Abstract
Flavonoids are secondary metabolites known to modulate plant growth and development. A primary function of flavonols, a subgroup of flavonoids, is thought to be the modification of auxin fluxes in the plant. Flavonols in the cell are glycosylated, and the repressor of lrx1 (rol1) mutants of Arabidopsis thaliana, affected in rhamnose biosynthesis, have a modified flavonol glycosylation profile. A detailed analysis of the rol1-2 allele revealed hyponastic growth, aberrant pavement cell and stomatal morphology in cotyledons, and defective trichome formation. Blocking flavonoid biosynthesis suppresses the rol1-2 shoot phenotype, suggesting that it is induced by the modified flavonol profile. The hyponastic cotyledons of rol1-2 are likely to be the result of a flavonol-induced increase in auxin concentration. By contrast, the pavement cell, stomata, and trichome formation phenotypes appear not to be induced by the modified auxin distribution. Together, these results suggest that changes in the composition of flavonols can have a tremendous impact on plant development through both auxin-induced and auxin-independent processes.
INTRODUCTION
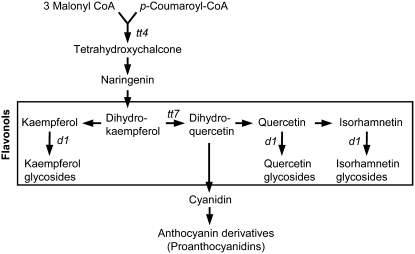
Flavonoids represent a highly diverse class of low molecular weight secondary metabolites, of which >6000 different compounds have been described. They are important for pigmentation and UV light protection. They serve as signals for pollinators and other beneficial organisms, participate in hormone signaling, and function as phytoalexins. A number of biological processes, such as transcriptional regulation, signal transduction, and cell-to-cell communication, are influenced by flavonoids. Due to their antioxidant activity, flavonoids appear to play an important role in the regulation of reactive oxygen species (ROS) in plant cells (Lepiniec et al., 2006; Peer and Murphy, 2006). In Arabidopsis thaliana, 24 mutants have been identified on the basis of aberrant flavonoid accumulation (Routaboul et al., 2006). Frequently, mutants are defective in the biosynthesis of proanthocyanidins (Figure 1), the subgroup of flavonoids responsible for seed coloring (Koornneef, 1990; Debeaujon et al., 2003; Lepiniec et al., 2006), and thus deviate from the typical brown seed color. The flavonols kaempferol, quercetin, and isorhamnetin constitute a subgroup of flavonoids that appears to be present in all tissues of Arabidopsis. They are O-glycosylated, mainly by glucose and rhamnose units at the C3 and C7 positions, resulting in a large number of different molecules (Veit and Pauli, 1999; Lepiniec et al., 2006).
Figure 1.
Biosynthesis of Flavonoids in Arabidopsis.
Flavonoids are made of three molecules of malonyl-CoA and one molecule of p-coumaroyl-CoA. Flavonols are a subgroup of flavonoids and commonly consist of glycosylated kaempferol, quercetin, and isorhamnetin derivatives. transparent testa (tt) mutants of Arabidopsis are flavonoid biosynthesis mutants characterized by a change in seed color due to the absence of proanthocyanidins. Mutants used in this study are indicated in the pathway; they are tt4 (defective in chalcone synthase), tt7 (defective in flavanone-3′-hydroxylase) and ugt78d1 (d1) (defective in UDP-rhamnose:flavonol-3-O-rhamnosyltransferase).
Biochemical experiments and analyses of auxin fluxes in flavonoid-deficient mutants suggest that flavonols negatively regulate auxin transport (Jacobs and Rubery, 1988; Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004). There is also evidence that flavonols directly influence cell growth. In petunia (Petunia hybrida) and maize (Zea mays), chalcone synthase mutants are blocked in the first step of flavonoid biosynthesis (Figure 1) and are defective in pollen tube growth. Kaempferol was identified as the pollen germination–inducing factor when applied to mutant stigma (Mo et al., 1992). In addition, the petunia mutant is defective in root hair development (Taylor and Grotewold, 2005).
Cell growth is largely determined by the extension rate of the cell wall, which is a complex structure composed of the polysaccharides cellulose, hemicellulose, and pectin, in addition to a number of structural proteins (Carpita and Gibeaut, 1993; Cassab, 1998). Epidermal leaf pavement cells provide a model system in which to study the processes of cell growth and determination of cell shape (Mathur, 2004). In Arabidopsis, pavement cells have a jigsaw puzzle–like shape in which lobes extend into neighboring cells. This pattern is based on coordinated outgrowth at a given point in one cell with inhibition of outgrowth in the adjacent cell. The underlying signaling network that regulates this process involves a complex activation–inactivation interaction between ROP2 and RIC1/RIC4 (Fu et al., 2005). ROP (for Rho-related GTPase of plants) proteins regulate the organization of cortical microtubules and actin microfilaments (Smith, 2003; Fu et al., 2005). Indeed, interfering with actin filament nucleation changes pavement cell shape (Frank et al., 2003; Mathur et al., 2003; Djakovic et al., 2006). Recently, the TREHALOSE-6-PHOSPHATE SYNTHASE mutant csp1 was shown to cause a pavement cell phenotype, although the function of trehalose-6-phosphate in this process remains unclear (Chary et al., 2008). The importance of the cell wall in cell shape determination is well known and is reflected by the fact that the cellulose synthase mutant rsw1 lacks lobe formation in pavement cells, resulting in straight cell boundaries (Williamson et al., 2001).
Previously, we identified two allelic rol1 (for repressor of lrx1) mutants, which suppress the Arabidopsis root hair mutant lrx1 (Diet et al., 2006). LRX1 encodes an extracellular Leu-rich repeat extensin protein that is specifically expressed in root hairs (Baumberger et al., 2001; Ringli, 2005). While lrx1 mutants develop defective root hairs, lrx1 rol1 double mutants show a suppressed lrx1 phenotype and form wild-type–like root hairs. The ROL1 locus encodes RHM1, one of three rhamnose synthase proteins that convert UDP-d-glucose to UDP-l-rhamnose in Arabidopsis (Diet et al., 2006; Oka et al., 2007). The RHM1 protein encoded by the rol1 mutants is unable to catalyze this conversion (Diet et al., 2006). Rhamnose is an important component of pectin (Ridley et al., 2001), and the rol1 mutants exhibit modifications in pectin structure (Diet et al., 2006), which may provide the molecular basis for the observed suppression of the lrx1 root hair phenotype. Seedlings of both rol1 alleles also develop cotyledons with an uneven surface and a peripheral zone that is bent upward. This is referred to as hyponastic growth and is the result of asymmetric growth of the adaxial and abaxial (upper and lower, respectively) surfaces of the cotyledon (Kang, 1979). In addition, the stronger rol1-2 allele develops slightly shorter roots and shorter root hairs than wild-type seedlings (Diet et al., 2006).
In this work, we show that the rol1 mutants contain a modified flavonol glycosylation profile. A detailed analysis of the rol1-2 allele revealed aberrations in cell shape, which result in oversized cotyledon pavement cells that lack typical lobes and have defective trichomes. Genetic experiments suggest that it is a change in the flavonol profile that induces the observed cotyledon and trichome phenotypes. Our results suggest that the shift in flavonol abundance results in a change of auxin concentration, inducing hyponastic growth in cotyledons. In addition, the modified flavonol profile appears to have a direct effect on cell formation that is independent of its effect on the auxin concentration. This suggests that flavonols can directly interfere with cell growth processes.
RESULTS
Flavonol Accumulation Is Modified in rol1 Mutants
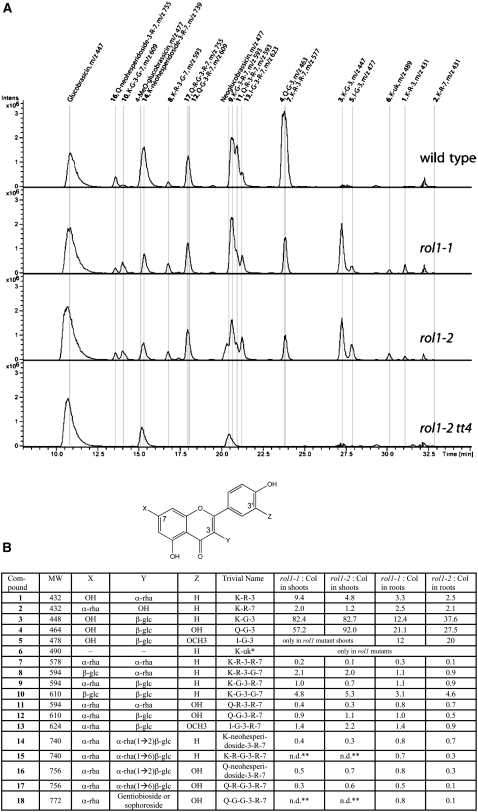
For the analysis of flavonol accumulation, wild-type, rol1-1, and rol1-2 seedlings were grown for 6 d on Murashige and Skoog (MS) agar plates in a vertical orientation. At this stage of seedling development, cotyledons are fully expanded and the first true leaves are about to form. Seedlings were cut in the hypocotyl, and shoot and root tissues were separately pooled and analyzed. The targeted metabolite analysis was performed by HPLC–mass spectrometry (MS). Most flavonol glycosides were identified by their UV light absorption spectra, MS/MS analysis, and comparison with reference compounds of known structure. Recently published data were also used to interpret results (Kerhoas et al., 2006; Le Gall et al., 2006; Stobiecki et al., 2006; Veit and Pauli, 1999). Flavonol levels were obtained by calculating the area below each HPLC peak per milligram dry weight of plant material. Figure 2A shows the elution profiles of shoot extracts. Several changes were observed between the wild type and the rol1 mutants. Some flavonols were less abundant than in wild-type tissues, while others were elevated in the rol1 mutants. Flavonol aglycones were not detected in any of the plant lines analyzed. Figure 2B shows the exact structure of each of the identified flavonol species and the induction/repression factor in the rol1 mutants compared with the wild type. The two rol1 alleles revealed comparable profiles. A reduction of up to 10-fold was found in rol1 mutants for flavonols glycosylated with several rhamnose units. Some flavonols containing single rhamnose units, such as kaempferol-3-O-rhamnose (Figure 2, peak 1), however, were increased severalfold. The clearest alteration in rol1 mutants was found for the 3-O-glucosides of kaempferol (kaempferol-3-O-glucose [K-G-3]), quercetin (quercetin-3-O-glucose [Q-G-3]), and isorhamnetin (isorhamnetin-3-O-glucose [I-G-3]) (Figure 2, peaks 3, 4, and 5, respectively). While the first two were strongly induced, particularly in the shoot (by a factor of 57 to 92), isorhamnetin-3-O-glucose newly accumulated in shoots and overaccumulated in roots of rol1 mutants. One flavonol (molecular weight 490; Figure 2, peak 6) has yet to be identified in Arabidopsis. Based on MS/MS analysis and the analysis of the rol1-2 tt7 double mutant (see below), this flavonol is a kaempferol and therefore is referred to as K-uk (for unknown kaempferol). According to the literature, it may be a kaempferol-3-O-(6″-acetyl-glucoside), which has been found in needles of Pinaceae species (Slimestad, 2003).
Figure 2.
Flavonol Accumulation Is Modified in rol1 Mutants.
(A) Flavonol elution profiles obtained from HPLC-(–)-electrospray ionization-MS analyses of shoots from 6-d-old seedlings. In order to improve the selectivity, the sums of the extracted ion chromatograms corresponding to flavonoid derivatives 1 to 16 are displayed (extracted ion chromatogram of m/z 431, 447, 463, 477, 489, 577, 593, 609, 623, 739, 755, and 771). Glucobrassicin derivatives (peaks with m/z 447 and 477) are nonflavonoid compounds that are indicated because they show the same mass spectra as some flavonoid derivatives.
(B) The flavonol structure is given. The groups at the C7, C3, and C3′ positions are listed in the table (X, Y, and Z, respectively). The substances are numbered according to their molecular weight (MW) and type of flavonol. The factors of induction in rol1 mutants compared with the wild type are listed separately for shoot and root tissue. The area under each peak was used as an arbitrary unit, and the amounts were calculated per milligram dry weight. For trivial names, K = kaempferol, Q = quercetin, I = isorhamnetin, G = Glc, and R = rhamnose. * This flavonol has not been described in Arabidopsis but may be a kaempferol-3-O-(6″-acetyl-glucoside); ** n.d., not detectable in either wild-type or rol1 mutant shoots.
The relative abundance of each flavonol compound in the different lines is shown in Supplemental Figure 1A online. Calculating the total amounts of shoot and root flavonols revealed a decrease by 25% in the shoot of the rol1 mutants and an increase by 30% (rol1-1) and 10% (rol1-2) in roots (see Supplemental Figure 1B online) compared with the wild type. Since the two rol1 alleles showed comparable changes in the flavonol profile, we decided to limit further analyses to the rol1-2 allele, which shows a stronger phenotype (Diet et al., 2006).
Flavonoids were visualized in vivo with diphenylboric acid-2-aminoethyl ester (DPBA), a flavonoid-specific stain, which binds flavonols (Buer et al., 2007). In agreement with previous reports (Peer et al., 2001), fluorescence was found in the cotyledonary node, the root/shoot transition zone, the root elongation zone, and very faintly in trichomes of wild-type plants. In rol1-2 mutants, increased fluorescence was detected in the root elongation zone and in trichomes (see Supplemental Figure 2 online). No increase in DPBA staining was observed in the cotyledonary node. To confirm that the observed fluorescence was due to the presence of flavonoids, a rol1-2 tt4 double mutant was generated and analyzed. tt4 plants carry a mutation in the CHALCONE SYNTHASE gene and are thus blocked in the first step of flavonoid biosynthesis (Shirley et al., 1995) (Figure 1). As shown in Figure 2A, the flavonol peaks are indeed absent in rol1-2 tt4 extracts. These plants also failed to produce fluorescence, other than a faint background staining in root tissue (see Supplemental Figure 2 online).
The rol1-2 Mutant Develops Several Shoot Phenotypes
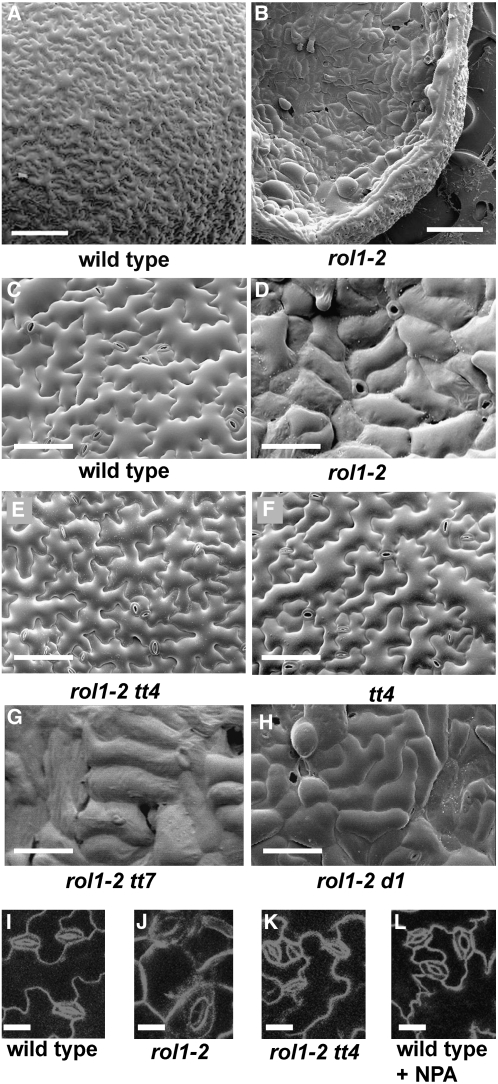
rol1-2 mutants show aberrant cotyledon development, with an uneven cotyledon surface and hyponastic growth. In addition, trichomes of the first rosette leaves are strongly deformed in rol1-2 (Diet et al., 2006) (Figures 3A and 3B). The cotyledon growth phenotype was investigated in more detail by scanning electron microscopy. Compared with the wild type, the size of adaxial pavement cells was frequently increased (Figures 4A and 4B) and the typical jigsaw puzzle–like cell shape of pavement cells was absent in rol1-2 mutants (cell borders were straight and the characteristic lobes were absent; Figures 4C and 4D). This phenotype was restricted to the adaxial side of the cotyledon. In the scanning electron microscopy analysis, stomatal distribution appeared to be affected in rol1-2 cotyledons. Stomata, therefore, were visualized by confocal microscopy using FM4-64, a membrane-specific fluorescent dye. rol1-2 plants had fewer stomata than wild-type plants (34 ± 8 versus 163 ± 26 per mm2), and the stomata were frequently larger than those of wild-type plants (Figures 4I and 4J).
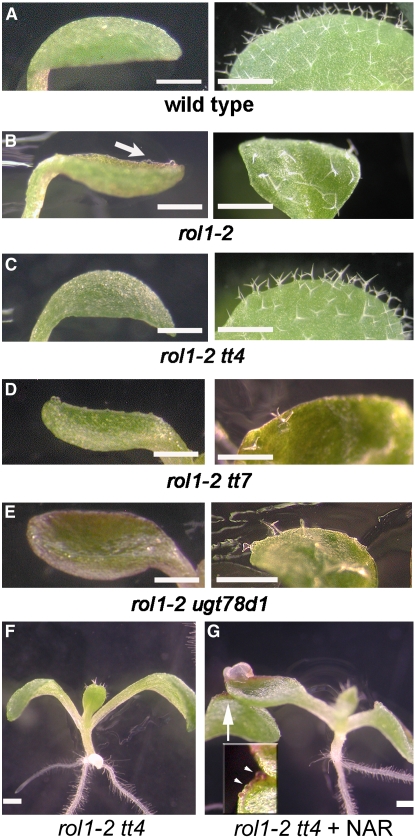
Figure 3.
Aberrant Cotyledon and Trichome Formation in rol1-2 Mutants.
(A) to (E) Cotyledons and leaves of 5- and 8-d-old Arabidopsis plants.
(A) Wild type.
(B) rol1-2. The periphery of cotyledons is bent upward, referred to as hyponastic growth. Also, the surface of rol1-2 mutant cotyledons is rough and contains bulging epidermal cells (arrow). Trichomes are frequently deformed.
(C) The features of the rol1-2 phenotype are suppressed by the tt4 mutation in rol1-2 tt4 double mutants.
(D) and (E) No suppression of the rol1-2 phenotype is observed in rol1-2 tt7 (D) or rol1-2 ugt78d1 (E) double mutants.
(F) and (G) The anthocyanin-less rol1-2 tt4 mutant (F) is chemically complemented by naringenin (G). The arrow in (G) indicates the area shown in the inset. Colocalization of anthocyanin and epidermal bulging is indicated by arrowheads in the inset.
Bars = 1 mm.
Figure 4.
Pavement Cell and Stomata Phenotypes in rol1-2 Mutants.
Scanning electron micrographs from 5-d-old cotyledons. Wild-type plants develop jigsaw puzzle–like pavement cells ([A] and [C]), whereas lobe formation is suppressed in rol1-2 mutants ([B] and [D]), resulting in brick-like cells. Also, cells are frequently oversized compared with those in the wild type. The pavement cell phenotype induced by the rol1-2 mutation is suppressed by the tt4 mutation (E). tt4 single mutants develop pavement cells that are indistinguishable from those in the wild type (F). tt7 and ugt78d1 do not suppress the rol1-2 phenotype in the corresponding double mutants ([G] and [H]). Compared with the wild type (I), stomata of cotyledons are larger in rol1-2 (J), an effect that is suppressed in the rol1-2 tt4 double mutant (K). Treatment of the wild type with NPA does not change the size of stomata (L). Bars = 250 μm in (A) and (B), 100 μm in (C) to (H), and 20 μm in (I) to (L).
The rol1-2 Shoot Phenotypes Are Flavonoid Dependent
To investigate the role of modified flavonoid accumulation in the development of the rol1-2 phenotype, the flavonoid-less rol1-2 tt4 double mutant was characterized. In the double mutant, the hyponastic growth and trichome formation phenotypes observed in the rol1-2 single mutants were fully suppressed (Figure 3C). This indicates that the rol1-2 mutant shoot phenotypes are related to the change in flavonol accumulation, a finding that was confirmed by scanning electron microscopy analysis. In rol1-2 tt4 double mutants, pavement cell shape was reverted to the wild type (Figure 4E). tt4 single mutant plants also developed wild-type–like pavement cells (Figures 4C and 4F). Furthermore, the size (Figure 4K) and density (142 ± 13 per mm2) of stomata in rol1-2 tt4 cotyledons were again comparable to those in the wild type.
To confirm cosegregation of the tt4 mutation with suppression of the rol1-2 mutant phenotypes, seedlings homozygous for rol1-2 but segregating for tt4 were grown. Of 460 seedlings analyzed, 126 showed a lack of anthocyanin accumulation, typical of homozygous tt4 mutants (Shirley et al., 1995), and suppression of the rol1-2 shoot phenotypes. This corresponds to the expected 3:1 wild type:mutant segregation of the recessive tt4 mutation (χ2 test, P = 0.05) and indicates a close linkage between the tt4 mutation and the locus that suppresses the rol1-2 mutant shoot phenotypes.
In a second approach, we tested the chemical complementation of the tt4 mutation in the rol1-2 tt4 double mutant using naringenin, a flavonoid precursor positioned downstream of tt4 (Figure 1) (Shirley et al., 1995). Growing rol1-2 tt4 double mutants in the presence of naringenin induced the accumulation of anthocyanin, confirming the uptake and metabolic processing of naringenin in the seedlings. At the same time, naringenin induced hyponastic growth and the irregular cell shapes characteristic of the rol1-2 mutation (Figures 3F and 3G), indicating that the effect of the tt4 mutation is suppressed in the presence of naringenin. Together, our genetic and chemical complementation experiments suggest that the tt4 mutation suppresses the rol1-2 mutation and, hence, that flavonols are involved in the development of the rol1-2 shoot phenotype.
The rol1-2 Phenotype Is Modified by Kaempferol Derivatives
In order to restrict flavonol accumulation more specifically, the rol1-2 mutation was crossed into the tt7 mutant background. The tt7 mutant is defective in flavanone-3′-hydroxylase, which converts dihydrokaempferol to dihydroquercetin, and thus inhibits the accumulation of quercetin and isorhamnetin while kaempferol accumulates to higher levels (Figure 1) (Peer et al., 2001). The effect of tt7 on flavonol accumulation was confirmed in rol1-2 tt7 double mutants (see Supplemental Figure 1A online), which showed almost no accumulation of quercetin or isorhamnetin. The increase in kaempferol accumulation was more pronounced in roots than in shoots, indicating that the tt7 mutation might have distinct effects on the activity of the flavonoid biosynthesis pathway in these tissues. rol1-2 tt7 seedlings developed a rol1-2 phenotype (Figures 3D and 4G), suggesting that the rol1-2 phenotype–inducing flavonols are kaempferols. The tt7 single mutant and the corresponding wild-type Landsberg erecta did not develop a rol1-2 phenotype (see Supplemental Figure 3 online), thus demonstrating that the increased accumulation of kaempferols, per se, does not cause the rol1-2 phenotype.
rol1-2 was also combined with the UDP-rhamnose:flavonol-3-O-rhamnosyltransferase mutant ugt78d1 (Jones et al., 2003). This double mutant contained reduced amounts of 3-O-rhamnosylated flavonols (see Supplemental Figure 1A online) but developed a rol1-2 like phenotype (Figures 3E and 4H).
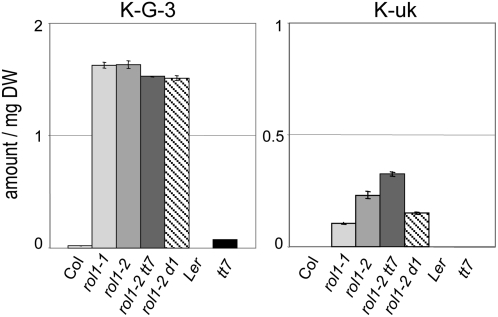
The flavonol(s) that cause the observed rol1-2 phenotype should be more abundant in the rol1 mutants than in wild-type plants, and they should also be present in the rol1-2 tt7 and rol1-2 ugt78d1 double mutants. Based on these assumptions, K-G-3 and K-uk are the best candidates as causal agents for the rol1-2 phenotype. In Landsberg erecta and tt7, these flavonols were absent or present in very small amounts compared with rol1-2 (Figure 5). Hence, the abundance of these two flavonol glucosides correlates with, and therefore may be responsible for, the observed rol1-2 phenotype.
Figure 5.
The Abundance of K-G-3 and K-uk Correlates with the rol1-2 Shoot Phenotype.
K-G-3 and K-uk [potentially kaempferol-3-O-(6″-acetyl-glucoside)] fulfill the predicted requirements for the flavonol species that cause the rol1-2 shoot phenotypes. Both are found in small amounts (K-G-3) or are absent (K-uk) in the wild type (Columbia and Landsberg erecta) and tt7 but are present in rol1, rol1-2 tt7, and rol1-2 ugt78d1. Mean ± se amounts of shoot flavonols are shown (n = 3). The area under the HPLC peak was used as the measurement for the amount of the flavonols.
The Modified Flavonol Profile of rol1-2 Does Not Influence Root Hair Formation
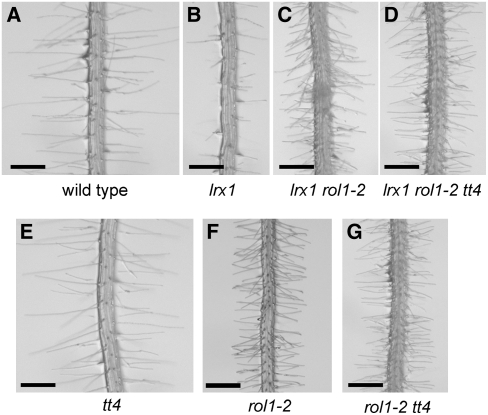
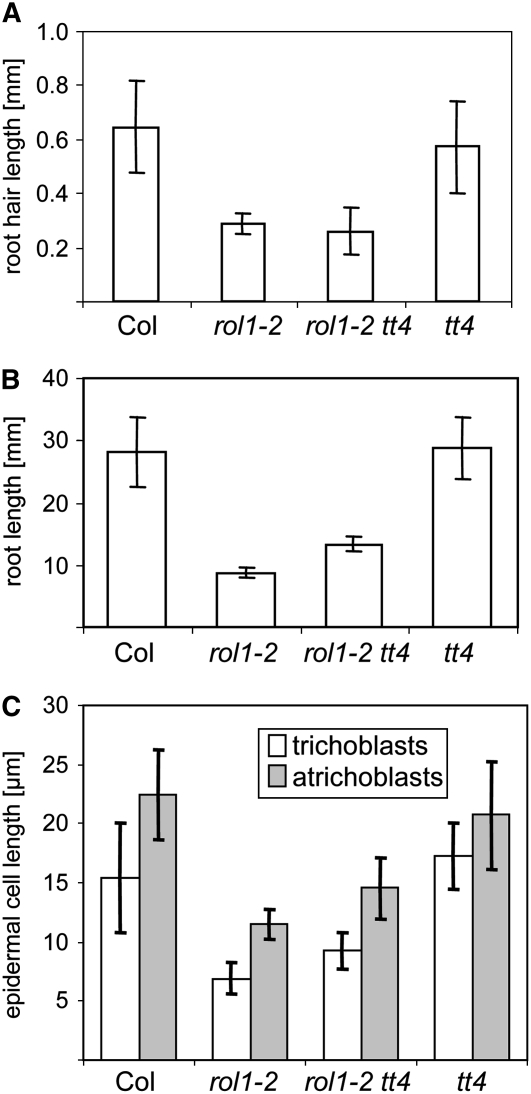
Suppression of the lrx1 root hair formation phenotype in the lrx1 rol1-2 double mutant was attributed to rol1-2–induced modifications in the cell wall (Diet et al., 2006). The drastic effect on cell growth caused by the modified flavonols prompted us to investigate their role in root hair development and the suppression of lrx1. The lrx1 mutant root hair phenotype (Figures 6A and 6B) (Baumberger et al., 2001) is suppressed in the lrx1 rol1-2 double mutant, in which root hairs form, although with reduced length compared with the wild type (Figure 6C) (Diet et al., 2006). The absence of flavonoids in the lrx1 rol1-2 tt4 triple mutant did not influence the suppression of lrx1 through rol1-2 (Figure 6D). To investigate the effect of tt4 on root development, the visual root hair phenotype, root length, and root hair length and density were determined in wild-type, rol1-2, tt4, and rol1-2 tt4 seedlings. tt4 mutants showed a wild-type–like root hair phenotype, and the tt4 mutation did not influence the rol1-2 root hair phenotype (Figures 6E to 6G). Measuring root hair length confirmed that neither the wild-type nor rol1-2 mutant root hair length is influenced by the tt4 mutation (Figure 7A). While root length was comparable between the wild type and tt4 mutants, the tt4 mutation partially (t test, P = 0.01) suppressed the short-root phenotype of the rol1-2 mutant (Figure 7B). The root hair density is reflected by the length of root hair–forming trichoblasts. rol1-2 mutants develop shorter trichoblasts compared with the wild type (Diet et al., 2006) (Figure 7C), resulting in a higher root hair density. While the length of trichoblasts of tt4 mutants and the wild type was comparable, the tt4 mutation partially (t test, P = 0.01) suppressed this aspect of the rol1-2 mutant. The same result was found for atrichoblasts, which lack root hairs (Figure 7C). Hence, the increase in root length of rol1-2 tt4 double mutants compared with rol1-2 was paralleled by an increased length of the root epidermal cells. Together, these results indicate that the root phenotype of rol1-2, including the suppression of the lrx1 phenotype, is largely determined by the effect of the rol1-2 mutation on the cell wall structure. The modified flavonol profile of rol1-2 seedlings has a small but significant influence on root development.
Figure 6.
Flavonoid Independence of Root Hair Development.
Seedlings were grown for 5 d in a vertical orientation. The lrx1 mutant root hair phenotype (compare [A] and [B]) is suppressed by the rol1-2 mutation (C). This suppression is not dependent on the flavonols in rol1-2, since the lrx1 phenotype is also suppressed in lrx1 rol1-2 tt4 triple mutants (D). The absence of flavonoids in the tt4 mutant does not have a significant effect on root hair formation (E) or on the rol1-2 mutant phenotype (compare [F] and [G]). Bar = 0.5 mm.
Figure 7.
Quantification of the Root Phenotypes in the Different Lines.
Five-day-old seedlings were used for the quantification of root hair length (A), root length (B), and epidermal cell length (C). Trichoblasts are root hair–forming root epidermal cells, and atrichoblasts are root hair–less root epidermal cells. Means ± se are shown (n = 25). Col, Columbia wild type.
Auxin Levels Are Increased in rol1-2 Mutants
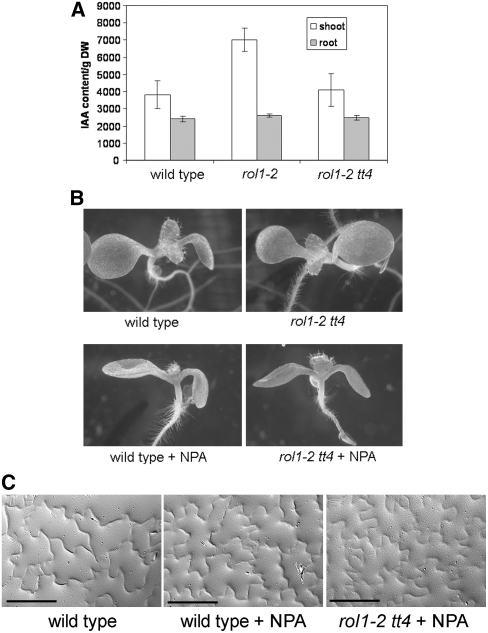
Flavonols are known to be negative regulators of auxin transport (Buer and Muday, 2004; Peer et al., 2004). In order to establish whether the modified flavonol profile in rol1-2 mutants influences auxin distribution, we measured free auxin concentrations in cotyledons and roots of 6-d-old seedlings. As shown in Figure 8A, the amount of auxin was increased in cotyledons of rol1-2 mutants compared with those of wild-type plants and was restored to wild-type levels in rol1-2 tt4 double mutants. In the roots of the same plants, no significant changes in auxin levels were observed.
Figure 8.
Auxin Levels Are Modified in the rol1-2 Mutant.
(A) Auxin concentration in the different plant lines was measured for shoots and roots of 6-d-old seedlings. Means ± se are shown (n = 3). The rol1-2 mutation causes a significantly increased concentration of auxin in the shoot compared with the Columbia wild type. This effect is reversed by the presence of the tt4 mutation. DW, dry weight.
(B) The hyponastic growth of rol1-2 cotyledons can be mimicked by growing wild-type or rol1-2 tt4 plants on 5 μM NPA (an auxin transport inhibitor).
(C) NPA does not influence pavement cell shape in wild-type or rol1-2 tt4 plants. Gel prints taken from the adaxial side of cotyledons are shown. Bars = 100 μm.
These results indicate that the increased levels of auxin in rol1-2 shoots correlate with the observed mutant phenotypes. To investigate this further, we inhibited auxin transport using 1-N-naphthylphthalamic acid (NPA) to determine whether this could phenocopy the rol1-2 mutation. Wild-type seedlings grown for 6 d in the presence of 5 μM NPA developed hyponastic cotyledons (Figure 8B), mimicking the effects of the rol1-2 mutation. Trichome and pavement cell shape were not affected by this treatment (Figures 8B and 8C). The size of the stomata was also unchanged (Figure 4L). However, the density of stomata was reduced (i.e., 119 ± 10 per mm2 for treated seedlings versus 163 ± 26 per mm2 for untreated seedlings), although not to the same degree as that observed in rol1-2 (34 ± 8 per mm2). These results suggest that the increase in auxin concentration affects stomatal density while having no effect on the shape of pavement cells, trichomes, or the size of stomata. In a control experiment using Columbia wild-type seedlings transformed with the auxin-sensitive DR5:GUS (for β-glucuronidase) construct (Ulmasov et al., 1997), treatment with 5 μM NPA resulted in hyponastic cotyledons and strongly increased GUS activity compared with untreated sibling plants (see Supplemental Figure 4 online).
To determine whether alterations in pavement cell shape and trichome formation in rol1-2 mutant plants are caused by the combination of the effect of the rol1-2 mutation on the cell wall structure (Diet et al., 2006) and auxin concentration, rol1-2 tt4 double mutants were grown in the presence of 5 μM NPA. The NPA treatment of these double mutants induced hyponastic cotyledons but did not affect trichome development or pavement cell shape (Figures 8B and 8C). These data suggest that the hyponastic cotyledon phenotype of rol1-2 mutants results from increased auxin levels caused by the altered flavonol profile of these plants. These changes to the flavonol profile affect trichome and pavement cell shapes, through an unknown mechanism, independent of auxin.
DISCUSSION
The Arabidopsis UDP-L-RHAMNOSE SYNTHASE mutant alleles of rol1 are affected in the flavonol glycosylation profile. Compared with the wild type, rol1 mutants contain strongly reduced amounts of flavonols glycosylated with multiple rhamnose units, while flavonols with single rhamnose units are, sometimes, even more abundant in rol1 mutants. The concomitant strong increase in monoglucosylated flavonols in rol1 seedlings might be a compensatory effect of the limited rhamnose availability. The metabolite analysis suggests that flavonol glycosylation is altered, with preferential conjugation at the C3 position and a redirection toward glucosylation. Compared with the wild type, there is a reduction in total amounts of flavonols in the shoots and an increase in the roots of the rol1 mutants. The increase of DPBA-induced fluorescence in rol1-2 trichomes seems to be at odds with the reduced amounts of flavonols in the shoot. This discrepancy can be explained by the small number of cells represented by trichomes compared with the total number of cells in the shoot. The increased amounts may reflect a difference in the biosynthesis or turnover rate of different flavonol species. Alternatively, flavonols may accumulate differentially in certain tissues by a directed transport mechanism (Buer et al., 2007). The possibility that the rol1-2 phenotype is induced by a nonflavonoid compound that is modified as a secondary indirect effect of rol1-2 cannot be ruled out. However, the suppression of rol1-2 by tt4 and chemical complementation by naringenin makes this scenario less plausible. Our data suggest that changes in the flavonol conjugation pattern in rol1-2 (i.e., an increase of one or several flavonol species) may interfere mainly with shoot development, while the complete removal of these conjugates may allow the plant to resume normal shoot development. Even though there is a measurable effect of the flavonols on rol1-2 root development, this process appears to be predominantly influenced by the modified cell wall composition of rol1-2 (Diet et al., 2006). Indeed, cell walls are known to be key determinants of cell expansion (Martin et al., 2001).
The changes in the pavement cell shape are observed only on the adaxial side of cotyledons, whereas the abaxial side appears to be unaffected. This may be due to overlapping expression patterns of the (assumed) functionally redundant genes RHM1 (i.e., ROL1) to RHM3 (Reiter and Vanzin, 2001). A very similar pavement cell phenotype is restricted spatially to the adaxial side of cotyledons in the ROP-interacting protein mutant icr1 (Lavy et al., 2007), suggesting a coordinated regulation of this process. It is an attractive hypothesis that, under natural conditions, plants have the potential to modify the flavonol glycosylation profile as a means to modulate growth and development. Interestingly, the absence of flavonoids appears to have no significant effect on cell growth or cell shape in Arabidopsis. This is in contrast with other plant species, such as maize, petunia, or tomato (Solanum lycopersicum), in which flavonoid deficiency leads to aberrant pollen tube growth, root hair growth, pavement cells, and stomata density (Mo et al., 1992; Liu-Gitz et al., 2000; Taylor and Grotewold, 2005; Schijlen et al., 2007).
The rol1-2 Phenotype Is Modified via Auxin-Dependent and -Independent Processes
Auxin is a phytohormone involved in a plethora of plant developmental processes. Modifying auxin transport directly affects plant growth (Friml, 2003). The rol1-2 mutation causes an increase in auxin concentration in cotyledons, which induces hyponastic growth and a reduction in stomatal density. This conclusion is based on the finding that treatment of wild-type plants with the auxin transport inhibitor NPA can at least partly mimic the rol1-2 mutant phenotype. A hyponastic growth phenotype is also found in the Arabidopsis mutants msg1 and cnr1, which are both impaired in responding to auxin (Watahiki and Yamamoto, 1997; Laxmi et al., 2006). Although the influence of auxin on stomata distribution is known (Vandenbussche and Van Der Straeten, 2007), the stomatal density is much more affected in rol1-2 than in the NPA-treated wild type. This suggests that flavonols also play an auxin-independent role in the patterning of stomata. The tt4 mutation, which blocks flavonoid biosynthesis (Shirley et al., 1995), neutralizes the effect of rol1-2 on auxin distribution and suppresses the rol1-2 cotyledon phenotype. Auxin transport has been shown to be negatively regulated by flavonols and is increased in tt4 mutants compared with the wild type (Jacobs and Rubery, 1988; Brown et al., 2001; Buer and Muday, 2004; Peer et al., 2004). Recent experiments in Arabidopsis have shown that unglycosylated kaempferol and quercetin are particularly able to compete with the auxin transport inhibitor NPA for a high-affinity binding site found in a protein complex containing PGP1, PGP2, and MDR1/PGP19. Among these proteins, roles in auxin transport for PGP1 and MDR1/PGP19 have been demonstrated (Noh et al., 2001; Murphy et al., 2002; Multani et al., 2003; Geisler et al., 2005). Flavonols are likely then to directly modulate auxin transport, a process that is reduced in the kaempferol-overaccumulating mutant tt7 (Peer et al., 2004). It is possible, therefore, that the change in auxin concentration observed in rol1-2 is caused by kaempferol-induced modulation of auxin transport.
Our data indicate that the aberrant size of stomata, pavement cell shape, trichome formation, and part of the stomatal density phenotype in rol1-2 plants are not caused by modified auxin distribution and that flavonols play a role in auxin-independent cell growth and development.
The Observed Mutant Phenotypes Are Specific for rol1-2
A number of flavonoid accumulation mutants have been described (Lepiniec et al., 2006). There are no reports of rol1-2–like phenotypes for tt mutants, suggesting that the aberrant development of rol1-2 seedlings is induced by the modified flavonol glycosylation profile or the combination of aberrant cell wall development (Diet et al., 2006) and the modified flavonol profile. K-G-3 and K-uk are the two flavonol glycosides whose accumulation in shoots correlates with the development of the rol1-2 phenotype. In lines developing a wild-type shoot phenotype, they are present in very low amounts or not at all, respectively. While K-G-3 has been identified in Arabidopsis, K-uk, which might be kaempferol-3-O-(6″-acetyl-glucoside), has so far only been found in needles of Pinaceae species (Slimestad, 2003). Unfortunately, it is not possible to provide these flavonols exogenously, since glycosylated flavonols are not taken up by the plant (Klein et al., 2000). These flavonols, therefore, cannot be defined as the rol1-2 phenotype–inducing compounds. It is possible that unglycosylated kaempferol, present in amounts below the detection limit, is the biologically active compound. The turnover rates of K-G-3 and K-uk might be different from those of other kaempferol glycosides, leading to abnormal levels of free aglycone kaempferol, which in turn affect plant development.
The mechanism(s) by which the flavonol(s) modify cell growth remains to be elucidated. A number of processes can be influenced by flavonoids (Peer and Murphy, 2006). Here, we show a change in shoot auxin concentration and provide evidence for an auxin-independent mechanism. One interesting aspect is the function of flavonoids in ROS homeostasis (Peer and Murphy, 2006), since ROS are known to influence cell growth (Gapper and Dolan, 2006). In mammalian cell systems, flavonoids have been shown to interfere with actin (Peer and Murphy, 2006). This is of interest because mutations affecting the actin cytoskeleton in Arabidopsis and maize have been shown to result in aberrant pavement cells (Frank et al., 2003; Mathur et al., 2003; Djakovic et al., 2006). Having established that flavonols can interfere with cell development, it will be necessary to unravel the mechanisms by which they influence these processes.
METHODS
Plant Material and Growth Conditions
The Arabidopsis thaliana lrx1, rol1, tt4 (2YY6 allele), and ugt78D1 mutants are Columbia accessions, and tt7 is a Landsberg erecta accession. These lines are described elsewhere (Shirley et al., 1995; Baumberger et al., 2001; Jones et al., 2003; Diet et al., 2006). The 2YY6 allele of tt4 contains a max4 mutation that affects auxin-dependent processes (Bennett et al., 2006). Using a molecular marker (see below), MAX4 tt4 single mutants were selected in the F2 generation of a backcross with a Columbia plant. Molecular markers for all mutations (see below) were used to establish the rol1-2 tt4, rol1-2 tt7, rol1-2 ugt78d1, rol1-2 lrx1, and rol1-2 lrx1 tt4 mutants. For tt4 and tt7, the lack of anthocyanin staining in young seedlings and the yellow seed color were used as visual markers for selecting homozygous mutants, which were confirmed using molecular markers. The transgenic line, accession Columbia, containing the auxin-sensitive DR5:GUS construct is described by Ulmasov et al. (1997).
For growth of plants in sterile conditions, seeds were surface-sterilized with 1% sodium hypochlorite and 0.03% Triton X-100, stratified for 3 to 4 d at 4°C, and grown for 5 d on half-strength MS medium containing 0.6% Phytagel (Sigma-Aldrich), 2% sucrose, and 100 mg/L myo-inositol with a 16-h-light/8-h-dark cycle at 22°C. For crosses and propagation of the plants, seedlings were transferred to soil and grown in growth chambers with a 16-h-light/8-h-dark cycle at 22°C.
For the chemical complementation of the rol1-2 tt4 double mutant, seedlings were grown for 7 d on agar plates as described above and complemented with 30 μM naringenin in a horizontal orientation. Since this amount of naringenin is not fully soluble in water, the exact final concentration in the agar medium is not certain.
For NPA treatment, seedlings were grown for 5 d in a vertical orientation on half-strength MS medium, as described above, containing 5 μM NPA.
Molecular Markers for tt4, tt7, and max4
Molecular markers for lrx1 and rol1-2 were described previously (Diet et al., 2004, 2006).
The point mutations in tt4, tt7, and max4 were confirmed by amplification of genomic DNA, comparing mutants with the wild-type DNA. Based on this information, cleaved-amplified polymorphic sequence markers could be established for all mutations. All PCR procedures were done as follows: annealing at 55°C for 30 s; extension at 72°C for 30 s; 40 cycles. For tt4 and max4, point mutations had to be introduced in one primer (underlined positions) to create a restriction site polymorphism. For tt4, tt4_F (5′-CCAACAGTGAACACATGACCGAC-3′) and tt4_R (5′-GTTTCCGAATTGTCGACTTAGCGC-3′), digested with Eco47III. For tt7, tt7_F (5′-CAAACCCAACACTATGGCAACTC-3′) and tt7_R (5′-GTTTGAAATCTTCGAGAGCTTTAG-3′), digested with MseI. For max4, max4_F (5′-GCGGGTGAGGTGTCGAAGTGGTACGT-3′) and max4_R (5′-AACCATCCATAAACTATGATCTAC-3′), digested with SnaBI. ugt78d1 is a T-DNA insertion mutant (Jones et al., 2003). The T-DNA insertion was identified by amplification of the border sequence between genomic DNA and the T-DNA using the gene-specific primer ugt78d1_F (5′-CTGGTAGATGTGAGTATGGAAGAG-3′) and the T-DNA–specific primer LbB1 (5′-GCGTGGACCGCTTGCTGCAACT-3′). Homozygous mutants were identified by the absence of a PCR product with a primer pair flanking the T-DNA insertion using ugt78d1_F and ugt78d1_R (5′-CATCGTCTTACCATCATCATGAAC-3′).
Microscopy and Root Measurements
Light microscopic observations were made using a Leica MZ125 stereomicroscope. For measurements of root epidermal cells, pictures were taken of fully elongated root sections of 6-d-old seedlings by differential interference contrast microscopy using an Axioplan microscope (Zeiss). Low-temperature scanning electron microscopy was performed as described by Baumberger et al. (2001).
Gel prints of epidermal cells were produced following an established protocol (Horiguchi et al., 2006) and observed by differential interference contrast microscopy using a Leica DMR microscope.
For root length and cell length measurements, seedlings were grown in a vertical orientation for 5 d. For root length, 25 seedlings per line were used. For root hair, trichoblast, and atrichoblast measurements, at least 25 cells originating from at least four different seedlings were used.
For the visualization of stomata, cotyledons were incubated for 60 min with 5 μM FM4-64, a membrane-specific fluorescent dye (Bolte et al., 2004), put on a glass slide, and analyzed. FM4-64 was excited at 488 nm, and emission was monitored at 600 to 645 nm using a confocal microscope (DMIRE2; Leica).
GUS and DPBA Staining
GUS staining was performed in 50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.5 mM K3Fe(CN)6, 0.5 mM K4Fe(CN)6, 0.1% Triton X-100, and 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid between 2 and 16 h at 37°C.
Flavonoids were visualized by the fluorescence of flavonoid-conjugated DPBA after excitation with blue light (Peer et al., 2001). Plants were grown for 6 d prior to staining. Fluorescent staining of whole seedlings was performed according to Buer and Muday (2004). Fluorescence was achieved by excitation with fluorescein isothiocyanate filters (450 to 490 nm, suppression long pass 515 nm) on a Leica DMR fluorescence microscope and 10× or 20× objectives. Digital images were captured with a Leica DC300 F charge-coupled device camera.
Flavonoid Analysis and Auxin Measurement
For the analysis of the flavonol accumulation profile, seedlings were grown in a vertical orientation for 6 d on half-strength MS as described above. One hundred intact seedlings were cut in the hypocotyl region, and roots and shoots were pooled separately, frozen in liquid nitrogen, and lyophilized to determine the dry weight. The dried material was incubated in 500 μL of 80% methanol overnight at 4°C and subsequently macerated with a pestle, followed by vigorous vortexing. After pelleting the cell debris by centrifugation, the supernatant was transferred to a fresh tube and evaporated in a Speed-Vac centrifuge, with the temperature being limited to a maximum of 43°C. After evaporation, the pellet was resuspended in 100 μL of fresh 80% methanol and used for analysis.
Flavonoid derivatives used as reference were commercially available (I-G-3 and Q-G-3; Extrasynthese) or obtained from M. Veit (K-G-3, K-G-3-G-7, K-G-3-R-7, and Q-R-G-3-R-7).
The flavonoid profile analyses were performed by HPLC-MS on an Agilent 1100 HPLC system (Agilent Technologies) fitted with a HTS PAL autosampler (CTC Analytics) and an ESQUIRE-LC ion-trap mass spectrometer (Bruker Daltonics). Chromatographic conditions were as follows: Nucleosil 100-3 C18 column (3 μm, 2 × 250 mm; Macherey-Nagel), with a flow rate 0.170 mL/min. Mobile phase was as follows: gradient within 25 min from 10 to 25% solvent B, and then within 10 min from 25 to 70% solvent B (solvent A was 0.1% [v/v] HCOOH in water, solvent B was 0.1% [v/v] HCOOH in acetonitrile). MS conditions were as follows: nebulizer gas, nitrogen, 40 p.s.i.; dry gas, nitrogen, 9 L/min; dry temperature, 300°C; HV capillary, 4000 V; high-voltage EndPlate offset, −500 V; capillary exit, −100 V; skimmer 1, −28.9 V; and trap drive, 53.4. The electrospray MS images were acquired in the negative mode at normal resolution (0.6 units at half peak height) under ion charge control conditions (ion charge control = 10,000) in the mass range from m/z 100 to 1000. MS/MS experiments were performed at 4 units isolation width, the fragmentation cutoff set by “fast calc,” and 0.9 V fragmentation amplitude in the “SmartFrag” mode.
For auxin measurement, 250 mg of root or hypocotyl/cotyledon tissue was collected, frozen in liquid nitrogen, macerated, suspended in 100% methanol, briefly warmed to 70°C, and kept for 30 min with continuous gentle shaking. Prior to warming to 70°C, 100 pmol of [2H]2-indole-3-acetic acid (IAA) was added as the internal standard. After centrifugation, the pellet was dried to determine the dry weight and the supernatant was used for auxin quantification. The gas chromatography–MS/MS analysis of IAA contents was performed according to Müller et al. (2002). In brief, the samples were precleaned by microscale solid-phase extraction on custom-made cartridges containing a silica-based aminopropyl matrix. After application of the samples and washing the microcolumn with 250 μL of CHCl3:2-propanol (2:1, v/v), the IAA fraction was eluted twice with 200 μL of diethyl ether containing 2% acetic acid. Thereafter, the samples were dried, redissolved in 20 μL of methanol, and treated with ethereal diazomethane. Subsequently, samples were transferred to autosampler vials and excessive diazomethane and solvent were removed in a gentle stream of nitrogen. The methylated samples were then taken up in 10 μL of chloroform. Aliquots of 1 μL of each sample were injected into the gas chromatography–MS system for separation and mass fragment analysis using the following autosampler and system. All spectra were recorded on a Varian Saturn 2000 ion-trap mass spectrometer connected to a Varian CP-3800 gas chromatograph equipped with a CombiPal autoinjector (Varian).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following gene identifiers: ROL1/RHM1, AT1G78570; TT4, AT5G13930; TT7, AT5G07990; UGT78D1, AT1G30530; LRX1, AT1G12040; MAX4, AT4G32810.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Graphical Representation of Flavonol Abundance in Wild-Type and rol1 Mutant Seedlings.
Supplemental Figure 2. Flavonoid Staining by DPBA in Arabidopsis Seedlings.
Supplemental Figure 3. The tt7 Mutation Does Not Develop rol1-2–Like Phenotypes.
Supplemental Figure 4. Effect of NPA on Seedling Development and DR5:GUS Activity.
Supplementary Material
Acknowledgments
We thank A. Schaeffner for the ugt78d1 mutant, Markus Geisler for [2H]2-IAA, and W. Heller (German Research Center for Environmental Health) and M. Veit (Kaufering) for providing us with flavonol standards that were invaluable for the flavonol structure analysis. We also thank Celia Jäger-Baroux, Petra Düchting, and Barbara Weder for technical assistance and Mark Curtis for critical reading of the manuscript. This work was supported by the Swiss National Science Foundation (Grants 31-61419.00 and 3100A0-103891 to C.R., R.-M.L., and A.D. and Grant 3100A0-116051 to M.K.), a Swiss National Center of Competence in Research Plant Survival Program grant to E. Martinoia (D.S.), the Julius-Klaus Stiftung (R.-M.L.), and the Forschungskommission der Universität Zürich (B.M.K.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Christoph Ringli (chringli@botinst.uzh.ch).
Online version contains Web-only data.
References
- Baumberger, N., Ringli, C., and Keller, B. (2001). The chimeric leucine-rich repeat/extensin cell wall protein LRX1 is required for root hair morphogenesis in Arabidopsis thaliana. Genes Dev. 15 1128–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, T., Sieberer, T., Willett, B., Booker, J., Luschnig, C., and Leyser, O. (2006). The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr. Biol. 16 553–563. [DOI] [PubMed] [Google Scholar]
- Bolte, S., Talbot, C., Boutte, Y., Catrice, O., Read, N.D., and Satiat-Jeunemaitre, B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214 159–173. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Rashotte, A.M., Murphy, A.S., Normanly, J., Tague, B.W., Peer, W.A., Taiz, L., and Muday, G.K. (2001). Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126 524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S., and Muday, G.K. (2004). The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16 1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buer, C.S., Muday, G.K., and Djordjevic, M.A. (2007). Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol. 145 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3 1–30. [DOI] [PubMed] [Google Scholar]
- Cassab, G.I. (1998). Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 281–309. [DOI] [PubMed] [Google Scholar]
- Chary, S.N., Hicks, G.R., Choi, Y.G., Carter, D., and Raikhel, N.V. (2008). Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 146 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Nesi, N., Perez, P., Devic, M., Grandjean, O., Caboche, M., and Lepiniec, L. (2003). Proanthocyanidin-accumulating cells in Arabidopsis testa: Regulation of differentiation and role in seed development. Plant Cell 15 2514–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diet, A., Brunner, S., and Ringli, C. (2004). The enl mutants enhance the lrx1 root hair mutant phenotype of Arabidopsis thaliana. Plant Cell Physiol. 45 734–741. [DOI] [PubMed] [Google Scholar]
- Diet, A., Link, B., Seifert, G.J., Schellenberg, B., Wagner, U., Pauly, M., Reiter, W.D., and Ringli, C. (2006). The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-L-rhamnose synthase. Plant Cell 18 1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic, S., Dyachok, J., Burke, M., Frank, M.J., and Smith, L.G. (2006). BRICK1/HSPC300 functions with SCAR and the ARP2/3 complex to regulate epidermal cell shape in Arabidopsis. Development 133 1091–1100. [DOI] [PubMed] [Google Scholar]
- Frank, M.J., Cartwright, H.N., and Smith, L.G. (2003). Three BRICK genes have distinct functions in a common pathway promoting polarized cell division and cell morphogenesis in the maize leaf epidermis. Development 130 753–762. [DOI] [PubMed] [Google Scholar]
- Friml, J. (2003). Auxin transport—Shaping the plant. Curr. Opin. Plant Biol. 6 7–12. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Gu, Y., Zheng, Z.L., Wasteneys, G., and Yang, Z.B. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120 687–700. [DOI] [PubMed] [Google Scholar]
- Gapper, C., and Dolan, L. (2006). Control of plant development by reactive oxygen species. Plant Physiol. 141 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler, M., et al. (2005). Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44 179–194. [DOI] [PubMed] [Google Scholar]
- Horiguchi, G., Fujikura, U., Ferjani, A., Ishikawa, N., and Tsukaya, H. (2006). Large-scale histological analysis of leaf mutants using two simple leaf observation methods: Identification of novel genetic pathways governing the size and shape of leaves. Plant J. 48 638–644. [DOI] [PubMed] [Google Scholar]
- Jacobs, M., and Rubery, P.H. (1988). Naturally-occurring auxin transport regulators. Science 241 346–349. [DOI] [PubMed] [Google Scholar]
- Jones, P., Messner, B., Nakajima, J.I., Schaffner, A.R., and Saito, K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278 43910–43918. [DOI] [PubMed] [Google Scholar]
- Kang, B.G. (1979). Epinasty. In Physiology of Movements, W. Haupt and M.E. Feinleib, eds (Berlin: Springer-Verlag), pp. 647–667.
- Kerhoas, L., Aouak, D., Cingoz, A., Routaboul, J.M., Lepiniec, L., Einhorn, J., and Birlirakis, N. (2006). Structural characterization of the major flavonoid glycosides from Arabidopsis thaliana seeds. J. Agric. Food Chem. 54 6603–6612. [DOI] [PubMed] [Google Scholar]
- Klein, M., Martinoia, E., Hoffmann-Thoma, G., and Weissenbock, G. (2000). A membrane-potential dependent ABC-like transporter mediates the vacuolar uptake of rye flavone glucuronides: Regulation of glucuronide uptake by glutathione and its conjugates. Plant J. 21 289–304. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1990). Mutations affecting the testa color in Arabidopsis. Arabidopsis Inf Serv 19 113–115. [Google Scholar]
- Lavy, M., Bloch, D., Hazak, O., Gutman, I., Poraty, L., Sorek, N., Sternberg, H., and Yalovsky, S. (2007). A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr. Biol. 17 947–952. [DOI] [PubMed] [Google Scholar]
- Laxmi, A., Paul, L.K., Raychaudhuri, A., Peters, J.L., and Khurana, J.P. (2006). Arabidopsis cytokinin-resistant mutant, cnr1, displays altered auxin responses and sugar sensitivity. Plant Mol. Biol. 62 409–425. [DOI] [PubMed] [Google Scholar]
- Le Gall, G., Metzdorff, S.B., Pedersen, J., Bennett, R.N., and Colquhoun, I.J. (2006). Metabolite profiling of Arabidopsis thaliana (L.) plants transformed with an antisense chalcone synthase gene. Metabolism 1 181–198. [Google Scholar]
- Lepiniec, L., Debeaujon, I., Routaboul, J.M., Baudry, A., Pourcel, L., Nesi, N., and Caboche, M. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57 405–430. [DOI] [PubMed] [Google Scholar]
- Liu-Gitz, L., Britz, S.J., and Wergin, W.P. (2000). Blue light inhibits stomatal development in soybean isolines containing kaempferol-3-O-2(G)-glycosyl-gentiobioside (K9), a unique flavonoid glycoside. Plant Cell Environ. 23 883–891. [Google Scholar]
- Martin, C., Bhatt, K., and Baumann, K. (2001). Shaping in plant cells. Curr. Opin. Plant Biol. 4 540–549. [DOI] [PubMed] [Google Scholar]
- Mathur, J. (2004). Cell shape development in plants. Trends Plant Sci. 9 583–590. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., and Hülskamp, M. (2003). Mutations in actin-related proteins 2 and 3 affect cell shape development in Arabidopsis. Plant Cell 15 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo, Y.Y., Nagel, C., and Taylor, L.P. (1992). Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. USA 89 7213–7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller, A., Düchting, P., and Weiler, E.W. (2002). A multiplex GC-MS/MS technique for the sensitive and quantitative single-run analysis of acidic phytohormones and related compounds, and its application to Arabidopsis thaliana. Planta 216 44–56. [DOI] [PubMed] [Google Scholar]
- Multani, D.S., Briggs, S.P., Chamberlin, M.A., Blakeslee, J.J., Murphy, A.S., and Johal, G.S. (2003). Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science 302 81–84. [DOI] [PubMed] [Google Scholar]
- Murphy, A.S., Hoogner, K.R., Peer, W.A., and Taiz, L. (2002). Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol. 128 935–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh, B., Murphy, A.S., and Spalding, E.P. (2001). Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13 2441–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka, T., Nemoto, T., and Jigami, Y. (2007). Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J. Biol. Chem. 282 5389–5403. [DOI] [PubMed] [Google Scholar]
- Peer, W.A., Bandyopadhyay, A., Blakeslee, J.J., Makam, S.I., Chen, R.J., Masson, P.H., and Murphy, A.S. (2004). Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16 1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., Brown, D.E., Tague, B.W., Muday, G.K., Taiz, L., and Murphy, A.S. (2001). Flavonoid accumulation patterns of transparent testa mutants of Arabidopsis. Plant Physiol. 126 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer, W.A., and Murphy, A.S. (2006). Flavonoids as signal molecules. In The Science of Flavonoids, E. Grotewold, ed (Berlin: Springer-Verlag), pp. 239–268.
- Reiter, W.D., and Vanzin, G.F. (2001). Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol. Biol. 47 95–113. [PubMed] [Google Scholar]
- Ridley, B.L., O'Neill, M.A., and Mohnen, D.A. (2001). Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 57 929–967. [DOI] [PubMed] [Google Scholar]
- Ringli, C. (2005). The role of extracellular LRR-extensin (LRX) proteins in cell wall formation. Plant Biosyst. 139 32–35. [Google Scholar]
- Routaboul, J.M., Kerhoas, L., Debeaujon, I., Pourcel, L., Caboche, M., Einhorn, J., and Lepiniec, L. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224 96–107. [DOI] [PubMed] [Google Scholar]
- Schijlen, E., de Vos, C.H.R., Martens, S., Jonker, H.H., Rosin, F.M., Molthoff, J.W., Tikunov, Y.M., Angenent, G.C., van Tunen, A.J., and Bovy, A.G. (2007). RNA interference silencing of chalcone synthase, the first step in the flavonoid biosynthesis pathway, leads to parthenocarpic tomato fruits. Plant Physiol. 144 1520–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley, B.W., Kubasek, W.L., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F.M., and Goodman, H.M. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8 659–671. [DOI] [PubMed] [Google Scholar]
- Slimestad, R. (2003). Flavonoids in buds and young needles of Picea, Pinus and Abies. Biochem. Syst. Ecol. 31 1247–1255. [Google Scholar]
- Smith, L.G. (2003). Cytoskeletal control of plant cell shape: Getting the fine points. Curr. Opin. Plant Biol. 6 63–73. [DOI] [PubMed] [Google Scholar]
- Stobiecki, M., Skirycz, A., Kerhoas, L., Kachlicki, P., Muth, D., Einhorn, J., and Mueller-Roeber, B. (2006). Profiling of phenolic glycosidic conjugates in leaves of Arabidopsis thaliana using LC/MS. Metabolism 2 197–219. [Google Scholar]
- Taylor, L.P., and Grotewold, E. (2005). Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8 317–323. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T.J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9 1963–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche, F., and Van Der Straeten, D. (2007). One for all and all for one: Cross-talk of multiple signals controlling the plant phenotype. J. Plant Growth Regul. 26 178–187. [Google Scholar]
- Veit, M., and Pauli, G.F. (1999). Major flavonoids from Arabidopsis thaliana leaves. J. Nat. Prod. 62 1301–1303. [DOI] [PubMed] [Google Scholar]
- Watahiki, M.K., and Yamamoto, K.T. (1997). The massugu1 mutation of Arabidopsis identified with failure of auxin-induced growth curvature of hypocotyl confers auxin insensitivity to hypocotyl and leaf. Plant Physiol. 115 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, R.E., Burn, J.E., Birch, R., Baskin, T.I., Arioli, T., Betzner, A.S., and Cork, A. (2001). Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215 116–127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.