Abstract
The DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) transcription factor controls water deficit–inducible gene expression and requires posttranslational modification for its activation. The activation mechanism is not well understood; however, the stability of this protein in the nucleus was recently found to be important for its activation. Here, we report the isolation of Arabidopsis thaliana DREB2A-INTERACTING PROTEIN1 (DRIP1) and DRIP2, C3HC4 RING domain–containing proteins that interact with the DREB2A protein in the nucleus. An in vitro ubiquitination assay showed that they function as E3 ubiquitin ligases and are capable of mediating DREB2A ubiquitination. Overexpression of DRIP1 in Arabidopsis delayed the expression of DREB2A-regulated drought-responsive genes. Drought-inducible gene expression was slightly enhanced in the single T-DNA mutants of drip1-1 and drip2-1. By contrast, significantly enhanced gene expression was revealed in the drip1 drip2 double mutant under dehydration stress. Collectively, these data imply that DRIP1 and DRIP2 function negatively in the response of plants to drought stress. Moreover, overexpression of full-length DREB2A protein was more stable in drip1-1 than in the wild-type background. These results suggest that DRIP1 and DRIP2 act as novel negative regulators in drought-responsive gene expression by targeting DREB2A to 26S proteasome proteolysis.
INTRODUCTION
Plants have evolved a number of mechanisms to adapt to environmental stresses such as low temperature, drought, and salinity. Studies of stress-responsive gene expression suggest that transcriptional modulation is one of the most important steps that plants undergo to adapt to stress conditions (Thomashow, 1999; Zhu, 2002; Chinnusamy et al., 2004; Bartels and Sunkar, 2005; Yamaguchi-Shinozaki and Shinozaki, 2006). Many stress-inducible genes have been characterized in Arabidopsis thaliana and serve as indicators for the perception and transmission of stress signals in plant cells. Promoter analysis has confirmed that several kinds of cis-acting elements play fundamental roles in controlling gene expression under stress conditions (Yamaguchi-Shinozaki and Shinozaki, 2005). The dehydration-responsive element (DRE), having the same core motif as the C-repeat and the low-temperature-responsive element, was identified from a subset of stress-inducible gene promoters (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Jiang et al., 1996; Thomashow, 1999).
DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) was isolated using a yeast one-hybrid screen for proteins that bind to the DRE sequence. Also isolated in this screen was DREB1A/CBF3, an AP2/ERF-type transcription factor that binds to the DRE sequence (Stockinger et al., 1997; Liu et al., 1998). The function of DREB1/CBF genes (DREB1A/CBF3, DREB1B/CBF1, and DREB1C/CBF2) has been well documented in the past decade. However, the function of DREB2A was only recently identified by generating a constitutively active form of DREB2A (DREB2A-CA) (Sakuma et al., 2006a). Overexpression of the full-length DREB2A gene did not result in any remarkable alterations in plant phenotype or the expression of downstream genes (Liu et al., 1998). It was hypothesized that a posttranslational modification was required for the activation of DREB2A. Deletion of a Ser- and Thr-rich 30–amino acid region adjacent to the ERF/AP2 DNA binding domain of DREB2A transformed DREB2A into DREB2-CA. As a result, this region was named the DREB2A negative regulatory domain (DREB2A-NRD) (Sakuma et al., 2006a). Importantly, the overproduced DREB2A protein was not stable under normal growth conditions. By contrast, the DREB2A-CA protein could stably accumulate in plant cells (Sakuma et al., 2006a). Transgenic plants overexpressing DREB2A-CA were more tolerant to drought as well as high-temperature stress, and specific target genes of DREB2A were identified (Sakuma et al., 2006a, 2006b). Although the generation of DREB2A-CA enabled the identification of an in planta functional role for DREB2A, the mechanism controlling its activation and degradation remained to be elucidated.
Ubiquitination and proteolysis by the 26S proteasome pathway is involved in a wide rage of biological processes, as is protein phosphorylation (reviewed in Hare et al., 2003; Vierstra, 2003; Moon et al., 2004). The general ubiquitin pathway conjugates either single or multiple ubiquitin molecules to the target protein, thus enabling the ubiquitin-labeled protein to be recognized by the 26S proteasome and targeted for degradation. Three enzymes catalyze this reaction and ultimately covalently attach a 76–amino acid ubiquitin protein to the target protein on the Lys residue. The three enzymes are E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase). For the E3 category, Arabidopsis is predicted to contain ∼700 F-box proteins and ∼400 RING domain–containing proteins, which may function as multiple-subunit or single-subunit E3 ligases, respectively (Gagne et al., 2002; Kosarev et al., 2002; Kuroda et al., 2002; Risseeuw et al., 2003; Stone et al., 2005). The RING domain is rich in Cys, and the eight metal ligand residues coordinate zinc ions in a cross-brace structure (Barlow et al., 1994; Borden and Freemont 1996; Borden 2000). Depending on whether the metal ligand at position 5 is a His or a Cys, RING domain proteins are divided into C3HC4 and C3H2C3 types (Freemont, 1993; Lovering et al., 1993).
Some single-subunit RING E3 ligases target transcription factors for degradation and thereby regulate gene expression in response to environmental or hormone signals. COP1 (for Constitutive Photomorphogenesis1) protein is a well-studied example that functions as a negative regulator of the light response (Deng et al., 1991; von Arnim and Deng, 1994). COP1 represses photomorphogenesis by targeting the activators of light-responsive genes for degradation, such as HY5, LAF1, and HFR1 (Hardtke et al., 2000; Osterlund et al., 2000; Seo et al., 2003; Yang et al., 2005). SINAT5 (for SINA of Arabidopsis5) is another single-subunit RING E3 ligase that interacts with NAC1 (for NAM/CUC-like protein1) and directs it to degradation for the attenuation of auxin signals (Xie et al., 2002). AIP2 (for ABI3 Interaction Protein2) is another single-subunit RING E3 ligase that targets a B3 domain transcription factor (ABI3) for proteolysis. This targeted degradation relieves the abscisic acid effect for germination after stratification (Zhang et al., 2005). HOS1 (for High Expression of Osmotically Responsive Gene1), another RING E3 ligase, was recently shown to target a MYC-like basic helix-loop-helix (ICE1) transcriptional activator to 26S proteasome degradation for the attenuation of cold stress in Arabidopsis (Dong et al., 2006). Therefore, it is interesting to speculate whether a ubiquitin E3 ligase might function in plant water deficit stress response through an interaction with key transcription factors signaling in such an important biological process.
In this study, we identified two C3HC4 RING–domain containing proteins, DREB2A-INTERACTING PROTEIN1 (DRIP1) and DRIP2, that interact with the key transcription factor DREB2A. These proteins were shown to function as E3 ubiquitin ligases and mediate DREB2A degradation as a mechanism to tightly control DREB2A protein abundance. The drip1 drip2 double mutant showed increased expression of stress-responsive genes and also a delay in plant development and growth. We hypothesize that DRIP1 and DRIP2 negatively regulate the response to water stress.
RESULTS
Isolation of DRIP1
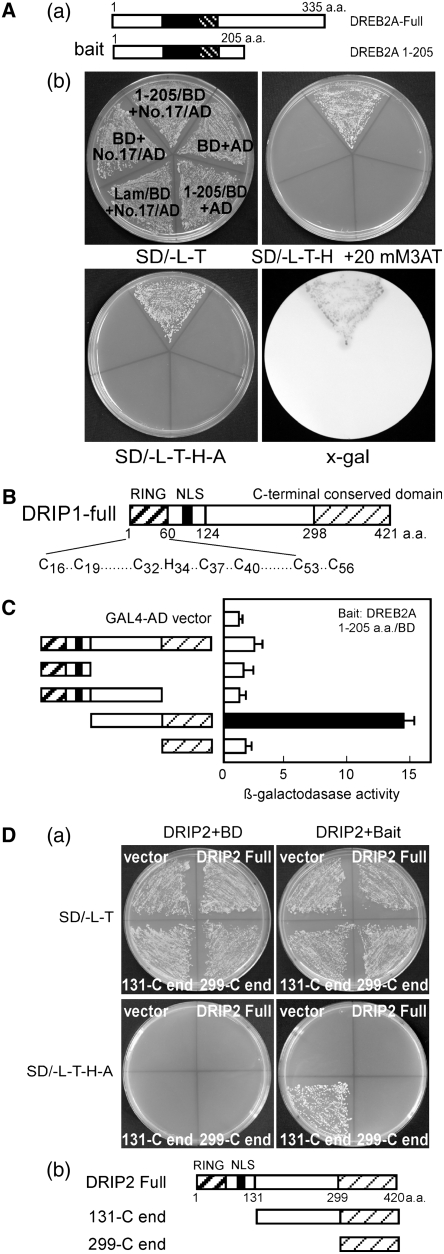
A yeast two-hybrid screen was performed to identify proteins that interact with DREB2A. The N-terminal region of DREB2A, between residues 1 and 205 (DREB2A 1-205), was fused to the yeast GAL4 DNA binding domain and used as a bait protein for screening against an Arabidopsis cDNA library fused to the yeast GAL4 activation domain (AD). Approximately 2.5 × 106 yeast transformants were screened on a synthetic defined (SD) medium lacking Leu, Trp, and His (SD/-T-L-H) plus 1.5 mM 3-amino-1,2,4-triazole. Forty-two positive clones were obtained, and β-galactosidase activity was individually assessed for all of these clones (see Supplemental Table 2 online). Sequence determination from the clone exhibiting the strongest activity identified a C3HC4 RING domain–containing protein of unknown function. Considering the high reporter gene activity and DREB2A protein instability, we further examined this clone and confirmed its interaction with DREB2A. As shown in Figure 1A, even under several selective conditions (SD/-T-L-H + 20 mM 3-amino-1,2,4-triazole or SD/-T-L-H-A [for adenine]), yeast cells transformed with the interacting clone and the bait plasmid could grow well. Conversely, the growth of other yeast cells was completely inhibited. The protein encoded by this gene was subsequently named DRIP1. A homolog of DRIP1 (At1g06770) in the Arabidopsis genome (At2g30580) was found to be closely related to DRIP1. Analysis with ClustalW revealed that At2g30580 shares 66% amino acid identity and 77% similarity with DRIP1. Consequently, this gene was designated DRIP2. Similar to DRIP1, the DRIP2 protein also contains a conserved N-terminal C3HC4-type RING domain of ∼60 amino acids. Both proteins contain a predicted nuclear localization signal (NLS) (Figure 1B). Moreover, orthologs of DRIP1 and DRIP2 were also found in the GenBank database from rice (Oryza sativa) and tomato (Solanum lycopersicum). Multiple sequence alignment revealed that these proteins share a region of high conservation in their C termini; however, no characterized motif could be identified within these regions (see Supplemental Figure 1 online).
Figure 1.
Identification of DRIP1 and DRIP2 Interaction with DREB2A via Yeast Two-Hybrid Analysis.
(A) Isolation of DRIP1 from yeast two-hybrid screening. (a) Diagram of the bait structure for DREB2A 1-205. Filled boxes indicate the AP2/ERF DNA binding domain; heavy striped boxes indicate the NRD domain. (b) The positive clone 17 (encoding DRIP1) growing on selective medium and exhibiting β-galactosidase activity. BD (for DNA binding domain) indicates pGBKT7 vector; AD indicates pGADT7 vector; Lam/BD indicates human lamin C fused to the pGBKT7 vector as a negative control.
(B) Illustration of DRIP1 and DRIP2 protein domain organization. Heavy striped boxes indicate the RING domain; filled boxes indicate the nuclear localization domain (NLS); light striped boxes identify the conserved C-terminal region.
(C) Localization of the DRIP1 interaction domain with yeast two-hybrid assays. β-Galactosidase activity was measured for each transformant. Error bars indicate se (n = 3). The enzyme activity of empty vector was defined as 1.0. The diagram for each construct is indicated (left) with domains indicated as in (B).
(D) Identification of DRIP2–DREB2A protein interaction. (a) Yeast cells growing on SD/-L-T plates or selective medium SD/-L-T-H-A. (b) Illustration of each construct, with domains indicated as in (B).
Localization of DRIP1 and DRIP2 Interacting Domains with DREB2A
We conducted additional yeast two-hybrid assays to localize the domain of DRIP1 that interacts with DREB2A. Based on the sequence alignment information, several DRIP1 fragments were generated and fused to the yeast GAL4-AD vector, and each construct was transformed into yeast cells containing the DREB2A bait fragment. The fragment excluding the RING and NLS domains strongly interacted with DREB2A and showed the highest β-galactosidase activity (Figure 1C). The fragment containing only the RING-NLS or lacking the C-terminal conserved region was not sufficient for mediating a protein–protein interaction (Figure 1C). Reporter gene expression of the bait cells transformed by full-length DRIP1 was lower than that of the N-terminal truncated DRIP1 fragment. These data were consistent with the fact that the clone isolated from screening was an N-terminal truncated cDNA and did not contain a full-length cDNA of DRIP1.
To determine whether DRIP2 also interacts with DREB2A, full-length DRIP2 and two different N-terminal truncated fragments were constructed in the yeast GAL4-AD vector and tested in yeast two-hybrid assays. When these constructs were transformed into yeast cells containing either empty pGBKT7 or the DREB2A bait construct, all cells could grow on SD/-L-T plates. However, on the selective medium, SD/-L-T-H-A, only bait cells transformed with the construct containing the DRIP2 N-terminal truncated fragment lacking the RING domain and NLS could grow well; other transformants could not survive (Figure 1D). Similar to DRIP1, these results suggest that the protein C-terminal region of DRIP2 is sufficient for the protein–protein interaction with DREB2A.
DRIP1 Interacts with DREB2A in Vitro and in Vivo
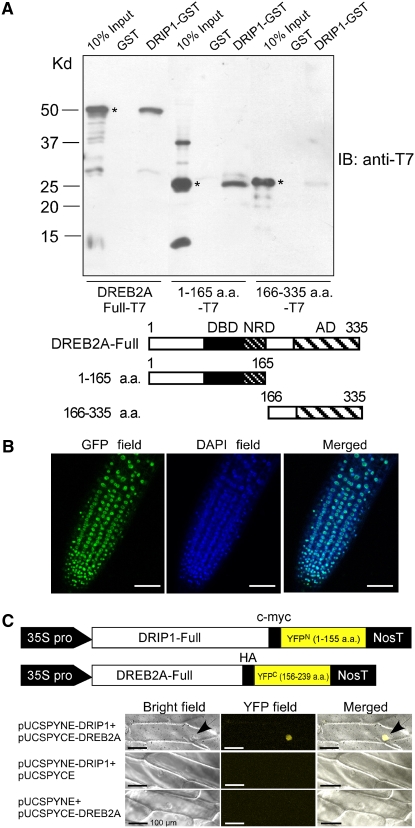
We further tested the interaction between the full-length DRIP1 protein and DREB2A with in vitro protein pull-down assays. Full-length DREB2A, the N-terminal region (amino acids 1 to 165, containing the DNA binding and NRD domains), and the C-terminal region (amino acids 166 to 335) were fused to the T7 tag in the pET28a vector. The three constructs were transformed into Escherichia coli, and the isopropylthio-β-galactoside–induced cell lysates were mixed with the DRIP1-GST (for glutathione S-transferase) fusion or GST protein and glutathione-Sepharose. After washing, the resultant glutathione-Sepharose was analyzed by anti-T7 immunoblotting. As a result, the full-length DREB2A-T7 and DREB2A 1-165-T7 proteins could be pulled down by DRIP1-GST, whereas DREB2A 166-335-T7 protein was barely retained by the bait DRIP1-GST protein (Figure 2A). These in vitro data indicate that DRIP1 interacts with the N-terminal region of DREB2A containing the DNA binding domain and NRD. Apparently, the AD of DREB2A is not involved in the interaction with DRIP1.
Figure 2.
In Vitro and in Vivo Interaction of the DRIP1 and DREB2A Proteins.
(A) In vitro pull-down assays of full-length or truncated DREB2A protein with GST or DRIP1-GST fusion protein. DRIP1-GST fusion protein was used as a bait to pull down the full-length or truncated DREB2A-T7 protein from the induced cell extracts. GST protein was assayed as a negative control. Immunoblot detection of prey protein is with a T7 antibody. Asterisks indicate the corresponding target proteins. The bottom panel diagrams the three kinds of target proteins.
(B) Nuclear localization of DRIP1-GFP fusion protein. Photomicrographs of transgenic Arabidopsis root harboring the 35S:DRIP1-GFP construct. GFP fluorescence (left), DAPI (for 4′,6-diamidine-2′-phenylindole dihydrochloride) staining (middle), and overlay images (right) are shown. Bars = 50 μm.
(C) Verification of in vivo DRIP1–DREB2A interaction with the BiFC system. The top panel illustrates constructs of pUCSPYNE-DRIP1 and pUCSPYCE-DREB2A. Bright-field, YFP fluorescence, and merged images are shown for each kind of transformation combination. Arrows indicate nuclei. Bars = 100 μm.
As a first step to confirm an in vivo interaction of DRIP1 with DREB2A, we confirmed that DRIP1 was a nuclear protein. Microscopic observations of transgenic plants carrying the 35S:DRIP1-GFP (for green fluorescent protein) construct confirmed its nuclear localization (Figure 2B). We then employed the bimolecular fluorescence complementation (BiFC) system to study their interaction in plant cells (Walter et al., 2004). Full-length DRIP1 cDNA was fused to the N-terminal region of the yellow fluorescent protein (YFPN1-155), and the DREB2A cDNA was fused to the C-terminal region of YFP (YFPC156-239). The two constructs were cobombarded into onion (Allium cepa) epidermal cells. In parallel, the empty vectors in combination with each fusion construct were also transformed into the cells. After an overnight incubation, YFP signals were observed with fluorescence microscopy. Samples cotransformed by pUCSPYNE-DRIP1 and pUCSPYCE-DREB2A yielded YFP fluorescence, whereas, all of the samples cotransformed by the empty vector with either pUCSPYNE-DRIP1 or pUCSPYCE-DREB2A failed to give any YFP signal (Figure 2C). These results indicate that DRIP1 and DREB2A colocalize and interact in plant cell nuclei.
DRIP1 and DRIP2 Potentially Function as E3 Ligases and Can Mediate DREB2A Ubiquitination
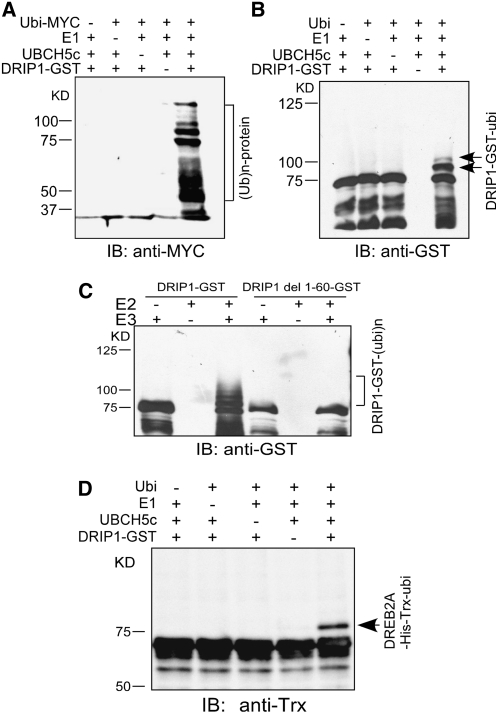
In vitro ubiquitination experiments were performed with DRIP1 protein to confirm its function as an E3 ligase. Purified DRIP1-GST fusion protein was mixed with myc-tagged ubiquitin (ubi-myc), rabbit E1, and human UBCH5c (E2). Immunoblot analysis with anti-myc showed that ubiquitinated proteins were detected in the presence of all these components (Figure 3A). Furthermore, when DRIP1 autoubiquitination activity was assessed with anti-GST, two additional higher molecular weight protein bands appeared when the DRIP1-GST protein was mixed with ubiquitin, E1, and E2. According to their molecular weights, these additional bands appear to result from the attachment of one or two ubiquitin monomers (Figure 3B). These results suggest that DRIP1 protein could be autoubiquitinated in the presence of E1 and E2 enzymes. To verify whether the RING domain was essential for DRIP1 protein ubiquitination ability, we deleted the first 60 amino acids of DRIP1, thereby removing the RING domain, and purified this truncated protein as a GST fusion (DRIP1 del 1-60-GST). In the presence of ubiquitin, E1, and E2 enzymes, the mutated DRIP1 protein was incapable of autoubiquitination (Figure 3C). These data confirm that the RING domain is required to facilitate protein ubiquitination.
Figure 3.
DRIP1 Functions as an E3 Ubiquitin Ligase and Mediates DREB2A Protein Ubiquitination.
(A) In the presence of the ubiquitin-myc, E1, and E2 enzymes, DRIP1-GST fusion proteins display ubiquitin E3 ligase activity. Protein bands with ubiquitin attached were detected by anti-myc immunoblot (IB) analysis (10% SDS-PAGE).
(B) Detection of DRIP1-GST autoubiquitination. DRIP1-GST fusion proteins were detected with a GST antibody, and shifted bands indicate the attachment of one or two ubiquitin molecules (6% SDS-PAGE).
(C) Assessment of E3 ubiquitin ligase activity. Wild-type and RING domain–deleted DRIP1-GST fusion proteins (DRIP1 del1-60-GST) were tested for E3 ubiquitin ligase activity in the presence of UBCH5c (E2), E1, and ubiquitin. A GST antibody was used to detect DRIP1-GST fusion protein (10% SDS-PAGE).
(D) DRIP1 mediates the ubiquitination of DREB2A protein. The full-length DREB2A protein was fused with His and Trx tags (DREB2A-His-Trx) and used as the substrate for the assay. Anti-Trx was used in the immunoblot analysis for the detection of Trx-tagged substrate protein (6% SDS-PAGE).
In order to determine whether DRIP1 could mediate DREB2A protein ubiquitination, the in vitro ubiquitination assay was conducted using DREB2A protein as a substrate. The DREB2A protein was fused with His and Trx tags, and the recombinant fusion protein was purified by the His tag affinity to a nickel-nitrilotriacetic acid agarose matrix. As shown in Figure 3D, in the presence of DRIP1, ubiquitin, E1, and E2, a higher molecular weight shifted band could be observed by anti-His immunoblot analysis. When any component of this assay was omitted from the mixture, only the original DREB2A-His-Trx protein band could be detected. According to the molecular weight, the shifted band corresponded to one additional ubiquitin monomer attached on the substrate. Similar results were obtained when a DRIP2-MBP fusion protein was used in the ubiquitination assays (see Supplemental Figure 2 online).
DRIP1 Is Expressed in Various Tissues and Is Not Induced by Stresses
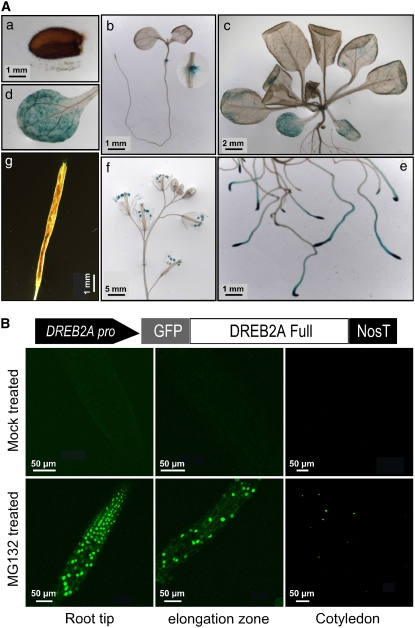
A 2.5-kb promoter sequence of DRIP1 was cloned and constructed in front of the β-glucuronidase (GUS) gene to facilitate the study of DRIP1 tissue-specific expression patterns. DRIP1pro:GUS transgenic plants were generated, and DRIP1 gene expression was followed developmentally. Figure 4A shows the representative GUS staining observed in a majority of the transgenic lines. GUS activity was observed in cotyledon nodes in 10-d-old seedlings (Figure 4A-b). In 3-week-old plants, GUS expression was detected in cotyledons and weakly in rosette leaves (Figures 4A-c and 4A-d). In roots, DRIP1 promoter activity was clearly exhibited in root tips and elongation zones (Figure 4A-e). In flowers, GUS expression was observed in the anthers of opened flowers (Figure 4A-f). After pollination, DRIP1 promoter activity was also exhibited in the stigma (Figure 4-g). DRIP1 expression was barely detected in mature dry seeds (Figure 4A-a). We also investigated the tissue-specific gene expression of DRIP1 and DRIP2 by quantitative RT-PCR analysis and revealed similar patterns of gene expression for DRIP1 and DRIP2 (see Supplemental Figure 3A online). DRIP1 and DRIP2 expression was also monitored under various stress conditions. No clear induction was observed for either gene when the plants were subjected to 250 mM NaCl, dehydration, or abscisic acid treatment. An exception to this was the minor induction of DRIP2 under dehydration stress (see Supplemental Figure 3B online). In general, DRIP1 and DRIP2 appear to be expressed in plants in only a subset of tissues, but at similar levels regardless of stress conditions.
Figure 4.
Expression Profiling of DRIP1 in Different Tissues, and the Degradation of DREB2A-GFP Protein Is Inhibited by the MG132 Proteasome Inhibitor.
(A) GUS staining of DRIP1pro:GUS transgenic plants from different growth stages. (a) Gene expression from mature seeds. (b) Ten-day-old seedlings growing on a GM agar plate. (c) Aerial portions of 3-week-old seedlings. (d) Higher magnification image of (c) for observing cotyledons. (e) Root tips and elongation zones from 3-week-old seedlings. (f) Flowers from 7-week-old plants. (g) Siliques from 7-week-old plants. Bars in each panel indicate actual lengths.
(B) A native DREB2A promoter sequence was constructed to drive GFP-DREB2A gene expression. Three-week-old plants treated (or mock-treated) with MG132 under dim light overnight were observed with fluorescence microscopy.
Potential Degradation of DREB2A Protein by the 26S Proteasome
We previously reported that the DREB2A gene was induced by drought stress but also showed low levels of expression under normal growth conditions, in contrast with undetectable levels of expression of DREB1A (Liu et al., 1998; Sakuma et al., 2002). DREB2A transcripts could be clearly detected in plant roots and weakly in the leaves under nonstressed conditions (Liu et al., 1998). We were interested to characterize DREB2A protein levels under normal conditions in plants. Transgenic Arabidopsis plants harboring a GFP-DREB2A construct driven by the DREB2A promoter (DREB2Apro:GFP-DREB2A) were generated in an attempt to determine DREB2A protein stability in cells. The DREB2A-GFP protein was not detected in either roots or leaves under normal growth conditions. However, when these plants were treated with a 26S proteasome inhibitor, MG132, clear GFP fluorescence was observed with the microscope in both roots and cotyledons (Figure 4B). A similar phenomenon was also observed when 35S:GFP-DREB2A transgenic plants were examined under similar conditions (see Supplemental Figure 4 online). Although the DREB2A gene was expressed under normal growth conditions, the DREB2A protein is apparently unstable and may be degraded by the 26S proteasome.
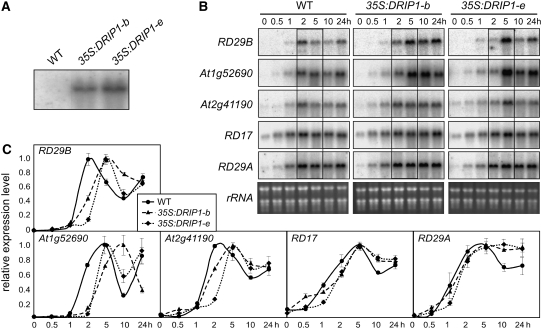
Overexpression of DRIP1 Delays Stress-Responsive Gene Expression under Dehydration Stress
Transgenic plants overexpressing DRIP1 were generated using an enhanced 35S promoter (Sakuma et al., 2006a). The growth of all transgenic lines was similar to that of wild-type plants, without any remarkable phenotypic changes (data not shown). Two transgenic lines with a similar level of transgene expression were further characterized (35S:DRIP1-b and 35S:DRIP1-e) (Figure 5A) and monitored for the expression of a set of stress-inducible marker genes. When subjected to dehydration stress, the expression of these stress marker genes in the transgenic plants was not obviously different from that in the wild type. However, upon a close comparison of gene expression under 2 and 5 h of dehydration stress between the transgenic and wild-type plants, the inductions of RD29B, At1g52690, and At2g41190, specific target genes of DREB2A (Sakuma et al., 2006a, 2006b), were delayed in the DRIP1 overexpressors relative to the wild type (Figure 5B). Under the same experimental conditions, maximum expression of these genes peaked earlier in the wild type than in the transgenics. Unlike specific target genes of DREB2A, it is interesting that the induction of genes targeted by both DREB1A and DREB2A, such as RD17 and RD29A, was not postponed. The expression level of these genes was also quantified by real-time PCR analysis; the data coincided with and supported the data obtained from RNA gel blot analysis (Figure 5C). This suggests that DRIP1 overexpression negatively affects DREB2A-regulated gene expression in an early dehydration stress response.
Figure 5.
Overexpression of DRIP1 in Plants Delays the Expression of DREB2A Target Genes in Response to Dehydration Stress.
(A) RNA gel blot analysis of two transgenic lines overexpressing DRIP1 to show the levels of DRIP1 mRNA expression. Transgene expression levels of lines b and e are shown.
(B) RNA gel blot analysis of some stress-responsive genes (indicated at left) under dehydration stress (times indicated above panels) in 3-week-old plants of the wild type and two overexpression lines (b and e). Changes in gene expression are highlighted by frames. Total RNA is shown to indicate equal loading.
(C) Quantitative RT-PCR analysis of the gene expression level in (B). The highest expression level in each type of plant was defined as 1.0. Error bars indicate se (n = 2).
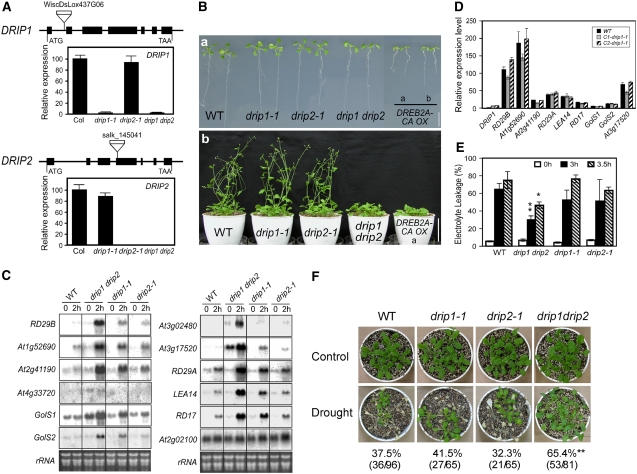
Identification of drip1-1, drip2-1, and drip1 drip2 Mutants
In order to further understand the physiological function of DRIP1 and DRIP2, we obtained T-DNA insertion mutants of drip1-1 and drip2-1 in the Columbia background. DRIP1 and DRIP2 gene expression was monitored in homozygous mutants with quantitative RT-PCR. DRIP1 expression was significantly reduced in the drip1-1 mutant, and DRIP2 expression was completely interrupted by T-DNA insertion in the drip2-1 mutant (Figure 6A). However, both single mutants did not give an apparent morphological phenotype (Figure 6B). Considering the high homology of DRIP1 and DRIP2 and their similar expression profiles, we generated a drip1 drip2 double mutant. Further RT-PCR analysis showed that the expression of both DRIP1 and DRIP2 genes in the double mutant was disrupted as in the single mutants (Figure 6A).
Figure 6.
Phenotypic and Gene Expression Studies of drip1-1, drip2-1, and drip1 drip2 Mutants.
(A) Relative DRIP1 and DRIP2 gene expression in drip1-1, drip2-1, and drip1 drip2 mutants as determined by quantitative RT-PCR. Error bars indicate se (n = 3). Expression of DRIP1 and DRIP2 in the wild type was determined as 100. The gene organizations of DRIP1 and DRIP2 and their T-DNA insertions are shown; exons are indicated as thick lines and introns as thin lines.
(B) Phenotypes of drip1-1, drip2-1, and drip1 drip2 mutants. (a) Growth of 10-d-old seedlings on agar plates. Growth of DREB2A-CA overexpression line b is also shown for comparison (Sakuma et al., 2006a). (b) Three-week-old plants were transferred into soil and photographed at 5 weeks. Growth of DREB2A-CA overexpressor line b is also shown.
(C) Stress-responsive gene expression analysis for the wild type and mutants in response to a 2-h dehydration stress. Total RNA is shown as a loading control.
(D) Complementation of the drip1-1 mutant. The expression level of DRIP1 was determined in the wild type and two independent complementation lines (C1-drip1-1 and C2-drip1-1) under normal conditions by quantitative RT-PCR. Relative expression levels of stress-responsive genes under a 2-h dehydration stress were also compared. For each gene, the expression level in the wild type under nonstressed conditions was defined as 1.0. Error bars indicate se (n = 3).
(E) Leaf electrolyte leakage of the wild type and drip1-1, drip2-1, and drip1 drip2 mutants before and after dehydration stress. Error bars indicate se of three replicates. * P < 0.05, ** P < 0.01, by t test.
(F) Survival rates of wild-type, drip1-1, drip2-1, and drip1 drip2 plants after being exposed to drought stress for 14 to 16 d, which is when the most significant difference was observed. Photographs were taken after a 1-week recovery period subsequent to rewatering. ** P < 0.01, by t test.
Plant development and growth of the drip1 drip2 double mutant were significantly delayed and prolonged compared with those of the wild type (Columbia) and the two single mutants. After 10 d, wild-type seedlings developed two rosette leaves in addition to two cotyledons, while most of the double mutant plants only had two cotyledon leaves or were just starting to produce the first true leaves. Moreover, after 5 weeks, both the wild type and the two single mutants already bolted and developed ∼12- to 13-cm-long inflorescences. Conversely, most of the drip1 drip2 plants still remained in the rosette stage or had just started bolting (Figure 6B-b). A similar retardation in plant development was also observed in DREB2A-CA overexpressors (Figure 6B-b). In addition, flowering and seeding of drip1 drip2 were also delayed compared with the wild type. However, if watering was maintained, the overall plant size of double mutants eventually exceeded that of wild-type plants (data not shown).
drip1 drip2 Mutation Enhances Stress-Responsive Gene Expression and Confers Tolerance to Dehydration Stress
We investigated alterations in the transcriptome between the drip1 drip2 mutant and wild-type plants under both normal and 2-h dehydration conditions with Agilent 22k microarray analyses. A massive transcriptomic change was revealed in the drip1 drip2 mutant under both normal and stressed conditions. Especially in 2-h dehydrated drip1 drip2 plants, the expression of many stress-inducible genes was significantly enhanced compared with the wild type. For the unstressed plants, 317 genes were upregulated more than twofold (fold change absolute > 2.0) in the mutant. Under nonstressed conditions, several stress-inducible genes were enhanced in the drip1 drip2 plants. Among the 317 upregulated genes, 28, 11, 43, and 26 genes were previously identified to be cold-, dehydration-, high salinity–, and abscisic acid–inducible, respectively (see Supplemental Data Set 2 online; a complete microarray data set is available at ArrayExpress [http://www.ebi.ac.uk/arrayexpress/] under accession number E-MEXP-1351). Thirty-four of the genes upregulated in drip1 drip2 were also found to be upregulated more than twofold in the DREB2A-CA overexpressor (Sakuma et al., 2006b). Also, 49 of the genes upregulated in drip1 drip2 contain DRE sequence(s) in their promoters. Among them, 12 genes were upregulated by DREB2A-CA. For the 2-h dehydrated plants, the expression of 369 genes was enhanced more than twofold in the double mutant relative to the wild type. Among the 369 upregulated genes, 121, 164, 202, and 125 genes were characterized to be cold-, dehydration-, high salinity–, and abscisic acid–inducible, respectively (see Supplemental Data Set 3 online; a complete microarray data set is available at ArrayExpress [http://www.ebi.ac.uk/arrayexpress/] under accession number E-MEXP-1352). Sixty-seven of the genes upregulated in drip1 drip2 under stress were also upregulated more than twofold in the DREB2A-CA overexpressor. Also, 134 of the genes upregulated in drip1 drip2 under stress contain DRE sequence(s) in the promoters (Sakuma et al., 2006b). Among them, 43 genes were upregulated by DREB2A-CA. These data indicate that DRIP1 and DRIP2 negatively function in DREB2A-regulated drought-inducible gene expression, especially under dehydration stress.
In order to verify the microarray results and to have a closer view of how DREB2A-regulated genes were affected in drip1 drip2, we investigated the expression of some stress-inducible genes by RNA gel blot analysis in wild-type, drip1-1, drip2-1, and drip1 drip2 plants. Expression of DREB2A-regulated genes, such as RD29B, At1g52690, At2g41190, and At4g33720, was significantly enhanced in the 2-h dehydrated double mutant compared with the wild type (Figure 6C) (Sakuma et al., 2006a). Prior analysis of the DREB2A-CA overexpressors placed GolS1, GolS2, At3g02480, and At3g17520 in the DREB2A regulon (Sakuma et al., 2006b). Similarly, expression of these genes was elevated in the 2-h dehydrated drip1 drip2 plants (Figure 6C). Expression of some genes was slightly enhanced in the unstressed plants relative to the wild type. Even in the drip1-1 and drip2-1 single mutants, the expression of most genes was also increased in the 2-h dehydrated plants, although the elevated levels were not very high (Figure 6C). The expression of genes regulated by both DREB1A and DREB2A, such as RD29A, LEA14, and RD17, was also verified to be upregulated in the mutants, but only under 2 h of dehydration stress (Figure 6C). However, the expression of a DREB1A-specific target gene, At2g02100, was not altered in the mutants (Figure 6C) (Sakuma et al., 2006a). Consistent with microarray results, the expression of some drought-responsive genes was mildly enhanced under nonstressed conditions. Additionally, many drought-responsive genes, especially DREB2A-regulated genes, were significantly upregulated under dehydration stress in the drip1 drip2 mutant relative to the wild type.
To confirm that the enhanced gene expression was caused by a disruption in the drip1 locus, we transformed the 35S:DRIP1 construct into the drip1-1 mutant as a means to complement the gene expression. From two independent complementation lines, none of the monitored stress-responsive genes exhibited higher expression relative to wild-type plants (Figure 6D). Recovery of DRIP1 gene function clearly complemented the mutant phenotype. DNA blot analysis suggested that the drip2-1 mutant was only a single-copy T-DNA insertional mutant (data not shown). Collectively, these data indicate that DRIP1 and DRIP2 function negatively and redundantly in drought-responsive gene expression, especially through affecting DREB2A-regulated gene expression.
In order to evaluate the comparative drought stress tolerance of three mutants and the wild type, we first monitored leaf electrolyte leakage subsequent to drought stress. Aerial parts of 3-week-old seedlings were removed from agar plates and subjected to dehydration stress on an empty plate. After a 3-h dehydration treatment, >64% of ions leaked from cells in the wild type, whereas ion leakage of the drip1 drip2 double mutant was less than 32%. Although a slight reduction of ion leakage was also observed in the drip1-1 and drip2-1 single mutants, these data did not meet the significance criteria (Figure 6E). A clear reduction of ion leakage was also observed in the drip1 drip2 mutant after a 3.5-h exposure to dehydration stress (Figure 6E). Drought stress tolerance was also evaluated by the survival rate in soil after plants stopped receiving water. When data were analyzed from several independent tests, 37.5% of the wild-type plants survived the drought stress treatment, and 41.5% and 32.3% of drip1-1 and drip2-1 plants, respectively, recovered from the stress after rewatering. Although the difference between the wild type and the single mutants was not statistically significant, the survival rate of the drip1 drip2 double mutant was 65.4%, which is significantly higher than that of the wild-type plants (Figure 6F). We also monitored the plant water content under dehydration stress at different time points. The standardized water content in the drip1 drip2 double mutant in response to water stress was slightly higher than that of wild-type plants. It is possible that this difference in water content might contribute to the increased plant dehydration stress tolerance in the double mutants (see Supplemental Figure 5 online).
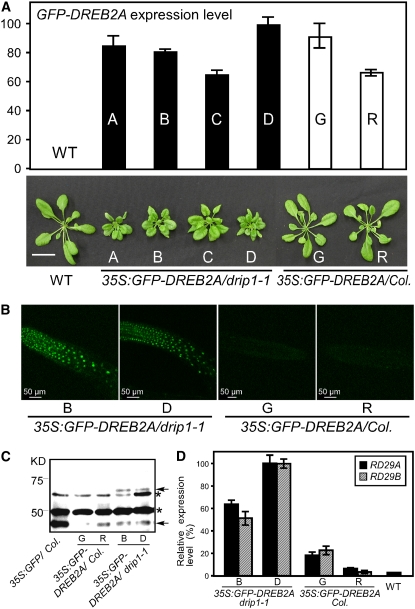
Overexpression of DREB2A in the drip1-1 Mutant Results in a Dwarf Phenotype That Mimics the DREB2A-CA Overexpressor
Since DREB2A is expressed at very low levels under nonstressed conditions, in order to understand DRIP1 function on DREB2A protein stability under normal conditions, we transformed the 35S:GFP-DREB2A construct into the drip1-1 mutant and obtained eight independent transgenic lines. Quantitative RT-PCR analysis revealed a relatively high level of transgene expression in four lines (Figure 7A). Interestingly, all four lines showed growth retardation and an overall dwarfed phenotype, resembling the DREB2A-CA overexpressor phenotype (Figure 7B). However, when the 35S:GFP-DREB2A construct was transformed into the wild-type Columbia plants, with a comparable level of transgene expression, normal growth was observed in transgenic plants (Figure 7B). Furthermore, stronger green fluorescent signals were observed in the roots of the 35S:GFP-DREB2A/drip1-1 plants relative to the 35S:GFP-DREB2A/Col plants. An anti-GFP immunoblot detection of GFP or GFP fusion proteins in 35S:GFP/Col, 35S:GFP-DREB2A/Col, and 35S:GFP-DREB2A/drip1-1 plants was performed. The predicted GFP-DREB2A molecular mass is ∼64.3 kD. With protein gel blot analysis, a protein band was detected in two independent 35S:GFP-DREB2A/drip1-1 transgenic lines (B and D) in the corresponding size range. However, no band was detected in 35S:GFP/Col or two 35S:GFP-DREB2A/Col transgenic lines, G and R (Figure 7C). The expression of two DREB2A-regulated genes (RD29A and RD29B) under normal conditions was compared in two separate lines from both 35S:GFP-DREB2A/drip1-1 and 35S:GFP-DREB2A/Col with quantitative RT-PCR analysis. As expected, even under nonstressed conditions, these genes were higher in the 35S:GFP-DREB2A/drip1-1 plants than in 35S:GFP-DREB2A/Col plants (Figure 7D). These results suggest that DRIP1 could mediate DREB2A protein degradation under nonstressed conditions and consequently may limit the amount of DREB2A to minimize its potential negative effects on plant growth.
Figure 7.
The Phenotype and Expression of Downstream Genes in 35S:GFP-DREB2A/drip1-1 Transgenic Plants.
(A) Relative transgene expression of GFP-DREB2A in 35S:GFP-DREB2A/drip1-1, 35S:GFP-DREB2A/Col, and wild-type plants as measured by quantitative RT-PCR. Error bars indicate se (n = 3). The highest gene expression level was designated as 100. The phenotypes for 4-week-old plants for all lines are shown at bottom. Bar = 2 cm.
(B) Confocal microscopic observation of GFP fluorescence in the roots of 35S:GFP-DREB2A/drip1-1-B and -D and 35S:GFP-DREB2A/Col-G and -R.
(C) Protein gel blot analysis of GFP or GFP-DREB2A protein levels in 35S:GFP/Col, 35S:GFP-DREB2A/drip1-1-B and -D, and 35S:GFP-DREB2A/Col-G and -R using anti-GFP antibody. The asterisks indicate unknown protein bands. The bottom arrow indicates the GFP protein band, and the top arrow indicates the GFP-DREB2A fusion protein band.
(D) Relative gene expression of RD29A and RD29B in 35S:GFP-DREB2A/drip1-1-B and -D and 35S:GFP-DREB2A/Col-G and -R and wild-type plants as measured by quantitative RT-PCR. Error bars indicate se (n = 3).
DISCUSSION
Signal transduction networks and the regulation of transcription factors affecting stress-responsive gene expression play a central role in stress responses. Among these transcription factors, DREB2A is one of the most important transcription factors controlling drought-responsive gene expression for several reasons: its expression is significantly induced by dehydration and high salt stresses; many stress-inducible genes containing DRE sequence(s) in their promoters are downstream target genes of DREB2A; and overexpression of DREB2A-CA confers strong drought stress tolerance to transgenic Arabidopsis (Sakuma et al., 2006a). Although the protein stability of DREB2A is known to be important for the regulation of DREB2A activity, the mechanism of its degradation has not been elucidated.
The single-subunit E3 ligase simultaneously docks ubi-E2 and intermediate and target substrate proteins for ubiquitination in a single protein (Moon et al., 2004; Stone et al., 2005). These two biological functions were hypothesized to be mediated by the RING domain and another protein–protein interactive domain. In this research, yeast two-hybrid analysis demonstrated that DRIP1 and DRIP2 proteins, both containing a C3H4 RING domain, interact with the DREB2A protein. Use of in vitro pull-down assays and the in vivo BiFC system confirmed the protein interaction of DRIP1 with DREB2A (Figures 2A and 2C). Moreover, the RING domains of DRIP1 and DRIP2 were found to be essential for the E3 ligase activity, and their C-terminal conserved regions were responsible for the protein interaction with DREB2A (Figures 1C, 1D, and 3; see Supplemental Figures 1 and 3 online). However, in yeast two-hybrid assays, the reporter gene activity of yeast cells transformed by the full-length DRIP1 construct and DREB2A bait was quite low (Figure 1C). One explanation is that in yeast cells, once the full-length DRIP protein associates with the DREB2A bait protein, it may also lead to the destabilization of the target protein. It is also possible that the full-length DRIP1 protein interaction with DREB2A is actually lower than that of the truncated protein. This would be reasonable to consider, since many protein–protein interaction studies suggest the possibility that the protein–protein interaction might be masked by some domain existing in the full-length protein. Thus, the truncated protein may be more interactive than its counterpart. For example, SPA1 interacts with COP1 through the coiled-coil domains that are contained in both proteins, and the interaction between the two coiled-coil domains was considerably higher than the interaction observed between the two full-length proteins (Saijo et al., 2003). The coiled-coil domain of the COP1 protein, rather than the full-length protein, also displayed a higher interaction with the HFR1 protein (Yang et al., 2005).
In vitro ubiquitination assays in the presence of ubiquitin, rabbit E1, and human UBCH5c (E2) showed that DRIP1 and DRIP2 proteins are clearly autoubiquitinated and display E3 ligase activity (Figures 3A and 3B). Deletion of the RING domain of DRIP1 abolished its autoubiquitination activity (Figure 3C). Four kinds of E2 enzymes from human, UBCH5c, UBCH10, UBCH12, and UBCH13, and one from Arabidopsis, UBC8, were tested with DRIP1- or DRIP2-GST fusion proteins in the in vitro ubiquitination assays. Only UBCH5c was capable of catalyzing DRIP1 and DRIP2 autoubiquitination (data not shown). Although DRIP1 and DRIP2 protein autoubiquitination was verified in the presence of UBCH5c as an E2 protein, the real interactive E2 protein for DRIP proteins in Arabidopsis remains unknown. DRIP1 and DRIP2 showed low activity mediating the monoubiquitination of the DREB2A in our in vitro ubiquitination assays (Figure 3D). However, future studies are necessary to study the in vivo DREB2A ubiquitination status in planta. It is likely that the functional Arabidopsis E2 enzyme may better enable DRIP1 to mediate DREB2A ubiquitination.
Overexpression of DRIP1 in Arabidopsis delayed the expression of some stress-inducible genes in response to dehydration stress. These data indicated that DRIP1 likely plays a negative role in drought stress signaling (Figure 5). Moreover, disruption of DRIP1 expression resulted in enhanced expression of drought-responsive genes in the drip1-1 mutant (Figure 6C). Similarly, disruption of DRIP2 also resulted in mildly elevated expression of the genes responsive to dehydration stress in the drip2-1 mutant (Figure 6C). Collectively, their sequence homology, similar tissue-specific expression patterns, and equivalent E3 ubiquitin ligase activities indicated that they might share a functional overlap in planta. A double mutant was created (drip1 drip2) and resulted in delay of growth and bolting, resembling the phenotype of the 35S:DREB2A-CA plants (Figure 6B). Moreover, microarray and RNA gel blot analyses revealed that disruption of both DRIP1 and DRIP2 significantly increased the expression of a large number of stress-responsive genes induced by dehydration stress, especially genes regulated by DREB2A (Figure 6C; see Supplemental Data Set 3 online). DRIP1 and DRIP2, therefore, negatively function in drought-responsive gene expression.
Tight control of the levels of regulatory proteins is critical for the homeostasis of plants. Overproduction of either DREB1A or DREB2A-CA proteins in plants resulted in deleterious effects on plant growth and development; transgenics in both cases showed overall dwarfed phenotypes (Liu et al., 1998; Sakuma et al., 2006a). In nature, DREB1A and DREB2A exhibit stress-inducible expression patterns in response to cold and water deficit, respectively. However, expression of the DREB1A gene was always absent before stress, whereas DREB2A transcription could be found in plant roots and weakly in leaves even under normal growth conditions (Liu et al., 1998; Sakuma et al., 2002). Although DREB2A mRNA was present, the protein was undetectable and may undergo proteolysis by the 26S proteasome under nonstressed conditions (Figure 4B). DRIP1 and DRIP2 are expressed at constant levels in various plant tissues (Figure 4A; see Supplemental Figure 2 online). Therefore, it is possible that they function to restrict DREB2A protein amounts prior to stress stimuli. DREB2A protein degradation mediated by DRIP1 and DRIP2 may minimize its negative effects on plant growth and development, since disruption of DRIP1 and DRIP2 expression in the double mutant significantly affected plant germination and bolting (Figure 6B). Although most of the drip1 drip2 mutant seeds are viable on germination medium, some seeds failed in germination or were completely arrested in development. It is possible that these observations might be due to seed immaturity. At present, it is also unclear whether the seed maturation program is affected in the double mutant. The ectopic expression of DREB2A in the drip1-1 mutant also resulted in a severe dwarfism in the transgenics, resembling the DREB2A-CA overexpressor (Figure 7).
Since overexpression of intact DREB2A in plants did not produce an evident phenotype, DREB2A protein was predicted to require stress signal–dependent activation for it to be active (Liu et al., 1998; Sakuma et al., 2006a). Here, we demonstrated that overexpressed, the full-length DREB2A protein is more stable in drip1-1 than in the wild-type background, indicating that elimination of DRIP1 protein results in the stability of DREB2A protein. Moreover, enhanced expression of downstream target genes was also detected in the 35S:GFP-DREB2A/drip1-1 plants, suggesting that stabilization of the protein contributed to the activity of DREB2A in plants. Due to the unavailability of an effective antibody, despite repeated efforts at antibody generation, we are currently unable to dissect the DREB2A ubiquitination status in planta. On the other hand, the possible activation mechanism of DREB2A, in addition to the stabilization of DREB2A under stress conditions, should not be ruled out. In the 2-h dehydrated drip1 drip2 plants, expression of DREB2A-regulated genes was significantly higher than in wild-type plants (Figure 6C; see Supplemental Data Set 3 online), indicating that the DREB2A response can be induced in the absence of DRIP1 and DRIP2. The enhanced expression of DREB2A-regulated genes may be attributed to the increased availability of DREB2A protein in plant cells. Additionally, DREB2A may be more accessible for activation upon dehydration stress in the double mutant. Moreover, in the 35S:GFP-DREB2A/drip1-1 plants, although expression of RD29A and RD29B was clearly elevated under normal conditions, higher expression of these genes could be achieved after stress treatment. These data indicate that the stress-dependent activation mechanism might also be necessary for the modulation of DREB2A activity (Figure 7C).
Some RING E3 ligases function to attenuate hormone or abiotic signals by modulating the abundance of transcription factors in cells, such as SINAT5 mediating NAC1 ubiquitination, AIP2 targeting ABI3, and HOS1 degrading ICE1 (Xie et al., 2002; Dong et al., 2006; Zhang et al., 2005). However, some results suggested that DRIP1 and DRIP2 did not function to attenuate stress signals in the late stage of drought stress, although DRIP1 and DRIP2 function negatively in stress-responsive gene expression. First, unlike SINAT5, AIP2, or HOS1, DRIP1 expression was not stress-inducible; instead, DRIP1 was constitutively expressed under stress and nonstressed conditions (see Supplemental Figure 3A online). These data cannot fit into an attenuation hypothesis. Second, overexpression of DRIP1 in Arabidopsis delayed the expression of drought-responsive genes at an early phase of the dehydration response but did not reduce the expression of these genes at a late phase of the stress. Third, we could not detect higher expression of stress-responsive genes in the drip1 drip2 double mutant at a later phase of drought stress (data not shown). DRIP1 may not function to attenuate drought stress signals but instead restricts DREB2A transactivation activity under nonstressed conditions.
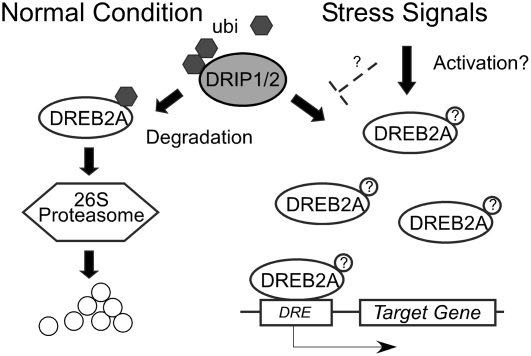
In conclusion, two novel proteins, DRIP1 and DRIP2, were identified and confirmed to interact with DREB2A. DRIP1 and DRIP2 function as RING E3 ligases and play negative roles in plant drought stress signaling, probably by mediating DREB2A proteolysis through 26S proteasome protein degradation. We hypothesized that under normal growth conditions, plants maintained low or leaky expression of DREB2A. However, the translated DREB2A protein was recognized and ubiquitinated by the constitutively expressed DRIP1 and DRIP2 and subjected to 26S proteasome proteolysis. Once stress signaling occurred, the degradation process might be blocked or inhibited directly by either stresses or a competition of a stress signal-dependent modification. Consequently, plant cells could quickly acquire a sufficient amount of the effective DREB2A protein to activate the expression of downstream stress-responsive genes (Figure 8).
Figure 8.
A Model of DRIP1 and DRIP2 Functioning in Dehydration Stress Signaling by Targeting DREB2A Degradation.
Under normal growth conditions, the DREB2A protein is expressed at low levels. To prevent the activation of a stress response in the absence of drought conditions, this protein is recognized and ubiquitinated by DRIP1 and DRIP2 protein and subjected to 26S proteasome proteolysis. Under stress, it is possible that the ubiquitination and proteolysis process is either blocked directly by stress-dependent signals or blocked via competition of a stress signal–dependent modification of DREB2A, indicated as question marks in circles. Thus, in drought conditions, plants are able to acquire a sufficient amount of effective DREB2A protein to activate downstream gene expression and produce a stress response.
METHODS
Plant Materials and Transgenic Plant Construction
Arabidopsis thaliana ecotype Columbia plants were grown on germination medium (GM) agar plates for 3 weeks, as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). They were subsequently transferred into vermiculite and grown under a 16-h-light/8-h-dark regime. T-DNA insertion lines of drip1 (WiscDsLox437G06) and drip2 (SALK_145041) were obtained from the ABRC. DRIP1- and DRIP2-specific primers were designed according to http://signal.salk.edu/tdnaprimers.2.html (see Supplemental Table 1 online). To generate 35S:DRIP1 and 35S:DRIP1-GFP constructs, DRIP1 coding sequence was inserted into pGKX and pGK-CsGFP vectors, respectively (see Supplemental Methods online), by a NotI site. The DRIP1 promoter was amplified from Columbia by the primer pair 5′-GGGGGATCCAGGAGATGACCTAGAGGAACC-3′ and 5′-GGGGGATCCCTTTCTCTTTATCTGTTGGGC-3′ using a high-fidelity PCR method. The generated fragment was constructed in front of the GUS gene by a BamHI site in the pGK-GUS vector (see Supplemental Methods online). The 35S:GFP-DREB2A plasmid was created by an insertion of DREB2A coding sequence into pGK-NsGFP (see Supplemental Methods online) by NotI-SacI sites. To prepare the DREB2Apro:GFP-DREB2A construct, the fragment of GFP-DREB2A-NosT was amplified from 35S:GFP-DREB2A with primers containing PstI-SalI linkers and constructed into pGK-GUS by these two sites. Then, the DREB2A promoter (Sakuma et al., 2006b) was inserted in front of the GFP-DREB2A-NosT fragment in a sense orientation by a PstI site. The constructed plasmid was introduced into Agrobacterium tumefaciens C58 cells. Plants were transformed as described previously (Liu et al., 1998).
Yeast Two-Hybrid Screening and Interaction Assay
An Arabidopsis cDNA library, which was prepared in the pGADT7 vector (MatchMaker GAL4 two-hybrid system 3; Clontech), was kindly donated by T. Umezawa (RIKEN Plant Functional Genomics Group). Yeast transformation was performed according to the supplier's instructions (Clontech). β-Galactosidase activity was determined in Y187 yeast report cells as described in the Yeast Protocols Handbook (Clontech).
RNA Preparation, Real-Time PCR, and RNA Gel Blot Analysis
Total RNA was isolated with Trizol reagent (Invitrogen) from 3-week-old plants. Procedures for RNA gel blot analysis were performed as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). One microgram of total RNA was used for cDNA synthesis with SuperScript III reverse transcriptase according to the supplier's instructions (Invitrogen). Real-time PCR analyses were performed using an Applied Biosystems 7500 real-time PCR system. The SYBR Premix Ex Taq kit (Takara) was used for the reactions according to the manufacturer's instructions. Arabidopsis 18S RNA amount was quantified as a loading control. All reactions were performed in duplicate or triplicate for technical repeats. For overexpression transgenic lines, at least two independent lines were analyzed for biological repeats. For T-DNA insertion lines, a representative result obtained from several independent samples was shown. The primers used for quantitative RT-PCR are listed in Supplemental Table 1 online. To obtain the quantitative data, three replicates were performed for characterization of expression data from each gene.
Fusion Protein Preparation
Recombinant GST fusion proteins were prepared as described previously (Liu et al., 1998). The coding region for DRIP1 (or DRIP del 1-60) was inserted into the pGEX-4T-3 vector by a NotI site. DRIP1-GST protein used for ubiquitination assays was dialyzed at 4°C overnight in the following buffer: 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 0.05 mM ZnCl2. The DRIP2 cDNA was inserted into the pMAL-c2x vector by a BamHI site. DRIP2-MBP fusion protein was purified according to the instruction book for the pMAL protein fusion and purification system (New England Biolabs) and dialyzed against the same buffer as DRIP1-GST protein. The full-length coding sequence of DREB2A was inserted into the pET48b vector (Novagen) between the SmaI and BamHI sites to generate an in-frame fusion to the N-terminal His and Trx tags. The plasmid was expressed in Rosetta (DE3) cells, and recombinant fusion proteins were purified by His tag affinity to nickel-nitrilotriacetic acid agarose matrix according to the Qiagen instruction book.
In Vitro Ubiquitination Assays and Protein Gel Blot Analysis
The ubiquitination assays were generally performed as described by Xie et al. (2002). Approximately 500 ng of DRIP-GST fusion protein was mixed with 100 ng of rabbit E1 (Boston Biochemicals), 200 ng of human E2 UBCH5C (Biomol), and 5 μg of ubiquitin-myc (Boston Biochemicals)/ubiquitin (Sigma-Aldrich). The reactions were made in the buffer containing 50 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 2 mM ATP, 2 mM DTT, 10 mM phosphocreatine, and 1 unit of creatine kinase (Sigma-Aldrich). After incubating at 30°C for 2 h, the reaction was stopped with 2× SDS sampling buffer by boiling at 100°C for 5 min. Ten microliters of each reaction was analyzed by SDS-PAGE and immunoblot analysis using a corresponding antibody. To confirm that DRIP1 mediates DREB2A ubiquitination, DREB2A-His-Trx fusion protein was affinity-purified and ∼200 ng of purified protein was incubated together with the ubiquitination mixture for 2 h. The mixture was then subjected to 6% SDS-PAGE and immunoblot analysis.
For anti-GFP immunoblot analysis, the nuclear fraction was prepared according to the methods of Busk and Pages (1997) with minor modifications. Eighteen micrograms of protein was loaded and blotted for immunodetection with a monoclonal anti-GFP primary antibody (Clontech).
Protein Pull-Down Assays
Full-length DREB2A, the DREB2A 1-165 fragment, or the DREB2A 166-335 fragment was inserted into pET-28a by an EcoRI site. Each construct was transformed into BL21 (DE3) Escherichia coli cells and grown in 50 mL of 2× YT broth until the OD600 reached 0.8. Cultures were then induced by adding 0.1 mM isopropylthio-β-galactoside and held at 16°C overnight. Harvested cells were lysed by adding 0.1 mg of lysozyme at 4°C for 1 h and were subsequently sonicated. After sonication, the disrupted cells were centrifuged and the supernatant was used as prey protein for pull-down assays. Forty microliters of supernatant was mixed with ∼1.2 μg of DRIP-GST or GST protein in the buffer (50 mM HEPES, pH 7.5, 1 mM EDTA, 150 mM NaCl, 0.1% Tween 20, 0.5 mM DTT, and 0.8% glycerol) in a total volume of 100 μL at room temperature for 1 h. Thirty microliters of a 50% slurry of glutathione-Sepharose 4B beads was added into the mixture and rotated at room temperature for 1 h. The mixture was subsequently washed three times with 1 mL of binding buffer, and washed beads were boiled in 30 μL of 2× SDS sampling buffer at 100°C for 5 min. Four microliters of soluble cell lysate (10% input) was loaded as a control to indicate the original prey protein amount.
Stress Treatment and Electrolyte Leakage Experiment
Three-week-old plants were removed from GM medium (or selective medium) and placed in empty Petri dishes and sealed with Parafilm. The dehydration treatment was performed as described previously (Liu et al., 1998). For the electrolyte leakage experiment, aerial portions of 3-week-old plants were dehydrated by sitting on a piece of Parafilm in empty Petri dishes under dim light conditions on a clean bench. The approximate RH and temperature for the corresponding time course treatment were 30% and 22 ± 2°C, respectively. Plants were subsequently dipped into 0.4 M mannitol solution and gently shaken at room temperature for 2 h. Electrolyte leakage was determined by measuring the conductivity of the solution using the DS-15 conductivity meter (Horiba). The total amount of electrolytes was determined after the solutions were autoclaved for 1 min. For the plant drought stress tolerance test, 3-week-old plants were transferred from agar plates to soil conditions. After a 1-week period of conditioning, they were subjected to a water stress by withholding watering. The plants were rewatered when significant differences in wilting were observed. Several independent experiments were performed under the conditions of 16 h/8 h of light/dark and RH of ∼60% to obtain statistically significant data.
BiFC Assays
According to Walter et al. (2004), DRIP1 and DREB2A full-length sequences were cloned into the pUCSPYNE or pUCSPYCE vectors with SpeI-SalI sites after the 35S promoter sequence, respectively. Analysis of transient expression in onion (Allium cepa) epidermal cells was performed as described previously (Fujita et al., 2005). YFP fluorescence was observed with a laser scanning confocal microscope (LSM510; Zeiss).
Histochemical GUS Staining and GFP Fluorescence Observation
Histochemical GUS staining was performed by soaking plants in solution containing 1 mM Glc, 50 mM phosphate buffer, pH 7.0, and 10% methanol at 37°C for ∼10 h. Three-week-old transgenic plants harboring the DREB2Apro:GFP-DREB2A constructs were removed from GMK agar plates and treated with either 50 μM MG132 (dissolved in DMSO; Calbiochem, lot B68556) or 50 μM DMSO solution (mock treatment) under dim light conditions overnight. GFP fluorescence was observed with a laser scanning confocal microscope (LSM510; Zeiss).
Microarray Analysis
Genome-wide expression studies using the Arabidopsis 2 Oligo Microarray (Agilent Technologies) were performed using 3-week-old wild-type and drip1 drip2 plants. Gene expression was compared between wild-type and drip1 drip2 plants under both unstressed and 2-h dehydration stress conditions. Total RNA (200 ng) isolated with Trizol reagent (Invitrogen) from each sample (pooled from six plants) was applied for the analysis. Biological replication was performed by analyzing the samples obtained from two independent treatments (for both control and 2-h dehydration stress). For each experiment, two slides were analyzed for a Cy3 and Cy5 dye swap, and eight slides were hybridized for the four experiments. All microarray data are available at http://www.ebi.ac.uk/arrayexpress/ with the accession numbers E-MEXP-1351 and E-MEXP-1352.
Statistical Analysis of the Microarray Data
Integration and normalization of each spot's signal intensities were performed using the Lowess method of the Feature Extraction 9.5 program (Agilent Technologies). Statistical analysis was performed using ArrayAssist software (Stratagene). The Welch t test was used for the parametric test, and the Benjamini and Hochberg false discovery rate for multiple testing corrections was used with a P value of <0.05 to filter reliable genes. By this statistical analysis, we generated lists of the genes that present significant differences in their expression between wild-type and drip1 drip2 plants under normal conditions and exposed to a 2-h dehydration stress. All genes that were considered to show significant expression differences by the previous tests were then filtered by a fold change absolute > 2.0. Such genes were considered to be upregulated in drip1 drip2 plants in relation to wild-type plants.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: DRIP1 (At1g06770), DRIP2 (At2g30580), RD29A (At5g52310), RD29B (At5g52300), LEA14 (At1g01470), RD17 (At1g20440), GolS1 (At2g47180), and GolS2 (At1g56600).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Multiple Sequence Alignment of DRIP1, DRIP2, and Their Orthologs.
Supplemental Figure 2. DRIP2 Functions as an E3 Ubiquitin Ligase and Mediates DREB2A Protein Ubiquitination.
Supplemental Figure 3. DRIP1 and DRIP2 Gene Expression Profiling under Various Stresses in Different Tissues.
Supplemental Figure 4. The Proteasome Inhibitor MG132 Inhibits GFP-DREB2A Fusion Protein Degradation When It Is Overexpressed by the 35S Promoter in Arabidopsis.
Supplemental Figure 5. Relative Water Content of Wild-Type Columbia and the drip1 drip2 Double Mutant under Dehydration Conditions.
Supplemental Table 1. Primers Used in Quantitative RT-PCR.
Supplemental Table 2. Positive Clones from Yeast Two-Hybrid Library Screening.
Supplemental Methods. Description of pGreen Vectors.
Supplemental Data Set 1. Text File Corresponding to the Alignment in Supplemental Figure 1 online.
Supplemental Data Set 2. Genes Upregulated in Nonstressed drip1 drip2 Mutant Plants.
Supplemental Data Set 3. Genes Upregulated in the drip1 drip2 Mutant Relative to Wild-Type Plants after a 2-h Dehydration Stress.
Supplementary Material
Acknowledgments
We thank E. Ohgawara, K. Amano, K. Yoshiwara, and H. Sado for their excellent technical support and M. Toyoshima for skillful editorial assistance. We are also grateful to Y. Osakabe for her helpful discussions. F.Q. was supported by a visiting research fellowship from the Japan International Research Center for Agricultural Sciences. This work was supported in part by the Program for the Promotion of Basic Research Activities for Innovative Biosciences, Core Research for Evolutional Science and Technology, and by a project grant from the Ministry of Agriculture, Forestry, and Fisheries, Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kazuko Yamaguchi-Shinozaki (kazukoys@jircas.affrc.go.jp).
Online version contains Web-only data.
References
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24 701–713. [DOI] [PubMed] [Google Scholar]
- Barlow, P.N., Luisi, B., Milner, A., Elliott, M., and Everett, R. (1994). Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237 201–211. [DOI] [PubMed] [Google Scholar]
- Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24 23–58. [Google Scholar]
- Borden, K.L. (2000). RING domains: Master builders of molecular scaffolds? J. Mol. Biol. 295 1103–1112. [DOI] [PubMed] [Google Scholar]
- Borden, K.L., and Freemont, P.S. (1996). The RING finger domain: A recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6 395–401. [DOI] [PubMed] [Google Scholar]
- Busk, P.K., and Pages, M. (1997). Microextraction of nuclear proteins from single maize embryos. Plant Mol. Biol. Rep. 15 371–376. [Google Scholar]
- Chinnusamy, V., Schumaker, K., and Zhu, J.K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 55 22–36. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Caspar, T., and Quail, P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5 1172–1182. [DOI] [PubMed] [Google Scholar]
- Dong, C.H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont, P.S. (1993). The RING finger. A novel protein sequence motif related to the zinc finger. Ann. N. Y. Acad. Sci. 684 174–192. [DOI] [PubMed] [Google Scholar]
- Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M.M., Seki, M., Hiratsu, K., Ohme-Takagi, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 3470–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, P.D., Seo, H.S., Yang, J.Y., and Chua, N.-H. (2003). Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 6 453–462. [DOI] [PubMed] [Google Scholar]
- Jiang, C., Iu, B., and Singh, J. (1996). Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol. Biol. 30 679–684. [DOI] [PubMed] [Google Scholar]
- Kosarev, P., Mayer, K.F., and Hardtke, C.S. (2002). Evaluation and classification of RING-finger domains encoded by the Arabidopsis genome. Genome Biol. 3 RESEARCH0016.1–RESEARCH0016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda, H., Takahashi, N., Shimada, H., Seki, M., Shinozaki, K., and Matsui, M. (2002). Classification and expression analysis of Arabidopsis F-box containing protein genes. Plant Cell Physiol. 43 1073–1085. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA-binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression in Arabidopsis. Plant Cell 10 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovering, R., Hanson, I.M., Borden, K.L., Martin, S., O'Reilly, N.J., Evan, G.I., Rahman, D., Pappin, D.J., Trowsdale, J., and Freemont, P.S. (1993). Identification and preliminary characterization of a protein motif related to the zinc finger. Proc. Natl. Acad. Sci. USA 90 2112–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Risseeuw, E.P., Daskalchuk, T.E., Banks, T.W., Liu, E., Cotelesage, J., Hellmann, H., Estelle, M., Somers, D.E., and Crosby, W.L. (2003). Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 34 753–767. [DOI] [PubMed] [Google Scholar]
- Saijo, Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. (2003). The COP1–SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17 2642–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y., Liu, Q., Dubouzet, J.G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold inducible gene expression. Biochem. Biophys. Res. Commun. 290 998–1009. [DOI] [PubMed] [Google Scholar]
- Sakuma, Y., Maruyama, K., Osakabe, K., Qin, F., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006. a). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006. b). Dual function of an Arabidopsis transcription factor DREB2A in water-stress- and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 103 18822–18827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.-H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423 995–999. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 571–599. [DOI] [PubMed] [Google Scholar]
- Vierstra, R.D. (2003). The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 8 135–142. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79 1035–1045. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.-H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10 88–94. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stress. Annu. Rev. Plant Biol. 57 781–803. [DOI] [PubMed] [Google Scholar]
- Yang, J., Lin, R., Sullivan, J., Hoecker, U., Liu, B., Xu, L., Deng, X.W., and Wang, H. (2005). Light regulates COP1-mediated degradation of HFR1, a transcription factor essential for light signaling in Arabidopsis. Plant Cell 17 804–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X., Garreton, V., and Chua, N.-H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.