EGR2 | early growth response 2 (Krox-20 homolog, Drosophila) | 2,10 |
| C20orf45 | chromosome 20 open reading frame 45 | 2,70 | BUB3 | BUB3 budding uninhibited by benzimidazoles 3 homolog (yeast) | 2,00 |
| RPS6KA3 | ribosomal protein S6 kinase, 90 kDa, polypeptide 3 | 3,07 | VEZATIN | transmembrane protein vezatin | 2,07 |
| RGS1 | regulator of G-protein signalling 1 | 4,62 | SPIN | spindlin | 2,36 |
| COL4A2 | collagen, type IV, alpha 2 | 3,09 | STS | steroid sulfatase (microsomal), arylsulfatase C, isozyme S | 2,74 |
| ZCCHC6 | zinc finger, CCHC domain containing 6 | 2,07 | CDC2 | cell division cycle 2, G1 to S and G2 to M | 2,17 |
| CNTN1 | contactin 1 | 2,03 | MT1B | metallothionein 1B (functional) | 2,44 |
| MGC13105 | hypothetical protein MGC13105 | 2,03 | PITX2 | paired-like homeodomain transcription factor 2 | 2,35 |
| SUHW2 | suppressor of hairy wing homolog 2 (Drosophila) | 2,48 | PRKCG | protein kinase C, gamma | 2,58 |
| ICAM1 | intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | 2,14 | FABP5 | fatty acid binding protein 5 (psoriasis-associated) | 3,03 |
| BAALC | brain and acute leukemia, cytoplasmic | 11,45 | EPHB1 | EphB1 | 2,13 |
| OSBPL8 | oxysterol binding protein-like 8 | 2,13 | EGR1 | early growth response 1 | 2,97 |
| RAB24 | RAB24, member RAS oncogene family | 2,19 | PIGF | phosphatidylinositol glycan, class F | 2,16 |
| LOC54103 | hypothetical protein LOC54103 | 2,16 | DYRK3 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 | 2,55 |
| HSPC195 | hypothetical protein HSPC195 | 2,24 | RANBP6 | RAN binding protein 6 | 2,26 |
| EHD3 | EH-domain containing 3 | 2,56 | C10orf36 | chromosome 10 open reading frame 36 | 2,92 |
| CFLAR | CASP8 and FADD-like apoptosis regulator | 2,02 | DHRS8 | dehydrogenase/reductase (SDR family) member 8 | 2,05 |
| ZFP28 | zinc finger protein 28 homolog (mouse) | 2,04 | P8 | p8 protein (candidate of metastasis 1) | 2,08 |
| GSTM3 | glutathione S-transferase M3 (brain) | 3,20 | SCD4 | stearoyl-CoA desaturase 4 | 3,23 |
| PPP3R1 | protein phosphatase 3 (formerly 2B), regulatory subunit B, | 2,07 | RAB27B | RAB27B, member RAS oncogene family | 3,45 |
| APMCF1 | APMCF1 protein | 2,18 | ZFP36L1 | zinc finger protein 36, C3H type-like 1 | 2,22 |
| N33 | Putative prostate cancer tumor suppressor | 2,05 | SLC2A3 | solute carrier family 2 (facilitated glucose transporter), member 3 | 2,16 |
| ALDH1A1 | aldehyde dehydrogenase 1 family, member A1 | 5,29 | THBS1 | thrombospondin 1 | 2,07 |
| NAP4 | Nck, Ash and phospholipase C binding protein | 2,13 | FLJ90440 | hypothetical protein FLJ90440 | 2,62 |
| LZTS1 | leucine zipper, putative tumor suppressor 1 | 2,00 | C5orf13 | chromosome 5 open reading frame 13 | 2,13 |
| P2RY2 | purinergic receptor P2Y, G-protein coupled, 2 | 2,11 | MT1F | metallothionein 1F (functional) | 2,10 |
| APPBP1 | amyloid beta precursor protein binding protein 1, 59 kDa | 2,35 | CAMK4 | calcium/calmodulin-dependent protein kinase IV | 2,07 |
| SLC5A6 | solute carrier family 5 (sodium-dependent vitamin transporter), member 6 | 2,18 | DCT | dopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) | 2,15 |
| CITED1 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain | 2,67 | EFG1 | mitochondrial elongation factor G1 | 2,61 |
| FYN | FYN oncogene related to SRC, FGR, YES | 2,25 | LDB2 | LIM domain binding 2 | 2,24 |
| FDFT1 | farnesyl-diphosphate farnesyltransferase 1 | 2,42 | COL9A3 | collagen, type IX, alpha 3 | 4,45 |
| GDF11 | growth differentiation factor 11 | 2,44 | BAIAP1 | BAI1-associated protein 1 | 2,02 |
| FOXO1A | forkhead box O1A (rhabdomyosarcoma) | 2,38 | ALDH1A3 | aldehyde dehydrogenase 1 family, member A3 | 3,65 |
| PSPC1 | paraspeckle component 1 | 2,04 | HSJ001348 | cDNA for differentially expressed CO16 gene | 3,79 |
| CLCN5 | chloride channel 5 (nephrolithiasis 2, X-linked, Dent disease) | 2,10 | IRAK1 | interleukin-1 receptor-associated kinase 1 | 2,02 |
| NMNAT2 | nicotinamide nucleotide adenylyltransferase 2 | 2,86 | |||
| MT1G | metallothionein 1G | 2,98 | |||
| PRKCA | protein kinase C, alpha | 2,27 | |||
The product of the BAALC gene has been previously discussed to be involved in cell dedifferentiation and motility. In differentiated cells BAALC is almost exclusively expressed in the central nervous system and in other neuroectodermal derived tissues. Moreover, BAALC expression has been described for CD34 positive hematopoietic progenitor cells from the bone marrow [18]. Notably, loss of expression of BAALC during differentiation of hematopoietic progenitor cells suggests that it might function in sustaining an undifferentiated state of these cells [19,20]. Interestingly, however, BAALC expression in both normal and malignant astrocytes is increased upon differentiation, suggesting complementary functions in different cell types [21]. Nevertheless, it has been postulated that a series of genes, frequently expressed in progenitor cells of the neuroectodermal and the haematopoietic system, maintain the proliferative capacity while inhibiting differentiation [22] and BAALC may belong to this group of genes. Recent studies performed in patients with leukaemic malignancies indeed suggest a role of BAALC in tumorigenesis. For example, blast cells from 28% of patients with acute myeloid leukaemia and 65% with acute lymphatic leukaemia displayed an overexpression of BAALC. In addition, high BAALC expression was identified as an independent risk factor in acute myeloid leukaemia [20]. The examination of 13 human tumor cell lines of non-hematopoietic origin, however, revealed BAALC expression in only five cell lines derived from glioblastoma while BAALC was not detectable in any of the other neoplastic cells indicating a specific function of BAALC in some tumors only [18]. Furthermore, the expression of BAALC in developing and mature muscle cells in mice suggests a possible role of BAALC in cell locomotion or adhesion [23].
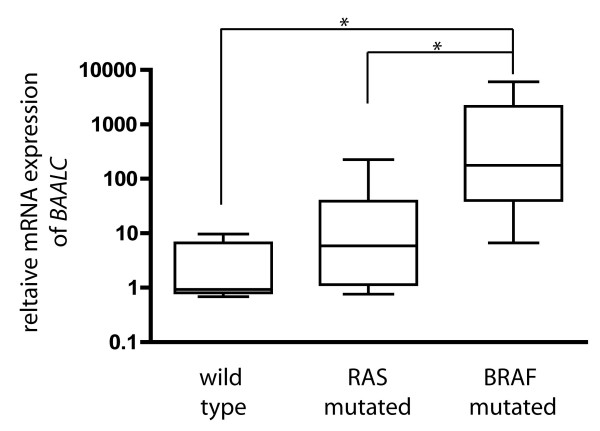
The distinct increase of BAALC expression in all three BRAFV600E mutated melanoma cell lines suggests a possible function of BAALC associated with activated BRAF mutations in melanoma and mediating cell dedifferentiation and motility. As mentioned before, complementary experiments using real time PCR analyses to measure the relative mRNA expression of BAALC have to be performed to confirm this correlation. To this end, BAALC expression was determined in 16 cell lines mutated for BRAF, 5 wild type BRAF melanoma cell lines as well as 9 melanoma cell lines in which the MAP kinase pathway was activated by RAS mutations. These analyses validated the first observations; BRAFV600E cell lines showed significantly (p < 0.01) elevated mRNA levels of BAALC (Fig. 1a) when compared to wild type cell lines. Notably, over-expression of BAALC was also significant when compared to RAS mutated cell lines (Fig. 1a). Importantly, most of the cell lines analyzed were short term cultures which should therefore resemble closely the parental tumor (table 1). Our results are in accordance with previous published data demonstrating that only a portion of regulated genes in cell lines with BRAF or NRAS mutations are common among the different mutations [10,11]. The different expression patterns might be ascribed to the differential capacity to receive input signals and to pass these on to various effectors.
Figure 1.
Relative mRNA expression of BAALC (brain and acute leukaemia, cytoplasmatic) in human malignant melanoma cell lines as revealed by real time PCR. 16 melanoma cell lines harboring BRAF mutation were compared to 9 cell lines with RAS mutation and 5 without any mutation in these genes. (* p < 0.01). As calibrator served a wild type melanoma cell line.
In summary, BAALC may function as an additional mediator of activating BRAF mutations. Future studies will have to clarify its exact role in malignant transformation of melanocytic lesions.
Contributor Information
David Schrama, Email: schrama_d@klinik.uni-wuerzburg.de.
Gunhild Keller, Email: keller_g@klinik.uni-wuerzburg.de.
Roland Houben, Email: houben_r@klinik.uni-wuerzburg.de.
Christian G Ziegler, Email: ziegler_c@klinik.uni-wuerzburg.de.
Claudia S Vetter-Kauczok, Email: vetter_c@klinik.uni-wuerzburg.de.
Selma Ugurel, Email: ugurel_s@klinik.uni-wuerzburg.de.
Jürgen C Becker, Email: becker_jc@klinik.uni-wuerzburg.de.
References
- Rodolfo M, Daniotti M, Vallacchi V. Genetic progression of metastatic melanoma. Cancer Lett. 2004;214:133–147. doi: 10.1016/j.canlet.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Kumar R, Angelini S, Snellman E, Hemminki K. BRAF mutations are common somatic events in melanocytic nevi. J Invest Dermatol. 2004;122:342–348. doi: 10.1046/j.0022-202X.2004.22225.x. [DOI] [PubMed] [Google Scholar]
- Gollob JA, Wilhelm S, Carter C, Kelley SL. Role of Raf kinase in cancer: therapeutic potential of targeting the Raf/MEK/ERK signal transduction pathway. Semin Oncol. 2006;33:392–406. doi: 10.1053/j.seminoncol.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Dong J, Phelps RG, Qiao R, Yao S, Benard O, Ronai Z, Aaronson SA. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res. 2003;63:3883–3885. [PubMed] [Google Scholar]
- Houben R, Becker JC, Kappel A, Terheyden P, Brocker EB, Goetz R, Rapp UR. Constitutive activation of the Ras-Raf signaling pathway in metastatic melanoma is associated with poor prognosis. J Carcinog. 2004;3:6. doi: 10.1186/1477-3163-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellbrock C, Ogilvie L, Hedley D, Karasarides M, Martin J, Niculescu-Duvaz D, Springer CJ, Marais R. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64:2338–2342. doi: 10.1158/0008-5472.CAN-03-3433. [DOI] [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, Horst CM van der, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Huang E, Ishida S, Pittman J, Dressman H, Bild A, Kloos M, D'Amico M, Pestell RG, West M, Nevins JR. Gene expression phenotypic models that predict the activity of oncogenic pathways. Nat Genet. 2003;34:226–230. doi: 10.1038/ng1167. [DOI] [PubMed] [Google Scholar]
- Pavey S, Johansson P, Packer L, Taylor J, Stark M, Pollock PM, Walker GJ, Boyle GM, Harper U, Cozzi SJ, Hansen K, Yudt L, Schmidt C, Hersey P, Ellem KA, O'Rourke MG, Parsons PG, Meltzer P, Ringner M, Hayward NK. Microarray expression profiling in melanoma reveals a BRAF mutation signature. Oncogene. 2004;23:4060–4067. doi: 10.1038/sj.onc.1207563. [DOI] [PubMed] [Google Scholar]
- Bloethner S, Chen B, Hemminki K, Muller-Berghaus J, Ugurel S, Schadendorf D, Kumar R. Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis. 2005;26:1224–1232. doi: 10.1093/carcin/bgi066. [DOI] [PubMed] [Google Scholar]
- Johansson P, Pavey S, Hayward N. Confirmation of a BRAF mutation-associated gene expression signature in melanoma. Pigment Cell Res. 2007;20:216–221. doi: 10.1111/j.1600-0749.2007.00375.x. [DOI] [PubMed] [Google Scholar]
- Zirn B, Hartmann O, Samans B, Krause M, Wittmann S, Mertens F, Graf N, Eilers M, Gessler M. Expression profiling of Wilms tumors reveals new candidate genes for different clinical parameters. Int J Cancer. 2006;118:1954–1962. doi: 10.1002/ijc.21564. [DOI] [PubMed] [Google Scholar]
- Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, Roth JA, Albelda SM, Davies H, Cox C, Brignell G, Stephens P, Futreal PA, Wooster R, Stratton MR, Weber BL. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- Kumar SM, Yu H, Edwards R, Chen L, Kazianis S, Brafford P, Acs G, Herlyn M, Xu X. Mutant V600E BRAF increases hypoxia inducible factor-1alpha expression in melanoma. Cancer Res. 2007;67:3177–3184. doi: 10.1158/0008-5472.CAN-06-3312. [DOI] [PubMed] [Google Scholar]
- Park T, Yi SG, Shin YK, Lee S. Combining multiple microarrays in the presence of controlling variables. Bioinformatics. 2006;22:1682–1689. doi: 10.1093/bioinformatics/btl183. [DOI] [PubMed] [Google Scholar]
- Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Comput Biol. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner SM, Austin JL, Leone G, Rush LJ, Plass C, Heinonen K, Mrozek K, Sill H, Knuutila S, Kolitz JE, Archer KJ, Caligiuri MA, Bloomfield CD, de la CA. BAALC, the human member of a novel mammalian neuroectoderm gene lineage, is implicated in hematopoiesis and acute leukemia. Proc Natl Acad Sci USA. 2001;98:13901–13906. doi: 10.1073/pnas.241525498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus CD, Tanner SM, Kusewitt DF, Liyanarachchi S, Choi C, Caligiuri MA, Bloomfield CD, de la CA. BAALC, a novel marker of human hematopoietic progenitor cells. Exp Hematol. 2003;31:1051–1056. [PubMed] [Google Scholar]
- Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, Caligiuri MA, Carroll AJ, Vardiman JW, Powell BL, Allen SL, Moore JO, Larson RA, Kolitz JE, de la CA, Bloomfield CD. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a cancer and leukemia group B study. Blood. 2003;102:1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- Moodbidri MS, Shirsat NV. Induction of BAALC and down regulation of RAMP3 in astrocytes treated with differentiation inducers. Cell Biol Int. 2006;30:210–213. doi: 10.1016/j.cellbi.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Ou J, Easterday MC, Dougherty JD, Jackson RL, Chen Z, Antoine H, Terskikh A, Weissman IL, Nelson SF, Kornblum HI. A genetic analysis of neural progenitor differentiation. Neuron. 2001;29:325–339. doi: 10.1016/S0896-6273(01)00209-4. [DOI] [PubMed] [Google Scholar]
- Satoskar AA, Tanner SM, Weinstein M, Qualman SJ, de la CA. Baalc, a marker of mesoderm and muscle. Gene Expr Patterns. 2005;5:463–473. doi: 10.1016/j.modgep.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Ugurel S, Thirumaran RK, Bloethner S, Gast A, Sucker A, Mueller-Berghaus J, Rittgen W, Hemminki K, Becker JC, Kumar R, Schadendorf D. B-RAF and N-RAS mutations are preserved during short time In Vitro propagation and differentially impact prognosis. PLoS ONE. 2007;2:e236. doi: 10.1371/journal.pone.0000236. [DOI] [PMC free article] [PubMed] [Google Scholar]