Abstract
Glycosyltransferases are involved in the biosynthesis of lipid-linked N-glycans. Here, we identify and characterize a mannosyltransferase gene from Arabidopsis thaliana, which is the functional homolog of the ALG3 (Dol-P-Man:Man5GlcNAc2-PP-Dol α1,3-mannosyl transferase) gene in yeast. The At ALG3 protein can complement a Δalg3 yeast mutant and is localized to the endoplasmic reticulum in yeast and in plants. A homozygous T-DNA insertion mutant, alg3-2, was identified in Arabidopsis with residual levels of wild-type ALG3, derived from incidental splicing of the 11th intron carrying the T-DNAs. N-glycan analysis of alg3-2 and alg3-2 in the complex-glycan-less mutant background, which lacks N-acetylglucosaminyl-transferase I activity, reveals that when ALG3 activity is strongly reduced, almost all N-glycans transferred to proteins are aberrant, indicating that the Arabidopsis oligosaccharide transferase complex is remarkably substrate tolerant. In alg3-2 plants, the aberrant glycans on glycoproteins are recognized by endogenous mannosidase I and N-acetylglucosaminyltransferase I and efficiently processed into complex-type glycans. Although no high-mannose-type glycoproteins are detected in alg3-2 plants, these plants do not show a growth phenotype under normal growth conditions. However, the glycosylation abnormalities result in activation of marker genes diagnostic of the unfolded protein response.
INTRODUCTION
In eukaryotes, secreted proteins may be modified on specific Asn residues by sugars upon entry into the endoplasmic reticulum (ER) in a process called N-glycosylation. The initial addition of N-glycan structure is to aid the folding process of the protein, and subsequent modifications of the N-glycans in the ER have a signaling function in the protein folding quality control mechanism. In mammals, N-glycans on glycoproteins that arise after further processing play crucial roles in many biological processes, and their structure and biosynthesis have been well studied (Kornfeld and Kornfeld, 1985; Fiedler and Simons, 1995; Ohtsubo and Marth, 2006). Plants also possess N-glycosylation, and the core structure of N-glycans in plants is similar to that found in mammals, but far less is known about their biosynthesis and biological function. N-glycan biosynthesis starts with the assembly of a precursor Man5GlcNAc2, linked to dolichol-phosphate (PP-dolichol) at the cytoplasmic side of the ER. Then, in a process not well understood, this precursor flips to the luminal side of the ER, where the residual four mannose and three glucose residues are added by distinctive glycosyl transferases in a stepwise manner (Snider et al., 1980; Helenius and Aebi, 2002). The lipid-linked Glc3Man9GlcNAc2-moiety is then transferred en bloc by the multisubunit oligosaccharyltransferase (OST) complex to the γ-amido group of selected Asn residues of nascent polypeptides during their translocation into the ER (Kornfeld and Kornfeld, 1985). By subsequent trimming reactions catalyzed by exoglycosidases of the ER and the Golgi apparatus, the so-called high-mannose-type (Man9GlcNAc2 to Man5GlcNAc2) glycans are generated. Removal of the glucose residues is part of a quality control process in the folding of newly synthesized glycoproteins (Parodi, 2000; Roth et al., 2003). Until the glycoprotein is correctly folded, the protein cycles between a deglucosylated and reglucosylated state by the concerted action of glucosidases, calnexin, calreticulin, and a UPD-Glc:glycoprotein glucosyltransferase (UGGT), which interacts with misfolded proteins. An accumulation of misfolded proteins in the ER triggers the unfolded protein response (UPR), through which, among others, genes encoding proteins that assist in protein folding are induced, such as protein disulfide isomerase (PDI), binding protein (BiP), calnexin, and calreticulin.
After removal of the three glucose residues and the four α(1,2)-linked mannose residues, the first obligatory step in complex-type glycan formation is catalyzed by the enzyme N-acetylglucosaminyltransferase (GnTI). In plants, complex-type glycans are characterized by a β(1,2)-xylose residue and/or an α(1,3)-fucose residue linked to the core glycan structure (Lerouge et al., 1998). A second terminal GlcNAc residue may be added to the mannose core; however, the terminal GlcNAc residues on N-glycans of plant glycoproteins, which are stored in the vacuoles, are removed by exoglycosidases, resulting in Man3XylFucGlcNAc2 complex-type glycans (Vitale and Chrispeels, 1984; Lerouge et al., 1998). Although these modifications of N-glycans seem to be well conserved within the plant kingdom, the biological function of these additions and trimmings of N-glycans in plants is not well understood. Since any of the above-described processing reactions may not go to completion, N-glycan structures, even on a single type of glycoprotein, can be heterogeneous and may include complex glycans and various high mannose structures (Sturm et al., 1987; Elbers et al., 2001).
Saccharomyces cerevisiae is often used as a model for the study of eukaryotic N-glycan processing, and mutants defective in the biosynthesis of N-glycans have lead to the characterization of ALG (for Asn-linked glycosylation) genes (Lehle et al., 2006). Many single alg mutations in S. cerevisiae do not result in a growth phenotype, but in combination with, for instance, a conditional defect in the OST activity, a growth phenotype may arise (Stagljar et al., 1994), which facilitates identification of the corresponding genes and pathway.
The gene ALG3 from S. cerevisiae encodes the enzyme Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase, which has been shown to be responsible for the first mannosylation step after the precursor glycan has flipped from the cytosolic face to the luminal side of the ER. The ALG3 mannosyltransferase uses Man-P-Dol as the glycosyl donor and converts Man5GlcNAc2-PP-Dol to Man6GlcNAc2-PP-Dol (Sharma et al., 2001). In an Δalg3 mutant of yeast, Man5GlcNAc2-PP-Dol accumulates (Huffaker and Robbins, 1983). However, the Man5GlcNAc2 can be transferred to the nascent polypeptide chain (Figure 1B) with low efficiency, resulting in an underglycosylation of secretory glycoproteins (Huffaker and Robbins, 1983; Verostek et al., 1993; Zufferey et al., 1995). Homologs of the yeast ALG3 gene have been identified in Pichia pastoris and Schizosaccharomyces pombe but also in the fruit fly Drosophila melanogaster and mammals (Huffaker and Robbins, 1982, 1983; Runge et al., 1984; Runge and Robbins, 1986; Kurzik-Dumke et al., 1997; Korner et al., 1999; Davidson et al., 2004). Little is known about lipid-linked oligosaccharide biosynthesis in plants, and no plant ALG-type genes involved in this part of N-glycosylation have been characterized to date. Here, we report the identification and characterization of a functional homolog of the S. cerevisiae ALG3 gene in Arabidopsis thaliana and provide a detailed analysis of the glycosylation pathway of an alg3 mutant plant.
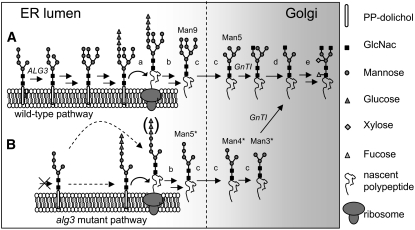
Figure 1.
Proposed N-Glycosylation in Arabidopsis, Starting with the Intermediate Man5GlcNAc2 Glycan at the Luminal Side of the ER.
(A) After extension by mannose and glucose residues and transfer of the oligosaccharide precursors by the OST complex onto specific Asn residues of nascent secretory proteins, N-linked glycans are further modified in the ER and Golgi by glycosidases and glycosyltransferases into more complex type plant N-glycans. Enzymes identified in plants to be involved in N-glycan biosynthesis are indicated by lowercase letters. a, OST subunits (DAD1, SST3, and DGL1); b, glucosidase I and II; c, mannosidase I; d, mannosidase II; e, N-acetylglucosaminyltransferase II, xylosyltransferase, and fucosyltransferase; ALG3, Dol-P-Man:Man5GlcNAc2-PP-Dol α1,3-mannosyl transferase; GnTI, N-acetylglucosaminyltransferase I (mutated in cgl plants).
(B) The Man5GlcNAc2 glycan in the alg3-2 mutant is either first glucosylated or directly transferred to the specific Asn residues of nascent secretory proteins (indicated by the dashed arrows). The asterisk indicates aberrant protein linked N-glycans. In the ER and Golgi, the aberrant Man5* glycan may be processed into regular complex-type plant N-glycans by glycosidases and glycosyltransferases.
RESULTS
Identification of the ALG3 Gene in Arabidopsis
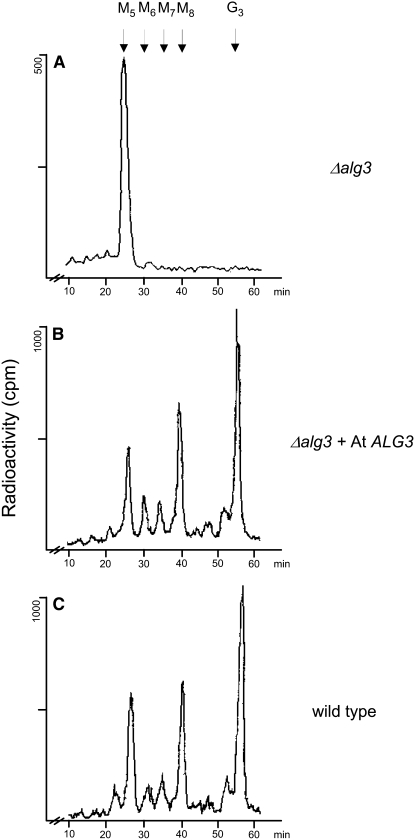
The ALG3 gene from S. cerevisiae and the human Not (for neighbor of tid) 56-like gene (Not56L) encode the α1,3-mannosyltransferase involved in the first lipid-linked glycan elongating step in the ER, resulting in the synthesis of Man6GlcNAc2-PP-Dol from Man5GlcNAc2-PP-Dol (Aebi et al., 1996; Korner et al., 1999; Sharma et al., 2001). Inspection of the Arabidopsis protein and genomic databases, based on amino acid sequences of the human Not56L and S. cerevisiae Alg3 proteins, resulted in the identification of a putative homolog encoded by the Arabidopsis genome. The cDNA encoding the putative ALG3 homolog was obtained from the ABRC, and its sequence was confirmed and designated as ALG3. To test whether the predicted At ALG3 open reading frame is indeed the functional homolog of yeast Alg3p and is able to replace it, we expressed the full-length At ALG3 cDNA under control of a constitutive promoter in a Δalg3 mutant strain of S. cerevisiae. This strain is characterized by the accumulation of Man5GlcNAc2-PP-Dol and an underglycosylation of glycoproteins, such as carboxypeptidase Y (CPY), that is ascribed to a reduced affinity of the yeast OST complex for the aberrant lipid-linked glycan form that accumulates in Δalg3. For analysis of lipid-linked glycans, yeast Δalg3 cells expressing At ALG3 or a control vector were metabolically labeled with [3H]mannose. The lipid-linked oligosaccharides were subsequently extracted and released from the lipid by acid hydrolysis for analysis by HPLC (Figure 2). In Δalg3 cells, lipid-linked oligosaccharides were mostly Man5GlcNAc2 (Figure 2A), as was shown previously (Aebi et al., 1996). By contrast, in cells expressing At ALG3, synthesis of lipid-linked saccharides occurred up to the fully assembled Glc3Man9GlcNAc2 (Figure 2B), similar to the lipid-linked glycan profile of wild-type yeast cells (Figure 2C), indicating that At ALG3 can complement the Δalg3 phenotype.
Figure 2.
Restoration of Lipid-Linked Oligosaccharide Biosynthesis in the Yeast Δalg3 Mutant by At ALG3.
(A) Analysis of lipid-linked oligosaccharides in Δalg3 yeast.
(B) Δalg3 transformed with the ALG3 homolog from Arabidopsis.
(C) Wild-type yeast cells.
Lipid-linked oligosaccharides were labeled by incorporation of 2-[3H]mannose. The [3H]oligosaccharides were released by mild acid hydrolysis and analyzed by HPLC. M5-8, Man5-8GlcNAc2; G3, Glc3Man9GlcNAc2.
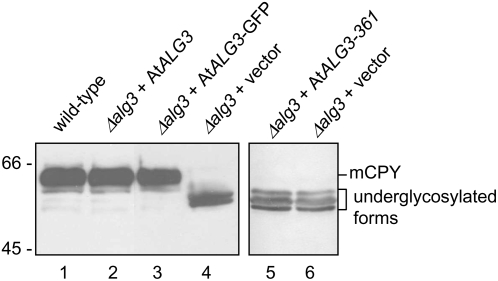
Since accumulation of the incomplete lipid-linked Man5GlcNAc2 precursor in Δalg3 cells results in inefficient transfer of glycans to proteins (Huffaker and Robbins, 1983; Verostek et al., 1993; Zufferey et al., 1995), the glycosylation status of CPY was analyzed in a Δalg3 strain, with and without complementation by At ALG3. Mature CPY in wild-type yeast carries four N-linked glycans migrating as a distinct band in SDS-PAGE. By contrast, glycosylation of CPY is severely reduced in the Δalg3 strain because of a reduced transfer of truncated oligosaccharides. The reduced N-glycosylation of CPY results in a lower mass and therefore higher mobility in SDS-PAGE gels (Figure 3, compare lanes 1 with 4). Expression of the At ALG3 cDNA, as well as a C-terminally fused green fluorescent protein (GFP)–tagged variant of At ALG3 in Δalg3 cells, restored CPY glycosylation (Figure 3, compare lanes 2 and 3 with lane 1), indicating that At ALG3 can also complement this Δalg3 phenotype.
Figure 3.
Restoration of CPY Glycosylation in the Yeast Δalg3 Mutant by At ALG3.
Protein gel blot analysis of CPY from wild-type yeast cells (lane 1) and Δalg3 cells transformed with either At ALG3 (lane 2), GFP-tagged At ALG3 (lane 3), truncated At ALG3 with a stop codon engineered directly after codon 361 (lane 5), or the vector control plasmid (lanes 4 and 6). The position of the mature form of CPY (mCPY) and of the underglycosylated forms is indicated on the right.
At ALG3 Localizes to the ER in Yeast and Arabidopsis
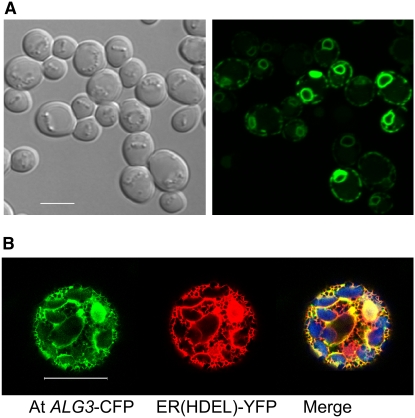
To determine the subcellular localization of At ALG3, we expressed At ALG3 fused to fluorescent protein in yeast and in Arabidopsis cells. In yeast, GFP fluorescence was observed around the nucleus and the plasma membrane (Figure 4A). This is typical of ER staining in yeast (Pichler et al., 2001) and suggests that At ALG3 localizes to the ER in yeast. For localization analysis in Arabidopsis, protoplasts were cotransfected with expression vectors containing cyan fluorescent protein (CFP)–tagged At ALG3 and YFP-HDEL, an established ER marker (Aker et al., 2006). A high degree of subcellular colocalization between the YFP and CFP fluorescence signals in these protoplasts indicates that the At ALG3 protein localizes to the ER also in plant cells (Figure 4B).
Figure 4.
Subcellular Localization of ALG3 in Yeast and Plants.
(A) Localization of the At ALG3-GFP fusion protein in yeast. Left panel, differential interference contrast optics (transmission); right panel, GFP fluorescence signal. Similar to ER staining, At ALG3-GFP fluorescence localizes to the nuclear envelope and below the plasma membrane. Bar = 5 μm.
(B) Colocalization of the At ALG3-CFP fusion protein with the ER(HDEL)-YFP marker in Arabidopsis protoplasts. Apparent labeling of the nucleus by both the At ALG3-CFP and the ER marker is caused by labeling of the ER around the nucleus. Left, fluorescence from the At ALG3-CFP construct; middle, fluorescence from the ER(HDEL)-YFP marker construct; right, overlay of CFP and YFP signals (yellow color combined with the chlorophyll signal [blue color]). Bar = 20 μm.
Sequence Analysis of ALG3
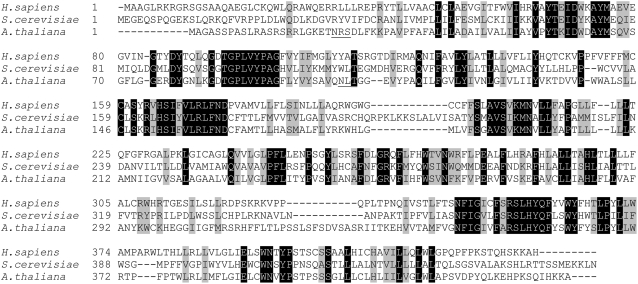
Complementing two glycosylation defects in the Δalg3 yeast strain provides evidence that At ALG3 is a functional homolog of Alg3p in S. cerevisiae. This is substantiated by the localization of At ALG3 to the ER in both yeast and Arabidopsis protoplasts. Comparison of the cDNA sequence of At ALG3 with the corresponding genomic sequence shows that the gene encodes a premessenger with 13 exons and 12 introns. The ALG3 mRNA encodes a protein of 439–amino acid residues. ALG3 has little sequence similarity with Alg3p from S. cerevisiae and Not56L from humans (29 and 40% identity, respectively), and sequence analysis reveals two potential glycosylation sites in ALG3 (Figure 5, underlined), while no glycosylation sites are present in the yeast and human homologs. Topology analysis of ALG3 using the TMHMM server (v2.0) predicts 10 possible transmembrane helices (see Supplemental Figure 1 online), which anchor ALG3 in the ER membrane. ALG3 contains a C-terminal KKA sequence, similar to the KKXX sequence motif (X = any amino acid) shown to be involved in retrieving escaped ER transmembrane proteins from the Golgi to the ER in mammalian cells and yeast (Benghezal et al., 2000).
Figure 5.
Alignment of ALG3 Amino Acid Sequences from S. cerevisiae, Homo sapiens, and Arabidopsis.
Amino acid sequences were aligned with ClustalW (http://www.ch.embnet.org/software/ClustalW.html). Identical and similar residues are shaded black and gray, respectively; putative N-glycosylation sites of At ALG3 are underlined.
Isolation and Characterization of a T-DNA–Tagged alg3 Mutant
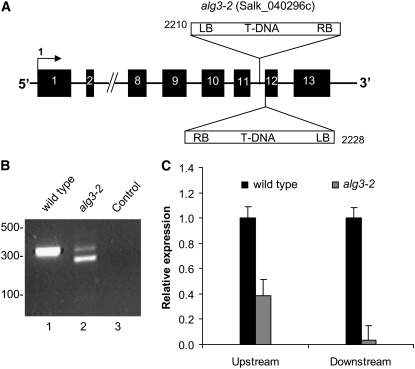
To study the function of the ALG3 gene in N-glycosylation of proteins in Arabidopsis, we selected a homozygous mutant line from the Salk Institute collection with a T-DNA insertion in the ALG3 locus (designated as mutant line alg3-2). Homozygosity was confirmed by PCR analysis of genomic DNA. T-DNA insertion in the ALG3 gene was analyzed by sequence analysis of PCR fragments, amplified using primers specific for the T-DNA and ALG3. We found that alg3-2 contains at least two T-DNA insertions in intron 11, one of which is precisely at the transition between intron 11 and exon 12 (Figure 6A). Lack of splicing after exon 11 would result in mRNA encoding a C-terminally truncated protein of 361–amino acid residues that lacks two putative membrane spanning domains. As was expected, such mRNA did not encode a functional enzyme, since a cDNA with a stop codon engineered immediately downstream of exon 11 was unable to restore glycosylation of CPY in the yeast Δalg3 mutant (Figure 3, lane 5).
Figure 6.
T-DNA Inserts in alg3-2 and ALG3 Expression Levels.
(A) Schematic representation of the ALG3 gene in the alg3-2 mutant. The insertion site of the left border of the T-DNAs in the alg3-2 mutant was determined by PCR and sequencing. Black boxes represent exons, and the numbers indicate the gene base number at which the left border of the T-DNA is inserted.
(B) Expression of ALG3 in the alg3-2 mutant. PCR analysis of cDNA from the wild type (lane 1) and alg3-2 (lane 2) using primers flanking the putative splice site of intron 11. Lane 3 represents the control experiment without cDNA template. Sequence analysis of the DNA fragment comigrating with the fragment amplified in the wild type revealed that correct splicing of intron 11 occurs.
(C) Quantification of the ALG3 transcript using real-time PCR. The results (mean ± se of three technical replicates using pooled sample of at least three plants) are expressed as the ratio of ALG3 versus the amount of actin transcript. Experiments were repeated twice with similar results.
Nevertheless, the mutation in alg3-2 did not result in a growth phenotype. We therefore investigated whether functional ALG3 mRNA is present in alg3-2, for instance, by simultaneous removal of intron 11 and the T-DNA sequences, through splicing of run-through transcripts. PCR analysis was performed on cDNA produced from RNA extracted from wild-type and alg3-2 mutant leaves, using primers specific for exon 11 and exon 13. For the mutant leaves, this resulted in two DNA fragments (Figure 6B, lane 2), one comigrating with the fragment amplified from wild-type cDNA and a shorter fragment (Figure 6B, lane 1). The sequence was determined of both cDNA fragments, and this revealed that intron 11, carrying the T-DNA sequences, was correctly spliced from the DNA fragment comigrating with the fragment amplified from the wild type. The smaller PCR fragment appeared to be the result of an aberrant splicing event, which couples exon 11 in frame to exon 13. This aberrant in-frame splicing, also known as exon skipping, is apparently triggered by the T-DNA insertion, since it is not observed in wild-type plants (Figure 6B, lane 1; see Supplemental Figure 2 online).
To quantify the amount of correctly spliced ALG3 mRNA in alg3-2, we performed real-time semiquantitative RT-PCR with two primer pairs on leaf cDNA from alg3-2 and wild-type plants. One pair amplified a DNA fragment at the 5′ end of the ALG3 mRNA, while the other amplified a fragment of exon 12, immediately downstream of the T-DNA insertion. The latter primer pair only detected correctly spliced ALG3 mRNA. We found that the ALG3-specific mRNA level in the homozygous mutant was ∼40% that of the wild type. However, the majority of this mRNA was truncated or the result of aberrant splicing, since only 4% appeared to be correctly spliced when quantified with the primer pair downstream of the T-DNA insert (Figure 6C).
N-Glycan Structures in alg3-2 Plants
The effect of ALG3 reduction on protein N-glycosylation in the alg3-2 mutant was analyzed. Proteins were isolated from young and old leaves of alg3-2 and wild-type plants. N-glycans were released by PNGase A treatment, and each N-glycan pool was analyzed by matrix-assisted laser-desorption ionization-time-of-flight (MALDI-TOF) (Table 1; see Supplemental Figure 3 online). The overall profiles of N-glycans isolated from young and old leaves were very similar, both from wild-type and mutant plants. Notably, the complex-type glycan profile in the mutant was similar to that of wild-type plants, indicating that the mutation in the early stage of the N-glycosylation pathway of alg3-2 has little effect on post-ER processing of glycans. However, in contrast with wild-type plants, no high-mannose-type glycans were detected in alg3-2 plants. Instead, Man3GlcNAc2 and Man4GlcNAc2 glycans accumulated in the mutant plant, consistent with the hypothesis that an aberrant Man5GlcNAc2 structure (Man5*; Figure 1B) is transferred to the nascent polypeptide and trimmed via Man4GlcNAc2 to Man3GlcNAc2 (Man4* and Man3*; Figure 1B) by the Golgi α(1,2)-mannosidase.
Table 1.
Relative Amounts of N-Glycans in Leaves from Wild-Type, alg3-2, and Complemented alg3-2 Plants
| Wild Type
|
alg3-2
|
alg3-2ALG3 YFP | ||||||
|---|---|---|---|---|---|---|---|---|
| Young | Old | Old +ManI | Young | Old | Old +ManI | |||
| m/z (M + Na)+ | Hybrid- and Complex-Type Structures | % of Total | % of Total | % of Total | % of Total | % of Total | % of Total | % of Total |
| 1065.4 | Man3XylGlcNAc2 | 8.3 | 5.0 | 1.7 | 6.8 | 7.3 | 4.7 | 4.7 |
| 1211.4 | Man3XylFucGlcNAc2 | 15.3 | 11.3 | 13.1 | 16.1 | 19.5 | 21.7 | 20.2 |
| 1268.5 | GlcNAcMan3XylGlcNAc2 | 8.5 | 7.7 | 3.5 | 5.8 | 6.8 | 4.6 | 2.2 |
| 1414.5 | GlcNAcMan3XylFucGlcNAc2 | 8.7 | 9.1 | 12.5 | 10.6 | 11.9 | 17.1 | 10.8 |
| 1617.6 | GlcNAc2Man3XylFucGlcNAc2 | 15.1 | 15.0 | 23.7 | 18.7 | 16.7 | 27.3 | 16.8 |
| (High) Mannose-Type Structures | ||||||||
| 933.3 | Man3GlcNAc2 (Man3*) | ND | ND | ND | 24.6 | 21.1 | 22.1 | ND |
| 1095.4 | Man4GlcNAc2 (Man4*) | ND | ND | ND | 11.8 | 11.4 | ND | ND |
| 1257.4 | Man5GlcNAc2 (Man5/Man*) | 26.7 | 31.7 | 45.5 | 5.6 | 5.4 | 2.6 | 21.6 |
| 1419.5 | Man6GlcNAc2 | 4.7 | 7.5 | ND | ND | ND | ND | 7.6 |
| 1581.6 | Man7GlcNAc2 | 4.5 | 5.7 | ND | ND | ND | ND | 6.5 |
| 1743.6 | Man8GlcNAc2 | 5.9 | 5.2 | ND | ND | ND | ND | 5.8 |
| 1905.7 | Man9GlcNAc2 | 2.2 | 1.8 | ND | ND | ND | ND | 2.9 |
The percentage is the total peak area divided by specific peak areas in MALDI-TOF mass spectra. +ManI, treated with α(1-2)-mannosidase; m/z, mass-to-charge ratio; ND, not detectable.
To distinguish between aberrant Man5* and the Man5 arising from the normal processing pathway, the N-glycan fractions isolated from wild-type and alg3-2 leaves were incubated with α(1,2)-mannosidase (Table 1). In the treated wild-type glycan fraction all high-mannose-type glycans with six or more mannose residues disappeared, while the relative level of Man5GlcNAc2 glycans increased, demonstrating the effectiveness of the α(1,2)-mannosidase treatment. In the α(1,2)-mannosidase–treated glycan fraction isolated from alg3-2, the Man4GlcNAc2 disappeared, proving that it contains an α-1,2 mannose residue and therefore is a Man4* isoform (Figure 1B). Despite the efficient in vitro trimming of high-mannose-type glycans (Man9-6GlcNAc2) and Man4* mannose-type glycans in alg3-2, only part of the Man5GlcNAc2 from mutant plants was subject to degradation by α(1,2)-mannosidase (Table 1). This suggests that this fraction of the Man5GlcNAc2 pool is α(1,2)-mannosidase resistant and is therefore derived from the normal N-glycosylation processing pathway (Figure 1A) as a result of low levels of ALG3 mannosyltransferase activity, due to leaky ALG3 expression.
Altogether, the accumulation of Man3GlcNAc2 and particularly of the α(1,2)-mannosidase sensitive isoforms of Man4GlcNAc2 (Man4*) and Man5GlcNAc2 (Man5*) on glycoproteins in the homozygous alg3-2 mutant demonstrates a block in elongation of normal biosynthetic Man5GlcNAc2-PP-Dol, which can be circumvented by an alternative route. We note that the proportion of complex-type glycans increased slightly after mannosidase I treatment for both the wild-type and mutant glycan samples (Table 1). We attribute this to the additional glycan purification step after the mannosidase I treatment, which may give a slightly biased yield for complex glycans.
No Underglycosylation in alg3-2 Plants
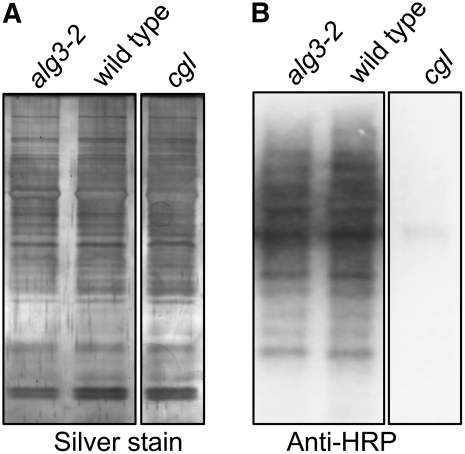
In yeast, the Δalg3 mutation causes accumulation of Man5GlcNAc2-PP-Dol and results in underglycosylation of glycoproteins (Huffaker and Robbins, 1983). We therefore investigated whether the overall efficiency of glycosylation is affected in alg3-2 plants by analyzing the level of glycosylation of proteins with complex-type glycans and of the ER resident protein PDI, which is modified by high-mannose-type glycans. Proteins from wild-type, alg3-2, and homozygous cgl mutant plants, which lack complex-type glycans because of a defect in GnTI (von Schaewen et al., 1993; Figure 1), were isolated from leaves and separated by SDS-PAGE. The presence of glycoproteins with complex-type glycans containing xylose and/or fucose can be detected by protein gel blot analysis with rabbit antihorseradish peroxidase (HRP) antibodies, which are mostly directed against the complex-type glycans on HRP. As shown in Figure 7, a similar intensity and profile of proteins with N-linked complex-type glycans was observed in the homozygous alg3-2 mutant strain and wild-type plants, indicating that the mutation does not negatively affect the level of complex glycans on glycoproteins. As expected, no complex-type N-glycans were detected in the homozygous cgl plants (Figure 7B).
Figure 7.
Protein and N-Glycosylation Analysis in Wild-Type, alg3-2, and cgl Mutant Plants.
(A) SDS-PAGE of total protein fraction from leaves of wild-type, alg3-2, and homozygous cgl plants by silver staining.
(B) Immunoblotting using polyclonal anti-HRP antibodies that specifically recognize complex N-glycans.
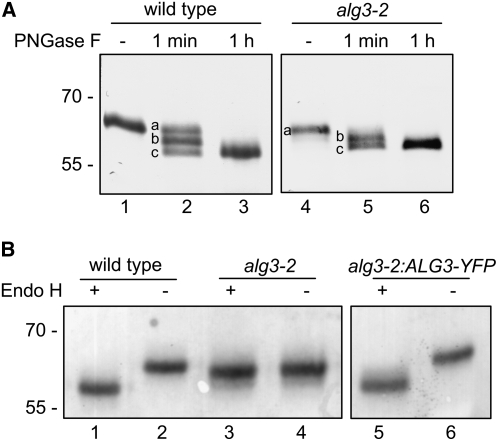
ER glycoproteins remain decorated with high-mannose-type glycans as they are not processed in the Golgi. Since glycosylation in the mutant differs mainly with respect to high-mannose-type N-glycans, we investigated the glycosylation of the ER-resident glycoprotein PDI, which contains two potential glycosylation sites. Both sites on PDI were efficiently glycosylated in both the wild-type and the alg3-2 plants, as visualized by the stepwise removal of the N-glycans after limited PNGase F treatment (Figure 8A).
Figure 8.
N-Glycan Analysis of PDI in Wild-Type and alg3-2 Plants.
(A) Protein gel blot analysis of PDI after limited treatment with PNGase F. Isolated proteins were incubated in the presence (+) or absence (−) of PNGase F for the indicated times (1 min; 1 h). Reactions were stopped and mixtures subjected to immunoblotting with anti-PDI. Both the wild type and the alg3-2 mutant PDI have two high-mannose-type glycans (PDI with two, one, or no glycan is indicated by lowercase letters a, b, and c, respectively).
(B) Protein gel blot analysis of PDI after treatment with Endo H. Isolated proteins were incubated overnight in the presence of Endo H (+) and subjected to immunoblotting with anti-PDI. As a control, the total homogenate was incubated in the absence of Endo H (−).
Efficient Transfer of Aberrant Glycans by the OST Complex
The data provided above show that both the alternative and normal pathways (Figure 1) are operating in alg3-2 plants and that glycosylation efficiency is hardly affected. However, from these data the relative flux of protein N-glycosylation through the mutant pathway cannot be determined. To determine the relative efficiency by which aberrant glycans are transferred, the identities of N-glycans on PDI were analyzed. High-mannose-type glycans are cleaved off by endo-β-N-acetylglucosaminidase H (Endo H), while Man5* oligosaccharides are resistant to Endo H (Huffaker and Robbins, 1983; Verostek et al., 1991, 1993; Zufferey et al., 1995). As visualized by an increased electrophoretic mobility (Figure 8B, compare lanes 1 and 2), the N-glycans of PDI from wild-type plants are removed by Endo H treatment, indicating that PDI contains exclusively high-mannose-type glycans. By contrast, N-glycans of PDI in mutant plants are almost entirely resistant to Endo H (Figure 8B, lanes 3 and 4) indicating that, at least for this ER resident protein, the N-glycans are almost entirely derived from the mutant glycosylation pathway.
It cannot be deduced whether complex-type glycans of mutant plants, which amount to ∼58% of the total glycan pool (Table 1), originate from the wild-type or mutant pathway, since aberrant glycans may also get processed to complex-type glycans (Figure 1B). To investigate the origin of glycans (wild-type or aberrant glycosylation pathway) on proteins that are destined to become complex-type glycoproteins, we prevented processing to complex-type glycans by combining the alg3-2 mutation with the cgl mutation in alg3-2 cgl double homozygous mutant plants. The cgl mutation blocks complex-type glycan formation because of a defect in GnTI, a Golgi enzyme essential in the initiation of complex glycan biosynthesis (von Schaewen et al., 1993). N-glycans were isolated from homozygous cgl mutant and homozygous alg3-2 cgl double mutant plants and analyzed by MALDI-TOF before and after treatment with α(1,2)-mannosidase (Table 2). The double mutant alg3-2 cgl completely lacks complex-type glycans and also lacks almost all of the wild-type Man5GlcNAc2 structures that accumulate in cgl plants (Strasser et al., 2005). Instead, 99% of the glycans are represented by Man3*, Man4*, and Man5* structures (Table 2) from the mutant pathway. The presence of some low-level wild-type Man5GlcNAc2 glycans after α(1,2)-mannosidase treatment shows that these double mutant plants also have leaky wild-type ALG3 activity.
Table 2.
Relative Amounts of N-Glycans in Leaves of cgl and alg3-2 cgl Double Mutant Plants
| cgl | alg3-2 cgl | alg3-2 cgl | ||
|---|---|---|---|---|
| m/z (M + Na)+ | Hybrid- and Complex-Type Structures | % of Total | % of Total | % of Total |
| 1065.4 | Man3XylGlcNAc2 | ND | ND | ND |
| 1211.4 | Man3XylFucGlcNAc2 | ND | ND | ND |
| 1268.5 | GlcNAcMan3XylGlcNAc2 | ND | ND | ND |
| 1414.5 | GlcNAcMan3XylFucGlcNAc2 | ND | ND | ND |
| 1617.6 | GlcNAc2Man3XylFucGlcNAc2 | ND | ND | ND |
| (High) Mannose-Type Structures | ||||
| 933.3 | Man3GlcNAc2 (Man3*) | ND | 78.4 | 98.8 |
| 1095.4 | Man4GlcNAc2 (Man4*) | ND | 15.3 | ND |
| 1257.4 | Man5GlcNAc2 (Man5/Man5*) | 84.4 | 6.4 | 1.2 |
| 1419.5 | Man6GlcNAc2 | 5.0 | ND | ND |
| 1581.6 | Man7GlcNAc2 | 4.3 | ND | ND |
| 1743.6 | Man8GlcNAc2 | 5.8 | ND | ND |
| 1905.7 | Man9GlcNAc2 | 0.5 | ND | ND |
The percentage is the total peak area divided by specific peak areas in MALDI-TOF mass spectra. +ManI, treated with α(1-2)-mannosidase; ND, not detectable.
Taken together, these data provide evidence that most glycosylation in alg3-2 occurs via the mutant pathway and the in vivo transfer efficiency of the aberrant glycans by the OST complex is indistinguishable from that of wild-type glycans. In addition, these aberrant glycans are recognized by mannosidase I and GnTI and are efficiently processed into complex-type glycans, as in wild-type plants.
UPR in alg3-2 Plants
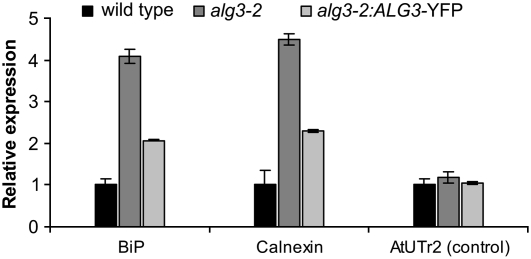
The ALG3 mutation particularly affects glycans on proteins in the ER, where protein folding quality control mechanisms are active (Huffaker and Robbins, 1983). We therefore investigated if aberrant glycans in alg3-2 affect N-glycan–dependent protein folding quality control and trigger the UPR. The level of UPR in wild-type and alg3-2 mutant plants was investigated by analyzing the expression of BiP, calnexin, and UDP-Galactose Transporter 2 (UTr2) by real-time semiquantitative RT-PCR. BiP and calnexin have both been implicated in alleviating ER stress and are frequently used as markers of the UPR. UTr2 is a nucleotide sugar transporter located in the Golgi apparatus that is capable of transporting UDP-galactose but is not involved in the ER stress response (Reyes et al., 2006). The results of the expression analysis show that mRNA levels of BiP and calnexin were induced 4.1-fold and 4.5-fold, respectively, in the homozygous alg3-2 mutant compared with wild-type plants, while the expression levels of the control gene UTr2 in the same samples was similar in both plants (Figure 9). The alg3-2 plants therefore seem to have an activated UPR.
Figure 9.
Expression of ER Chaperones in Wild-Type, alg3-2 Mutant, and Complemented alg3-2 Plants.
BiP, calnexin, and UTr2 transcripts were quantified by real-time PCR. The results (mean ± se of three technical replicates using a pooled sample of at least three plants) are expressed as the ratio of BiP, calnexin, and UTr2 versus the amount of actin transcript. Experiments were repeated twice with similar results.
Complementation of alg3-2 Plants
To establish whether disruption of the ALG3 gene was responsible for the observed phenotypes, alg3-2 was transformed with Agrobacterium tumefaciens carrying a binary vector with the ALG3 cDNA fused to YFP, under control of the cauliflower mosaic virus 35S promoter, and multiple independent transformants were selected on hygromycin. Individual T2 offspring plants from independent transformants were screened for restoration of high-mannose-type glycans on PDI. From this initial screen, one alg3-2:ALG3-YFP line was selected for detailed analysis. Leaves from three sibling plants were pooled for protein extraction and glycan characterization. In contrast with the N-glycans of PDI from alg3-2 mutant plants, but similar to those from wild-type plants, the N-glycans of PDI from the alg3-2:ALG3-YFP line was sensitive to Endo H (Figure 8B, lanes 5 and 6). Furthermore, complementation of the alg3-2 line also restored the N-glycan profile (Table 1). Finally, ER stress was markedly reduced as shown by the expression analysis of BiP and calnexin, although not to the level observed in wild-type plants (Figure 9).
DISCUSSION
This study identifies a plant gene (ALG3) that is involved in the lipid-linked phase of N-glycan biosynthesis and that codes for the Dol-P-Man:Man5GlcNAc2-PP-Dol α1,3-mannosyl transferase. Biochemical characterization of a homozygous alg3 mutant reveals substrate promiscuity of several subsequent enzymatic reactions as well as effects on the UPR in plants. These findings have implications on various steps in protein N-glycosylation.
A Normal and Alternative N-Glycosylation Pathway Operating in the alg3-2 Mutant
We identified an alg3-2 mutant with T-DNA insertions in intron 11 of ALG3 (Figure 6A). Despite the T-DNA insertions, we demonstrated the presence of low levels of functional ALG3 mRNA, which is likely the result of removal of the T-DNA sequences through splicing from a low percentage of primary transcripts (Figure 6C). In addition to these very low levels of correctly spliced mRNA, most of the transcripts originate from an aberrantly spliced mRNA (Figures 6B and 6C). However, this mRNA encodes a truncated ALG3 protein lacking at least two putative membrane spanning domains that is most likely nonfunctional. The strongly reduced levels of functional ALG3 mRNA block extension of the Man5GlcNAc2-PP-Dol to the full-length lipid-linked oligosaccharide. As a result, aberrant glycans in alg3-2 are transferred from lipids to proteins via one of two pathways (Figure 1B). In addition, the presence of mannosidase-resistant Man5 glycans provides evidence that a wild-type biosynthesis pathway is also operating in this mutant plant due to the presence of some low level of functional alg3 mRNA.
Properties of Glycosylation Enzymes as Revealed by the alg3-2 Mutant Plant
Although both a mutant and a wild-type pathway operate in the alg3-2 plant, the flux through the mutant pathway is far greater. This was shown by the almost exclusive presence of aberrant glycans on PDI and the almost complete lack of wild-type Man5 glycans in the alg3-2 cgl double mutant (Figure 8B, Table 2). Apparently, lipid-linked glycan extension is almost completely blocked in the alg3-2 mutant. The resulting Man5* structure does not appear to be a substrate for the plant homolog of the yeast ALG12, which adds α1,6-mannose residues to the lipid-linked glycans after the action of ALG3. Similarly, elongation of the lipid-linked oligosaccharide in yeast is also dependent on prior ALG3 activity (Burda et al., 1999).
It is remarkable that the lipid-linked Man5* intermediate glycans are efficiently transferred from lipid to protein by the plant OST complex; despite the observation that 99% of the flux proceeds through the mutant pathway, no underglycosylation of proteins was detected (Figure 8A). This is in sharp contrast with the substrate characteristics of the yeast and human OST complexes. In the S. cerevisiae Δalg3 strain and also in human patients, who have a mutation in the ALG3 gene, a profound underglycosylation of secretory glycoproteins was demonstrated (Huffaker and Robbins, 1983; Aebi et al., 1996; Korner et al., 1999).
Normally in yeast, mammals, and plants, the lipid-linked glycan that is transferred to proteins is the glucosylated Glc3Man9GlcNAc2-PP-Dol (Helenius and Aebi, 2002). There is no evidence that the Δalg3 mutant in S. cerevisiae makes Glc3Man5GlcNAc2-PP-Dol (Burda et al., 1999). However, triglucosylated Man5GlcNAc2 oligosaccharides have been detected on proteins from a mutant CHO cell line (Ermonval et al., 1997; Foulquier et al., 2002). Therefore, it could be that at least part of the Man5GlcNAc2-PP-Dol pool is glucosylated in alg3-2 plants before transfer to proteins. Alternatively, the aberrant lipid-linked Man5GlcNAc2 saccharides are a direct substrate for the plant OST activity, as is the case in the yeast mutant.
Once transferred from lipid to protein, the aberrant Man5* glycans in the mutant plant are efficiently converted to complex-type glycans. The size and composition of the complex-type glycan pool is very similar in alg3-2 and in wild-type plants (Table 1, Figure 7). The conversion to complex glycans in the ER and/or early Golgi starts with trimming of Man5* to Man4* and Man3* structures, presumably by α(1,2)-mannosidase (ManI). These trimming reactions generate the putative substrate Man3* glycan for GnTI. Indeed, in vitro experiments have shown that, although Man5 is the preferred substrate for both plant and mammalian GnTI, Man3* can also be used as substrate (Schachter et al., 2003; Strasser et al., 2005). The normal amounts of complex-type glycans in the alg3-2 mutant (Table 1, Figure 6) show that the overall conversion of Man3* by GnTI in vivo is indistinguishable from the conversion of its normal wild-type Man5 substrate. The GnTI activity in the mutant presumably renders the glycan structure GlcNAcMan3GlcNAc2, which is also produced in the wild-type pathway after the action of GnTI and ManII (Figure 1A). Thus, contrary to the wild type, ManII is not required for the downstream processing to complex-type glycans in alg3-2 plants. The GlcNAcMan3GlcNAc2 glycans are apparently rapidly used by subsequent glycosyl transferases like GnTII, XylT, and FucT, as only xylosylated and/or fucosylated GlcNAc(2)Man3GlcNAc2 glycans are detected in both mutant and wild-type plants (Table 1).
UPR in alg3-2 Plants
Despite the fact that no underglycosylation was observed in alg3-2 plants (Figures 7 and 8A), the induction of UPR-related genes reflects an increase in ER stress in the mutant plants (Figure 9). This may be explained by the presence of aberrant glycans on glycoproteins in the ER of alg3-2 plants. N-glycans on misfolded proteins are normally subject to glucose addition by UGGT and trimming activity of glucosidase I and II (calreticulin/calnexin cycle), as part of the protein folding quality control mechanism. Evidence from CHO cells suggests that aberrant Man5* glycans may also be subject to this glucosylation- and deglucosylation-mediated quality control. However, it was suggested that UGGT in CHO cells does not recognize the aberrant Man4* or Man3* glycans (Foulquier et al., 2002). Therefore, if ManI trims the terminal mannose residue of Man5*, the glycan is no longer a substrate for UGGT; thus, the glycoprotein is removed from the quality control cycle. In addition, the lectins that are part of the ER-associated misfolded glycoprotein degradation pathway of quality control may have reduced affinity for the aberrant glycans, resulting in inefficient removal of misfolded glycoproteins. Both factors would contribute to elevated levels of misfolded proteins in alg3-2, explaining the elevated UPR. It has not been investigated whether UPR is induced in the ALG3 mutants of yeast and in CHO cells.
Comparison with Other N-Glycosylation Mutants
To date, only a few Arabidopsis genes involved in N-glycan assembly or early N-glycan processing have been described in the literature. Orthologs of OST subunits characterized in yeast have been characterized in Arabidopsis: DAD1 (Gallois et al., 1997), STT3a and SST3b (Koiwa et al., 2003), and DGL1 (Lerouxel et al., 2005), corresponding to Ost2p, Stt3p, and Wbp1p, respectively. From these, the stt3a stt3b double mutant and the dgl1-2 mutant both display an embryo lethal phenotype. Mutants altered in Arabidopsis α-glucosidase I and II also display an embryo lethal phenotype (Boisson et al., 2001; Burn et al., 2002; Gillmor et al., 2002). All these mutants show that functional early steps in N-glycosylation are important for normal embryo development. In humans, a defect in the ALG3 gene causes congenital disorder of glycosylation-type Id. At present, only four patients have been described in the literature (Stibler et al., 1995; Korner et al., 1999; Denecke et al., 2004, 2005; Schollen et al., 2005; Sun et al., 2005). All known patients are homozygous for the mutation in the ALG3 gene, but all were shown to have leaky ALG3 expression. It is striking that the described Arabidopsis alg3-2 mutant also displays leaky ALG3 expression; however, we note that complete inactivation of the ALG3 gene in S. cerevisiae and P. pastoris is not lethal (Aebi et al., 1996; Davidson et al., 2004).
In summary, we identified a plant mannosyltransferase involved in the initial phase of lipid-linked glycan assembly in the ER. Characterization of the mutant alg3-2 plant provides information on in vivo substrate specificity of several of the enzymes involved in subsequent steps in N-glycan biosynthesis and shows a tolerance of the Arabidopsis OST complex for aberrant glycan substrates. In addition, the mutation results in the induction of the UPR in plants, a stress response that has not been investigated in alg3 mutants in other organisms.
METHODS
Materials
Yeast strain Δalg3 (MATα ade2 his 3 ura3 tyr1 Δalg3∷ HIS3) was used. Cells were grown in standard yeast media: either YEPD (1% Bacto yeast extract, 2% Bacto peptone, and 2% dextrose) or YNBD (0.67% yeast nitrogen base and 2% dextrose) supplemented with amino acid residues and nucleotides as necessary.
Plant Materials and Growth Conditions
The alg3-2 cgl double mutant was obtained by crossing homozygous alg3-2 with homozygous cgl plants. The F2 generation was screened for homozygous cgl plants by ELISA using rabbit anti-HRP antibodies that bind very strongly and specifically to typical plant complex-type N-glycans. Genomic DNA from homozygous cgl plants was then screened by PCR analysis for t for the alg3-2 mutation as described below for homozygous alg3-2 T-DNA insertion. Seeds of Arabidopsis thaliana lines were sown on 9-cm 0.8% Daishun agar Petri dishes and placed in a cold room at 4°C for 2 d in the dark to promote uniform germination. Germination and plant culture were performed in a climate chamber (20°C/15°C day/night temperatures; 250 μmol light m−2 s−1 at plant level during 12 h/d and 75% relative humidity).
Analysis of Lipid-Linked Oligosaccharides with [2-3H]Mannose
Formation and determination of the composition of lipid-linked oligosaccharides was performed as previously described (Knauer and Lehle, 1999).
Analysis of Glycosylation of Carboxypeptidase
Yeast cells were grown in YNBD at 30°C to mid log phase, harvested, and lysed with glass beads as described previously (Knauer and Lehle, 1999). The soluble fraction obtained by centrifugation at 48,000g for 30 min was applied to SDS-PAGE (8% gel), blotted to nitrocellulose, and decorated with anti-CPY antiserum.
Identification and Genotyping of T-DNA Insertion Lines
The Arabidopsis T-DNA line alg3-2 from the SALK institute was obtained via the Nottingham Arabidopsis Stock Centre. alg3-2 was screened by PCR on genomic DNA using the forward primer Alg3-F2.2 (5′-CGTTCAGTGATGTATCAGCCTCACGG-3′) and the reverse primer Alg3-R2 (5′-GGATTTAGGGTGTTCTTTGAGCTGATAAGG-3′) as well as the T-DNA–specific LBb1 primer. PCR products were sequenced to determine the exact insertion site.
Cloning of Arabidopsis ALG3 and Construction of CFP/YFP-Tagged ALG3
Based on sequence homology of ALG3 homologs, a cDNA clone containing the putative Arabidopsis ALG3 gene was ordered from The Arabidopsis Information Resource website. ALG3 was PCR amplified using Platinum Pfx polymerase (Invitrogen) and primers flanking the coding region. The forward primers Alg3-pMonF (5′-GTGACAGATCTATGGCGGGCGCCTCATCACC-3′) or AtALGup (5′-CTAGAAGCTTATGGCGGGCGCCTCATCACC-3′) were used containing a BglII or HindIII site, respectively. The reverse primer AtALGepi (5′-GTCAGGATCCTGCTT TTTTGTGTATTTGGGA-3′) was used that contains a BamHI site that deletes the original stop codon. For construction of the CFP/YFP-tagged ALG3, the full-length At Alg3 cDNA was inserted into pMON999-CFP or pMON999-YFP (Monsanto) using BglII and BamHI or a yeast expression vector using HindIII and BamHI. The 35S-ALG3-YFP cassette was isolated from the pMON999 vector by HindIII and SmaI digestion and ligated into a pUCM2 vector. This pUCM2-ALG3-YFP vector was digested with PacI and AscI to isolate the 35S-ALG3-YFP cassette, which was ligated into the PacI and AscI sites of the binary pBinplus vector.
Isolation of Genomic DNA
Plant material was collected in Eppendorf tubes and ground with a pestle in liquid nitrogen and 400 μL DNA isolation buffer (5 M urea, 0.3 M NaCl, 50 mM Tris-HCl, pH 7.5, 20 mM EDTA, 2% N-lauroyl sarcosine, 0.5% SDS, and 5% phenol, pH 8.0) and 400 μL phenol:chloroform:isoamylalcohol (25:24:1) solution. The supernatant containing DNA was precipitated with isopropanol, washed twice in 70% ethanol, and redissolved in 50 μL milliQ water containing 10 μg/mL of RNase A.
Arabidopsis Protoplast Isolation and Transfection
Arabidopsis mesophyll protoplasts were prepared and transfected as described by Aker et al. (2006). The different expression constructs were introduced pairwise into Arabidopsis protoplasts, and the subcellular localization of the resulting YFP and CFP fluorescence signal was determined 24 h after transfection, using a confocal laser scanning microscope 510 (Carl Zeiss) excited at 458 and 514 nm with an argon laser. The fluorescence was detected via a band-pass filter (CFP, 470 to 500 nm; YFP, 535 to 590 nm). Chlorophyll was detected using a 650-nm long-pass filter.
Transformation of Arabidopsis
pBinplus constructs containing the ALG3-YFP cassettes were transferred to Agrobacterium tumefaciens AGL-0 by triparental mating using the Escherichia coli pRK2013 helper plasmid. Arabidopsis plants were transformed by the standard floral dip method (Clough and Bent, 1998). Seeds from transformed plants were selected by growing them on plates containing Murashige and Skoog powder (4.4 g/L), sucrose (10 g/L), and Daishin agar (8 g/L), supplemented with hygromycin (50 μg/mL) as the selective marker.
SDS-PAGE and Immunoblotting
Plant material was ground in liquid nitrogen, resuspended in 10 μL PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH 7.4) per mg of plant material, and centrifuged. An aliquot of the supernatant was immediately mixed with SDS-PAGE loading buffer, denatured at 95°C for 5 min, and subjected to SDS-PAGE (8 or 12.5%) under reducing conditions. Protein gel blotting was performed using polyvinylidene difluoride membranes, blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline (20 mM Tris-HCl, pH 7.6, and 137 mM NaCl) with 0.05% Tween 20. The membranes were probed with either anti-HRP (1:2000; Sigma-Aldrich) or anti-PDI (1:5000; Rosebiotech). Detection of bound primary antibodies was performed with either BCIP/NBT or Lumi-Light and Lumi-Imager (Roche Diagnostics) after incubation with goat anti-rabbit antibodies. PNGase F and Endo H were purchased from New England Biolabs for glycosylation analysis of PDI and used according to the suggested methods along with the manufacturer's recommended buffers. The reactions were terminated as indicated in the figures by boiling in SDS loading buffer.
RNA Isolation and RT-PCR Analysis
RNA was extracted from alg3-2, wild-type, and cgl mutant leaves using TriPure isolation reagent (Roche), and the concentration was measured with a NanoDrop ND-1000 UV-Vis Spectrophotometer. One microgram of RNA was used to make cDNA with Taqman reverse transcript reagent (Applied Biosystems). Quantitative real-time PCR was performed using the Bio-Rad iQSYBR Green Supermix single color detection system. Briefly, after a 3-min denaturation at 94°C, 40 cycles of 15 s at 94°C, and 30 s at 60°C were followed by a melting curve gradient. No template controls served as blanks, and β-Actin was used as a reference gene. Samples were run in triplicate and averaged, and relative gene expression was calculated using the 2−δδt method (Livak and Schmittgen, 2001). All values are presented as mean ± se. Primers were designed using Beacon Designer (Biosoft International) and ordered from Sigma-Genosys. The following pairs of primers were used: for actin, 5′-GGTAACATTGTGCTCAGTGGTGG-3′ and 5′-AACGACCTTAATCTTCATGCTGC-3′; for BiP, 5′-ATGGCTCGCTCGTTTGGAGC-3′ and 5′-AAGTTTCCTGTCCTTTTGAA-3′; for calnexin, 5′-ATGAGACAACGGCAACTATT-3′ and 5′-TTCCTGAGGACGGAGGTACT-3′; for UTr2, 5′-CACATTTATCGGTCAAGTCTCCGTT-3′ and 5′-TCGCAGGAGGCGATGGTGATAGAG-3′; for upstream of the T-DNA insertion, 5′-CGCACTTATTATCGCCTATGTTC-3′ and 5′-GCCCTGTATCGCCTTTCAAG-3′; for downstream of the T-DNA insertion, 5′-ACTTCTATTCGCTACCTTATCTAC-3′ and 5′-GGAGGACGGTGTTGATGG-3′. For PCR analysis of alg3-2 and wild-type splicing of intron 12, the Alg3-F2.2 and Alg3-R2 primers were used.
Isolation of N-Glycans and α(1,2)-Mannosidase Treatment
Young leaves of ∼1.5 cm long and old leaves (longer than 3 cm) of 4-week-old Arabidopsis plants were used for N-glycan analysis. Proteins for N-glycan purification were extracted from 250 mg of leaf material from at least three individual plants and digested with pepsin. Bound N-glycans were released by PNGase A (Roche) as described previously (Bakker et al., 2006). Aspergillus satoi α(1,2)-mannosidase (10 mU/mL; Pro-enzyme, Glyko) digestions were performed in the supplied buffer for 24 h at 37°C. Reactions were then diluted with 0.5 mL of milliQ water and desalted by passage through a 500-mg C18 column (Varian).
N-Glycan Analysis
Purified N-glycans were dissolved in 5 mM NaAc and mixed with an equal volume of 1% 2,5-D in 50% acetonitrile. One-microliter aliquots were spotted onto a stainless steel sample plate and dried under a stream of air at room temperature. Positive-ion MALDI-TOF spectra of [M+Na]+ adducts were recorded on an Ultraflex mass spectrometer (Bruker) fitted with delayed extraction and a nitrogen laser (337 nm). A maltodextrin series was used as an external molecular weight standard. Spectra were generated from the sum of at least 300 laser pulses.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: U14893 (ALG3 cDNA clone), At2g47760 (ALG3 locus), Salk_040296c (ALG3 T-DNA insertion line), AAA75352 (yeast ALG3), CR616285 (human ALG3), and AAC63631 (Arabidopsis ALG3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. At ALG3 Topology.
Supplemental Figure 2. Aberrant In-Frame Splicing of alg3-2 Mutant mRNA.
Supplemental Figure 3. MALDI-TOF Spectra of Released N-Linked Glycans from Wild-Type and alg3-2 Mutant Plants.
Supplemental Data Set 1. Text File of Alignment Corresponding to Figure 5.
Supplementary Material
Acknowledgments
We thank Henk Kieft, Dion Florack, Jan Willem Borst, Bas Heinhuis, Ineke Braakman, Linus van der Plas, and Titti Mariani for their useful comments and contribution to experiments. This work was supported by the Netherlands Proteomics Centre, the Graduate School “Experimental Plant Sciences,” the Centre for Biosystems Genomics (Netherlands Genomics Initiative), and grants from the Deutsche Forschungsgemeinschaft and the Körber-Stiftung.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Dirk Bosch (dirk.bosch@wur.nl).
Online version contains Web-only data.
References
- Aebi, M., Gassenhuber, J., Domdey, H., and te Heesen, S. (1996). Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. Glycobiology 6 439–444. [DOI] [PubMed] [Google Scholar]
- Aker, J., Borst, J.W., Karlova, R., and de Vries, S. (2006). The Arabidopsis thaliana AAA protein CDC48A interacts in vivo with the somatic embryogenesis receptor-like kinase 1 receptor at the plasma membrane. J. Struct. Biol. 156 62–71. [DOI] [PubMed] [Google Scholar]
- Bakker, H., Rouwendal, G.J., Karnoup, A.S., Florack, D.E., Stoopen, G.M., Helsper, J.P., van Ree, R., van Die, I., and Bosch, D. (2006). An antibody produced in tobacco expressing a hybrid beta-1,4-galactosyltransferase is essentially devoid of plant carbohydrate epitopes. Proc. Natl. Acad. Sci. USA 103 7577–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal, M., Wasteneys, G.O., and Jones, D.A. (2000). The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12 1179–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson, M., Gomord, V., Audran, C., Berger, N., Dubreucq, B., Granier, F., Lerouge, P., Faye, L., Caboche, M., and Lepiniec, L. (2001). Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda, P., Jakob, C.A., Beinhauer, J., Hegemann, J.H., and Aebi, M. (1999). Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology 9 617–625. [DOI] [PubMed] [Google Scholar]
- Burn, J.E., Hurley, U.A., Birch, R.J., Arioli, T., Cork, A., and Williamson, R.E. (2002). The cellulose-deficient Arabidopsis mutant rsw3 is defective in a gene encoding a putative glucosidase II, an enzyme processing N-glycans during ER quality control. Plant J. 32 949–960. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Davidson, R.C., Nett, J.H., Renfer, E., Li, H., Stadheim, T.A., Miller, B.J., Miele, R.G., Hamilton, S.R., Choi, B.K., Mitchell, T.I., and Wildt, S. (2004). Functional analysis of the ALG3 gene encoding the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase enzyme of P. pastoris. Glycobiology 14 399–407. [DOI] [PubMed] [Google Scholar]
- Denecke, J., Kranz, C., Kemming, D., Koch, H.G., and Marquardt, T. (2004). An activated 5′ cryptic splice site in the human ALG3 gene generates a premature termination codon insensitive to nonsense-mediated mRNA decay in a new case of congenital disorder of glycosylation type Id (CDG-Id). Hum. Mutat. 23 477–486. [DOI] [PubMed] [Google Scholar]
- Denecke, J., Kranz, C., von Kleist-Retzow, J., Bosse, K., Herkenrath, P., Debus, O., Harms, E., and Marquardt, T. (2005). Congenital disorder of glycosylation type Id: Clinical phenotype, molecular analysis, prenatal diagnosis, and glycosylation of fetal proteins. Pediatr. Res. 58 248–253. [DOI] [PubMed] [Google Scholar]
- Elbers, I.J., Stoopen, G.M., Bakker, H., Stevens, L.H., Bardor, M., Molthoff, J.W., Jordi, W.J., Bosch, D., and Lommen, A. (2001). Influence of growth conditions and developmental stage on N-glycan heterogeneity of transgenic immunoglobulin G and endogenous proteins in tobacco leaves. Plant Physiol. 126 1314–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermonval, M., Cacan, R., Gorgas, K., Haas, I.G., Verbert, A., and Buttin, G. (1997). Differential fate of glycoproteins carrying a monoglucosylated form of truncated N-glycan in a new CHO line, MadIA214214, selected for a thermosensitive secretory defect. J. Cell Sci. 110 323–336. [DOI] [PubMed] [Google Scholar]
- Fiedler, K., and Simons, K. (1995). The role of N-glycans in the secretory pathway. Cell 81 309–312. [DOI] [PubMed] [Google Scholar]
- Foulquier, F., Harduin-Lepers, A., Duvet, S., Marchal, I., Mir, A.M., Delannoy, P., Chirat, F., and Cacan, R. (2002). The unfolded protein response in a dolichyl phosphate mannose-deficient Chinese hamster ovary cell line points out the key role of a demannosylation step in the quality-control mechanism of N-glycoproteins. Biochem. J. 362 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois, P., Makishima, T., Hecht, V., Despres, B., Laudie, M., Nishimoto, T., and Cooke, R. (1997). An Arabidopsis thaliana cDNA complementing a hamster apoptosis suppressor mutant. Plant J. 11 1325–1331. [DOI] [PubMed] [Google Scholar]
- Gillmor, C.S., Poindexter, P., Lorieau, J., Palcic, M.M., and Somerville, C. (2002). Alpha-glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius, J., and Aebi, M. (2002). Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 13 171–178. [DOI] [PubMed] [Google Scholar]
- Huffaker, T.C., and Robbins, P.W. (1982). Temperature-sensitive yeast mutants deficient in asparagine-linked glycosylation. J. Biol. Chem. 257 3203–3210. [PubMed] [Google Scholar]
- Huffaker, T.C., and Robbins, P.W. (1983). Yeast mutants deficient in protein glycosylation. Proc. Natl. Acad. Sci. USA 80 7466–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauer, R., and Lehle, L. (1999). The oligosaccharyltransferase complex from Saccharomyces cerevisiae. Isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J. Biol. Chem. 274 17249–17256. [DOI] [PubMed] [Google Scholar]
- Koiwa, H., Li, F., McCully, M.G., Mendoza, I., Koizumi, N., Manabe, Y., Nakagawa, Y., Zhu, J., Rus, A., Pardo, J.M., Bressan, R.A., and Hasegawa, P.M. (2003). The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. Plant Cell 15 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner, C., Knauer, R., Stephani, U., Marquardt, T., Lehle, L., and von Figura, K. (1999). Carbohydrate deficient glycoprotein syndrome type IV: Deficiency of dolichyl-P-Man:Man(5)GlcNAc(2)-PP-dolichyl mannosyltransferase. EMBO J. 18 6816–6822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld, R., and Kornfeld, S. (1985). Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54 631–664. [DOI] [PubMed] [Google Scholar]
- Kurzik-Dumke, U., Kaymer, M., Gundacker, D., Debes, A., and Labitzke, K. (1997). Gene within gene configuration and expression of the Drosophila melanogaster genes lethal(2) neighbour of tid [l(2)not] and lethal(2) relative of tid[l(2)rot]. Gene 200 45–58. [DOI] [PubMed] [Google Scholar]
- Lehle, L., Strahl, S., and Tanner, W. (2006). Protein glycosylation, conserved from yeast to man: A model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 45 6802–6818. [DOI] [PubMed] [Google Scholar]
- Lerouge, P., Cabanes-Macheteau, M., Rayon, C., Fischette-Laine, A.C., Gomord, V., and Faye, L. (1998). N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol. Biol. 38 31–48. [PubMed] [Google Scholar]
- Lerouxel, O., Mouille, G., Andeme-Onzighi, C., Bruyant, M.P., Seveno, M., Loutelier-Bourhis, C., Driouich, A., Hofte, H., and Lerouge, P. (2005). Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J. 42 455–468. [DOI] [PubMed] [Google Scholar]
- Livak, K.J., and Schmittgen, T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Ohtsubo, K., and Marth, J.D. (2006). Glycosylation in cellular mechanisms of health and disease. Cell 126 855–867. [DOI] [PubMed] [Google Scholar]
- Parodi, A.J. (2000). Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 69 69–93. [DOI] [PubMed] [Google Scholar]
- Pichler, H., Gaigg, B., Hrastnik, C., Achleitner, G., Kohlwein, S.D., Zellnig, G., Perktold, A., and Daum, G. (2001). A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268 2351–2361. [DOI] [PubMed] [Google Scholar]
- Reyes, F., Marchant, L., Norambuena, L., Nilo, R., Silva, H., and Orellana, A. (2006). AtUTr1, a UDP-glucose/UDP-galactose transporter from Arabidopsis thaliana, is located in the endoplasmic reticulum and up-regulated by the unfolded protein response. J. Biol. Chem. 281 9145–9151. [DOI] [PubMed] [Google Scholar]
- Roth, J., Ziak, M., and Zuber, C. (2003). The role of glucosidase II and endomannosidase in glucose trimming of asparagine-linked oligosaccharides. Biochimie 85 287–294. [DOI] [PubMed] [Google Scholar]
- Runge, K.W., Huffaker, T.C., and Robbins, P.W. (1984). Two yeast mutations in glucosylation steps of the asparagine glycosylation pathway. J. Biol. Chem. 259 412–417. [PubMed] [Google Scholar]
- Runge, K.W., and Robbins, P.W. (1986). A new yeast mutation in the glucosylation steps of the asparagine-linked glycosylation pathway. Formation of a novel asparagine-linked oligosaccharide containing two glucose residues. J. Biol. Chem. 261 15582–15590. [PubMed] [Google Scholar]
- Schachter, H., Reck, F., and Paulsen, H. (2003). Use of synthetic oligosaccharide substrate analogs to map the active sites of N-acetylglucosaminyltransferases I and II. Methods Enzymol. 363 459–475. [DOI] [PubMed] [Google Scholar]
- Schollen, E., Grunewald, S., Keldermans, L., Albrecht, B., Korner, C., and Matthijs, G. (2005). CDG-Id caused by homozygosity for an ALG3 mutation due to segmental maternal isodisomy UPD3(q21.3-qter). Eur. J. Med. Genet. 48 153–158. [DOI] [PubMed] [Google Scholar]
- Sharma, C.B., Knauer, R., and Lehle, L. (2001). Biosynthesis of lipid-linked oligosaccharides in yeast: the ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. Biol. Chem. 382 321–328. [DOI] [PubMed] [Google Scholar]
- Snider, M.D., Sultzman, L.A., and Robbins, P.W. (1980). Transmembrane location of oligosaccharide-lipid synthesis in microsomal vesicles. Cell 21 385–392. [DOI] [PubMed] [Google Scholar]
- Stagljar, I., te Heesen, S., and Aebi, M. (1994). New phenotype of mutations deficient in glucosylation of the lipid-linked oligosaccharide: Cloning of the ALG8 locus. Proc. Natl. Acad. Sci. USA 91 5977–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stibler, H., Stephani, U., and Kutsch, U. (1995). Carbohydrate-deficient glycoprotein syndrome – A fourth subtype. Neuropediatrics 26 235–237. [DOI] [PubMed] [Google Scholar]
- Strasser, R., Stadlmann, J., Svoboda, B., Altmann, F., Glossl, J., and Mach, L. (2005). Molecular basis of N-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex N-glycans. Biochem. J. 387 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm, V., et al. (1987). Stereotactic percutaneous single dose irradiation of brain metastases with a linear accelerator. Int. J. Radiat. Oncol. Biol. Phys. 13 279–282. [DOI] [PubMed] [Google Scholar]
- Sun, L., Eklund, E.A., Chung, W.K., Wang, C., Cohen, J., and Freeze, H.H. (2005). Congenital disorder of glycosylation id presenting with hyperinsulinemic hypoglycemia and islet cell hyperplasia. J. Clin. Endocrinol. Metab. 90 4371–4375. [DOI] [PubMed] [Google Scholar]
- Verostek, M.F., Atkinson, P.H., and Trimble, R.B. (1991). Structure of Saccharomyces cerevisiae alg3, sec18 mutant oligosaccharides. J. Biol. Chem. 266 5547–5551. [PubMed] [Google Scholar]
- Verostek, M.F., Atkinson, P.H., and Trimble, R.B. (1993). Glycoprotein biosynthesis in the alg3 Saccharomyces cerevisiae mutant. I. Role of glucose in the initial glycosylation of invertase in the endoplasmic reticulum. J. Biol. Chem. 268 12095–12103. [PubMed] [Google Scholar]
- Vitale, A., and Chrispeels, M.J. (1984). Transient N-acetylglucosamine in the biosynthesis of phytohemagglutinin: Attachment in the Golgi apparatus and removal in protein bodies. J. Cell Biol. 99 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen, A., Sturm, A., O'Neill, J., and Chrispeels, M.J. (1993). Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey, R., Knauer, R., Burda, P., Stagljar, I., te Heesen, S., Lehle, L., and Aebi, M. (1995). STT3, a highly conserved protein required for yeast oligosaccharyl transferase activity in vivo. EMBO J. 14 4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.