Abstract
The rice (Oryza sativa) E3 ligase SPOTTED LEAF11 (SPL11) negatively regulates programmed cell death and disease resistance. We demonstrate here that SPL11 also regulates flowering via interaction with SPIN1 (for SPL11-interacting protein1), a Signal Transduction and Activation of RNA family member. SPIN1 binds RNA and DNA in vitro and interacts with SPL11 in the nucleus. Spl11 mutants have delayed flowering under long-day conditions. Spin1 overexpression causes late flowering independently of daylength; expression analyses of flowering marker genes in these lines suggested that SPIN1 represses flowering by downregulating the flowering promoter gene Heading date3a (Hd3a) via Hd1-dependent mechanisms in short days and by targeting Hd1-independent factors in long days. Both Spin1 and Spl11 are regulated diurnally in opposing phases. SPL11 negatively regulates Spin1 transcript levels, while SPIN1 also affects Spl11 expression. Moreover, we show that coincidence of high accumulation of Spin1 mRNA with the light in the morning and early evening is needed to repress flowering. SPIN1 is monoubiquitinated by SPL11, suggesting that it is not targeted for degradation. Our data are consistent with a model in which SPIN1 acts as a negative regulator of flowering that itself is negatively regulated by SPL11, possibly via ubiquitination.
INTRODUCTION
Flowering in plants is triggered by both endogenous and environmental cues. Molecular genetic studies in Arabidopsis thaliana have identified at least four major pathways that regulate flowering, each perceiving and processing different signals (Simpson and Dean, 2002; Boss et al., 2004; Putterill et al., 2004). The photoperiodic and vernalization pathways mediate responses to changes in daylength and temperature associated with seasonal variation, respectively, whereas the autonomous and gibberellin pathways transduce internal developmental and physiological signals to promote flowering (Simpson and Dean, 2002; Putterill et al., 2004). These signaling pathways form a multilevel regulatory complex in which input signals ultimately converge at a small set of key regulatory genes known as the floral pathway integrators (Simpson and Dean, 2002; Boss et al., 2004; Putterill et al., 2004). For example, the flowering activators FT (for FLOWERING LOCUS T) and SOC1 (for SUPPRESSOR OF OVEREXPRESSION OF CO1) are not only positively regulated via the photoperiodic pathway by CO (for CONSTANS) (Samach et al., 2000; Suarez-Lopez et al., 2001; Yanovsky and Kay, 2002) but also are negatively regulated by the floral repressor FLC (for FLOWERING LOCUS C) through the vernalization and autonomous pathways (Lee et al., 2000; Boss et al., 2004; Henderson and Dean, 2004).
In Arabidopsis, flowering is promoted under long-day (LD) conditions. By contrast, rice (Oryza sativa) flowers earlier under short-day (SD) conditions. Despite this obvious difference in response to photoperiod, molecular genetic studies in rice have revealed the existence of both conserved and unique components in the daylength response pathway between the two species (Izawa et al., 2003; Hayama and Coupland, 2004; Izawa, 2007). The rice genes Heading date1 (Hd1), Hd3a, and Hd6 are orthologs of Arabidopsis CO, FT, and the α-subunit of kinase CK2, respectively (Takahashi et al., 1998; Yano et al., 2000; Kojima et al., 2002). Moreover, the homolog of GIGANTEA (GI), a gene regulating both circadian clock inputs and the control of flowering (Fowler et al., 1999), and the homolog of the floral pathway integrator SOC1 have also been isolated in rice (Hayama et al., 2002, 2003; Tadege et al., 2003). In the current model for flowering control in rice, phytochromes (light receptor proteins) are essential in determining the photoperiod sensitivity (Izawa et al., 2000, 2002). Under noninductive LD conditions, the coincidence of phytochrome signaling with Hd1 expression in the light makes Hd1 a repressor of the floral activator Hd3a (Izawa et al., 2002; Hayama et al., 2003; Hayama and Coupland, 2004). This is in contrast with Arabidopsis, in which overexpression of CO promotes flowering independently of daylength (Onouchi et al., 2000). On the other hand, high-level expression of Hd1 occurs in the dark under SD conditions, which in turn activates Hd3a to promote flowering (Izawa et al., 2002; Kojima et al., 2002; Hayama et al., 2003; Hayama and Coupland, 2004). Hence, through fine-tuning of the activation/accumulation of Hd3a via Hd1 in response to photoperiod, flowering is delayed under LD and induced under SD in rice.
Ubiquitination is one of the major types of posttranslational modifications in the cell (Nalivaeva and Turner, 2001). It is achieved by a cascade of enzymatic reactions that involve coordinated activities of three enzymes: ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) (Ciechanover, 1998). Ubiquitination regulates various aspects of plant life, including disease resistance (Zeng et al., 2006), hormone signaling (Itoh et al., 2003), and many developmental processes (Moon et al., 2004) such as flowering time control. In Arabidopsis, CO is degraded via the 26S proteasome in darkness (Valverde et al., 2004) by the E3 ligase CONSTITUTIVELY PHOTOMORPHOGENIC1 (COP1) (Liu et al., 2008), while its transcriptional repressor CYCLING DOF FACTOR1 (CDF1) is degraded by the F-box protein FKF1 (Imaizumi et al., 2005). Thus, by means of directly targeting CO or its negative regulators through ubiquitination, tight regulation of CO levels is achieved in response to daylength variations. Whether ubiquitination functions to control rice flowering time remains to be elucidated.
In addition to ubiquitination, RNA processing also plays an important role in the regulation of plant flowering time. The autonomous pathway genes FCA and FPA encode proteins containing RNA recognition motifs (Macknight et al., 1997; Schomburg et al., 2001). The FLOWERING LOCUS K (FLK) gene also encodes a protein with putative RNA processing function (Lim et al., 2004; Mockler et al., 2004). The FLK protein contains three K homology (KH) domains, a widely found motif involved in RNA binding activity (Adinolfi et al., 1999). FCA, FPA, and FLK were shown to negatively regulate FLC expression (Macknight et al., 1997; Schomburg et al., 2001; Lim et al., 2004; Mockler et al., 2004). Recent evidence suggests that FPA and FCA mediate small interfering RNA-directed DNA methylation of FLC chromatin (Baurle et al., 2007; Liu et al., 2007).
A recent bioinformatic analysis revealed that E3 ligase proteins containing domains associated with RNA binding could be found in animals, plants, and fungi (Lucas et al., 2006), suggesting that the link between ubiquitination and RNA metabolism is an ancient, evolutionarily conserved mechanism. The rice gene Spotted leaf11 (Spl11), encoding a U-box domain/ARM repeats–type E3 ubiquitin ligase, was previously shown to negatively regulate programmed cell death (PCD) and disease resistance (Yin et al., 2000; Zeng et al., 2004). Here, we present evidence that Spl11 also controls flowering time via its interaction with a novel RNA/DNA binding KH domain protein called SPIN1 (for SPL11-interacting protein1). The interaction between SPL11 and SPIN1 occurs in the nucleus, suggesting that such interaction may be related to RNA processing. SPL11 both ubiquitinates SPIN1 in vitro and negatively regulates Spin1 mRNA levels. Overexpression of Spin1 significantly delays rice flowering under both SD and LD conditions, while mutation of Spl11 only delays flowering in LD. Our data suggest that the SPL11–SPIN1 interaction regulates a posttranscriptional mechanism involved in flowering time control in rice.
RESULTS
Flowering Time Phenotypes of the spl11 Mutation
The original spl11 allele isolated is an ethyl methanesulfonate–induced mutation in cv IR68 (spl11-IR68) that causes a premature stop codon (Zeng et al., 2004). Two additional γ-ray alleles in the IR64 background (GR5612 and GR5717) have also been described (Zeng et al., 2004); for this study, only the GR5717 allele was used.
The mutant shows enhanced, non-race-specific resistance to both Magnaporthe oryzae and Xanthomonas oryzae pv oryzae, the pathogens that cause rice blast and bacterial blight diseases, respectively (Yin et al., 2000). A more detailed analysis revealed that the spl11 mutant showed alterations in flowering time in addition to previously reported phenotypes.
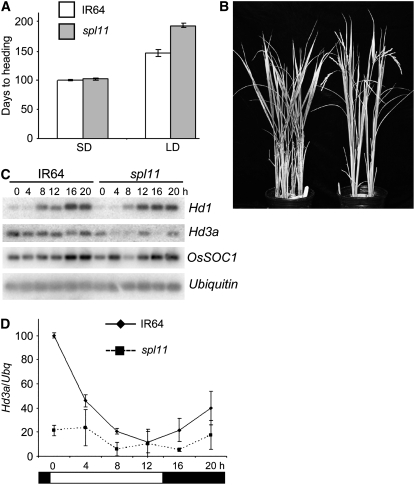
Under SD, no significant difference in flowering time between wild-type IR64 and the spl11 mutant GR5717 was observed (Figure 1A).
Figure 1.
Flowering Time Phenotype of the spl11 Mutation.
(A) Days to heading in SD and LD for IR64 (wild type) and IR64-GR5717 (spl11). Error bars indicate sd; n = 5 to 10 plants.
(B) Delayed flowering phenotype of spl11 mutant (left) compared with the wild type (right) in LD.
(C) RT-PCR/DNA gel blot analysis of Hd1, Hd3a, SOC1, and Ubq in IR64 and spl11 under LD in 55-d-old plants.
(D) Quantification of mRNA abundance relative to Ubq of the Hd3a blot shown in (C). White and black rectangles represent light and dark conditions over a 24-h period, respectively. Error bars indicate sd of three independent experiments.
However, under noninductive LD conditions, flowering was significantly delayed in the spl11 mutant compared with the wild type (Figures 1A and 1B). Flowering in rice is promoted under SD by activating the CO homolog Hd1, which in turn activates Hd3a, the rice homolog of FT (Hayama et al., 2003). Under LD, Hd1 negatively regulates flowering by repressing Hd3a (Hayama et al., 2003). To test whether the late-flowering phenotype of the spl11 mutant in LD is due to changes in flowering time gene expression, a quantitative RT-PCR/DNA gel blot (Yanovsky and Kay, 2002) analysis was conducted in wild-type and spl11 plants over a 24-h period. Both IR64 and spl11 showed similar diurnal regulation of mRNA levels for Hd1 and SOC1 (Figure 1C). However, Hd3a transcript levels were significantly reduced in the mutant compared with the wild type in both light and dark growth periods (Figures 1C and 1D). Moreover, Spl11 showed a diurnal expression pattern under SD conditions that is typical of many flowering regulators, with mRNA levels peaking in the dark and decreasing in midafternoon (see Figure 5C, right). Interestingly, the diurnal regulation of Spl11 mRNA level was abolished in LD (see Figure 5D, right). Taken together, these results confirm a role for the Spl11 gene in the regulation of flowering time.
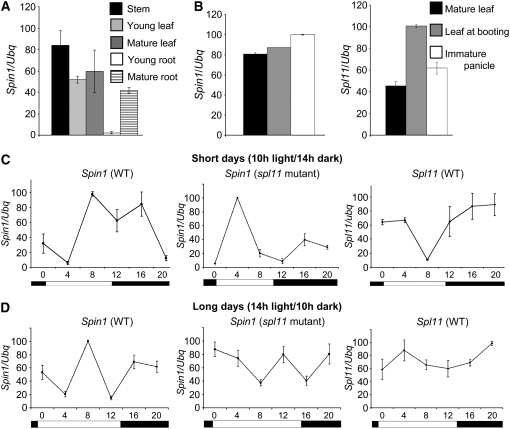
Figure 5.
Expression Analysis of Spin1 and Spl11.
(A) Quantitative RT-PCR of Spin1 expression in vegetative rice tissues.
(B) RT-PCR analysis of Spin1 and Spl11 expression patterns in mature leaf and flowering tissues.
(C) RT-PCR of Spin1 and Spl11 expression in leaves under SD over a 24-h period.
(D) RT-PCR of Spin1 and Spl11 expression in leaves under LD over a 24-h period.
White and black rectangles in (C) and (D) represent light and dark conditions over a 24-h period, respectively. Error bars indicate sd of three independent experiments.
Identification and Subcellular Localization of SPIN1
To elucidate the molecular basis by which Spl11 controls flowering time and/or PCD, yeast two-hybrid screens were performed using the ARM domain region of the SPL11 protein as the bait. In total, 29 positive clones were isolated representing eight SPL11-interacting proteins. Homolog searches against the GenBank databases using the BLAST2 algorithm (http://www.ncbi.nlm.nih.gov/) revealed significant hits for six of the eight candidates (see Supplemental Table 1 online).
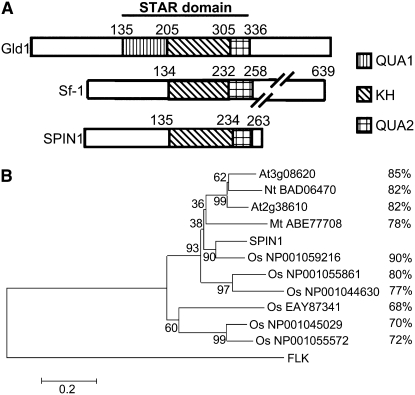
One of the putative interactors, named SPIN1, accounted for nearly one-third (11 of 29) of the positive clones identified in the screens. The Spin1 full-length cDNA was obtained from the library used for the yeast two-hybrid analysis and consisted of an 846-bp open reading frame encoding a 281–amino acid protein with a theoretical pI of 9.48. Searches in various protein databases using the deduced amino acid sequence revealed that Spin1 encodes a novel protein containing a single KH domain (Figure 2). Sequence similarity searches using the BLAST algorithm on the National Center for Biotechnology Information website showed that SPIN1 belongs to a subfamily of KH domain–containing proteins known as STAR (for Signal Transduction and Activation of RNA) (Figure 2A; see Supplemental Figure 1 online) (Vernet and Artzt, 1997). The STAR domain is a tripartite motif with a single KH domain flanked by two subdomains termed QUA1 and QUA2 (Figure 2A) (Vernet and Artzt, 1997). RNA binding proteins in the STAR family include proteins involved in the regulation of key developmental processes in animals, such as Gld-1 in Caenorhabditis elegans, How in Drosophila melanogaster, and Sam-68 in mouse and human (Vernet and Artzt, 1997; Ryder et al., 2004). The sequence homology between SPIN1 and the animal STAR family members is limited to the STAR region (see Supplemental Figure 1 online). The SPIN1 N-terminal region shows no homology to any known domain present in the databases. The STAR domain in SPIN1 is most similar to mammalian Splicing factor1 (Sf1) (Berglund et al., 1997), which lacks the QUA1 subdomain (Figure 2A), with 59% similarity and 42% identity at the amino acid level.
Figure 2.
SPIN1 Is a Member of the STAR Family of RNA Binding Proteins.
(A) Schematic representation of the STAR domain. Gld-1 is from C. elegans, and Sf1 is from human. Note that similar to Sf1, SPIN1 lacks the QUA1 subdomain.
(B) Phylogenetic analysis of plant SPIN1 homologs from rice (Os), Arabidopsis (At), tobacco (Nt), and M. truncatula (Mt). The dendogram was constructed using the MEGA3.1 software with the neighbor-joining algorithm; Arabidopsis FLK, a KH domain–containing protein unrelated to the SPIN1-like sequences, was used as an outgroup. Amino acid percentage similarity to SPIN1 is shown.
Several SPIN1-like sequences were identified in rice and other plant species, including Arabidopsis, tobacco (Nicotiana tabacum), and Medicago truncatula, when SPIN1 was used as a query in database mining (Figure 2B; see Supplemental Figure 2 and Supplemental Data Set 1 online). The existence of six other Spin1-like genes in the rice genome suggested that Spin1 belongs to a small gene family. Phylogenetic analysis indicated that SPIN1 has highest similarity to one of its paralogs (Os NP_001059216), and the similarity between SPIN1 and its dicot orthologs is higher than that of the rest of its rice homologs (Figure 2B; see Supplemental Data Set 1 online). Sequence alignment revealed that a high level of amino acid similarity spanning the whole protein sequence exists among the SPIN1-like homologs, suggesting that these proteins are evolutionarily conserved (see Supplemental Figure 2 online).
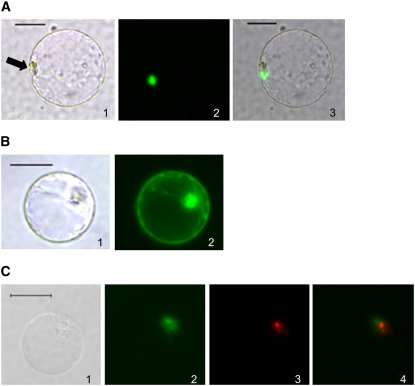
A protein's localization in the cell is usually closely related to its biological function. Green fluorescent protein (GFP)–SPIN1 and GFP-SPL11 fusions were expressed in rice protoplasts and their subcellular localizations were determined. While GFP-SPIN1 was localized strictly in the nuclear region (Figure 3A), GFP-SPL11 accumulated all over the cell (Figure 3B). These data suggest that SPIN1 may function at the level of nucleus-related RNA metabolism such as transcription or splicing, while SPL11 may be involved in a broader range of cellular functions. Interestingly, the fluorescence signal for GFP-SPIN1 did not cover the entire nucleus (Figure 3A), suggesting a distinct subnuclear localization pattern for SPIN1.
Figure 3.
Subcellular Localization of SPIN1 and SPL11 in Rice Protoplasts.
(A) SPIN1-GFP localizes to the nuclear region. Panel 1, bright field; panel 2, green fluorescence; panel 3, bright and green merged. Arrow indicates the nucleus.
(B) SPL11-GFP is localized in the entire cell. Panel 1, bright field; panel 2, green fluorescence.
(C) Subnuclear localization of the SPIN1–SPL11 interaction using BiFC. Green fluorescence represents SPL11-NYFP + SPIN1-CYFP; red fluorescence represents the nucleolar protein PRH75-RFP. Panel 1, bright field; panel 2, green fluorescence; panel 3, red fluorescence; panel 4, green and red merged.
Bars = 20 μm.
SPIN1 Interacts with SPL11 in Vitro and in Rice Protoplasts
To validate the yeast two-hybrid data, both in vitro and in vivo experiments were performed to test the SPIN1–SPL11 interaction. Recombinant glutathione S-transferase (GST) fusion proteins containing the SPL11 ARM domain or full-length SPL11 were purified from Escherichia coli and incubated with in vitro translated and biotinylated SPIN1 protein. Results from the GST pull-down assay showed that SPIN1 was present in reactions containing either GST-ARM or GST-SPL11 but not GST, confirming the in vitro interaction between SPIN1 and SPL11 (Figure 4A). To determine which region of the SPIN1 protein is minimally required for the interaction with SPL11, an interaction domain–mapping assay using the yeast two-hybrid approach was used. Different domains of SPIN1 were coexpressed in yeast with the ARM region of SPL11 (Figure 4C). Only the N-terminal region of SPIN1 interacted with the ARM domain of SPL11, as seen by a positive X-Gal assay (Figure 4C). As the SPIN1 N-terminal region shows no homology to any known domain in the existing protein databases, these results suggested that the N-terminal region of SPIN1 encodes a novel protein–protein interaction domain.
Figure 4.
SPIN1 Interacts with SPL11 Both in Vitro and in Vivo.
(A) GST pull-down assay shows in vitro interaction of GST-SPL11 (full length) and GST-ARM (ARM domain only) with SPIN1. In vitro translated and biotinylated SPIN1 was incubated with GST fusion proteins in glutathione beads. SPIN1 was pulled down in beads containing GST-SPL11 and GST-ARM but not GST. The SDS-PAGE gel at right is shown as a protein loading reference.
(B) BiFC detection of the SPIN1–SPL11 interaction in rice protoplasts. Cells were observed on a fluorescence microscope under bright and UV lights to detect cells and green fluorescence, respectively.
(C) SPIN1 interaction domain mapping with SPL11-ARM in yeast. SPL11-ARM and different domains of SPIN1 were fused to the GAL4 DNA binding and activation domains, respectively. Blue colonies in an X-Gal assay indicate interaction in yeast.
Testing of the interaction in living cells was performed by bimolecular fluorescence complementation (BiFC) in rice protoplasts (Bracha-Drori et al., 2004; Chen et al., 2006). Yellow fluorescent protein (YFP) fluorescence was reconstituted when full-length SPIN1 and SPL11, ARM and SPIN1, or SPIN1-N and SPL11 proteins were coexpressed in protoplasts (Figure 4B), indicating that these proteins interact in vivo. Control experiments coexpressing the SPIN1 C terminus with SPL11, or the unrelated protein β-glucuronidase (GUS) with either SPIN1 or SPL11, showed no fluorescence (Figure 4B), confirming that the SPIN1–SPL11 interaction is specific. Moreover, the BiFC technique allowed us to clearly identify the subcellular localization of the SPL11–SPIN1 interaction in the nucleus (Figure 4B). As an attempt to more clearly define this interaction, the spinach nucleolar protein PRH75 (Lorkovic et al., 1997) was used as a marker to pinpoint the subnuclear localization of the SPIN1–SPL11 interaction relative to the nucleolus. As shown in Figure 3C, the SPIN1–SPL11 interaction occurs in the vicinity of, and partially overlapping with, the nucleolus.
Spin1 Is Ubiquitously Expressed in Rice, Shows Diurnal Expression Patterns, and Its Expression Is Negatively Regulated by Spl11 in the Light
RT-PCR analyses revealed that Spin1 is expressed in most tissues from stems to old leaves (Figure 5A) and flowers (Figure 5B). Spin1 transcript level was highest in stems and mature leaves and almost undetectable in young roots, although it was present in mature roots (Figure 5A). Interestingly, both Spin1 and Spl11 seem to be upregulated at flowering, since their expression levels were higher in mature leaves at booting stage compared with leaves prior to booting (Figure 5B). The booting stage represents the swelling of the flag leaf sheath caused by the growth of the inflorescence. This increase in expression at booting was followed by an increased expression in immature panicles for Spin1 but not for Spl11 (Figure 5B).
Similar to Spl11, Spin1 expression showed a diurnal regulation under SD over a 24-h period (Figure 5C). However, high Spl11 expression tended to correlate with decreased Spin1 levels, and highest Spin1 expression occurred when Spl11 levels were the lowest, especially during the light period (Figure 5C, left and right). Surprisingly, in the spl11 mutant background in SD, Spin1 maintained its diurnal regulation, but at a phase opposite to the one observed in the wild-type IR64 (Figure 5C, middle). These results suggest that Spl11 negatively regulates Spin1 transcript levels. The Spin1 gene expression phase change between IR64 and spl11 during light was also observed in LD (Figure 5D, left and middle). While Spl11 mRNA accumulation was aphasic and remained constant throughout the day, its expression peaked at dawn under LD conditions (Figure 5D, right). Importantly, the elevated Spl11 transcript levels in LD correlated with a decrease in Spin1 mRNA accumulation that coincided with the extended light period during morning and early evening (Figure 5D, left and right, time points 0, 4, and 12). This decrease of Spin1 expression in LD in IR64 plants at morning and early evening was reversed in the spl11 mutant (Figure 5D, middle), which demonstrated that it is under the control of Spl11.
Taken together, the expression analyses of Spin1 revealed the ubiquitous expression of this gene in rice tissues and developmental stages as well as its negative regulation in both SD and LD by Spl11, especially during light periods. The aphasic Spl11 transcript accumulation in LD suggests that a photoperiod-sensitive clock component controls Spl11 expression levels.
SPL11 Targets SPIN1 for Monoubiquitination
SPL11 was shown to possess E3 ubiquitin ligase activity (Zeng et al., 2004). The interaction between SPL11 and SPIN1 both in vitro and in vivo suggests that SPL11 might target SPIN1 for ubiquitination. To test this possibility, an in vitro ubiquitination assay was performed using recombinant GST-SPIN1-HA (for hemagglutinin), GST-SPL11, E1, E2, and ubiquitin (Ub) (Figure 6). Ubiquitination was observed as higher molecular mass bands compared with free GST-SPIN1-HA when GST-SPL11 was present in the reaction (Figure 6, lane 4). An unrelated E3 ligase, Os SINAT5, homologous to Arabidopsis SINAT5 (Xie et al., 2002) failed to ubiquitinate SPIN1 (Figure 6, lane 8), indicating that SPL11 specifically targets SPIN1 for ubiquitination. Importantly, the size of the ubiquitinated protein suggested monoubiquitination or diubiquitination of SPIN1 by SPL11.
Figure 6.
Ubiquitination of SPIN1 by SPL11 and TAP-SPIN1 Accumulation in SD and LD.
SPL11 monoubiquitinates SPIN1 in vitro. The arrowhead indicates the position of monoubiquitinated SPIN1; the arrow indicates unubiquitinated SPIN1. The unrelated E3 ligase Os SINAT5 was used as a negative control (At UBC7 was used as the E2 enzyme in the Os SINAT5 reaction, while At UBC9 was used in the SPL11 reaction).
SPIN1 Has Both RNA and DNA Binding Activities in Vitro
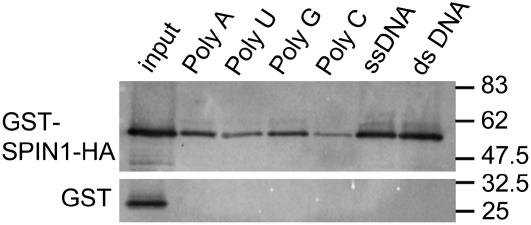
Our bioinformatic analyses indicated that SPIN1 is a member of the STAR family of RNA binding proteins (Figure 2). To confirm its biochemical function, recombinant GST-SPIN1-HA protein was incubated in vitro with ribohomopolymer-bound beads representing poly(A), poly(U), poly(G), and poly(C) as well as single- and double-stranded calf thymus DNA. Protein gel blot analysis following incubation and washing of beads revealed that GST-SPIN1-HA bound to all RNA and DNA polymers tested (Figure 7). As a control, GST did not show any RNA or DNA binding activity. These results confirmed that SPIN1 has RNA/DNA binding activity in vitro.
Figure 7.
SPIN1 Has Both RNA and DNA Binding Activities in Vitro.
GST-SPIN1-HA binds to ribohomopolymers and both single- and double-stranded calf thymus DNA in vitro. GST-SPIN1-HA was purified from E. coli and incubated with beads containing different nucleic acid polymers. Binding of SPIN1 to RNA or DNA was confirmed by a protein gel blot using anti-HA antibodies. The bottom panel shows a GST negative control, anti-GST WB.
Overexpression of Spin1 Causes Late Flowering under Both SD and LD
To functionally characterize Spin1, RNA interference (RNAi) and overexpression transgenic lines were generated in the rice japonica cv Nipponbare (NPB). Twenty-six RNAi lines successfully silencing Spin1 and 13 lines overexpressing the gene with an N-terminal TAP (for tandem affinity purification) tag (Spin1-OX) were generated. As an example of successful silencing and overexpression of the transgenes, RT-PCR analysis indicating reduction or increase of Spin1 expression in some of the transgenic lines is shown in Supplemental Figure 3 online.
Neither Spin1-RNAi nor Spin1-OX lines showed signs of lesion mimic or cell death–related phenotypes. Moreover, no enhanced resistance or susceptibility was observed when both Spin1-silenced and overexpressed lines were inoculated with virulent strains of M. oryzae and X. oryzae pv oryzae compared with nontransformed NPB (see Supplemental Figure 4 online). These results suggested that SPIN1 might not be involved in the cell death and disease resistance signaling that was reported to be associated with Spl11. Nevertheless, examination of Spin1-OX lines revealed that they showed delayed flowering time compared with the RNAi and NPB plants (Figure 8). Under both SD and LD conditions, no significant difference in the appearance of the first panicle was observed between Spin1-RNAi lines and NPB or segregant transgenic lines not carrying the transgene (Figure 8B). However, flowering time was significantly delayed in Spin1-OX lines under the same conditions (Figure 8B). The delayed flowering phenotype correlated with the overexpression of the TAP-SPIN1 protein in T4 transgenic lines (Figure 8A) and was due to the overexpression of the Spin1 gene, since transgenic lines overexpressing the TAP tag alone in the Kitaake background had no differences in flowering time compared with nontransformed plants (see Supplemental Figure 5 online).
Figure 8.
Overexpression of Spin1 Causes Delayed Flowering under Both SD and LD Conditions.
(A) Protein gel blot detection of the accumulation of TAP-SPIN1 protein in a T4 line overexpressing Spin1-TAP. Line 32-3-5 is segregating for flowering time in SD.
(B) Days to heading under SD and LD in T4 RNAi and overexpressing Spin1 (Spin1-TAP) lines and on the untransformed recipient cultivar NPB. + and – indicate siblings from the same T3 family that do and do not contain the RNAi or overexpressing construct. The asterisk indicates a plant that did not flower after 230 d.
(C) RT-PCR/DNA gel blot analysis of Hd1, Hd3a, SOC1, and Ubq in Spin1-TAP line 11-15 and NPB under SD and LD over a 24-h period in 55-d-old plants.
(D) to (G) Quantification of gene expression relative to Ubq of the Hd1 and Hd3a blots shown in (C). White and black rectangles represent light and dark conditions over a 24-h period.
Error bars indicate sd from three independent experiments.
The expression of Hd1, Hd3a, and SOC1 was monitored over a 24-h period in both Spin1-OX and NPB plants under SD and LD conditions. Higher expression of Hd1 at midmorning and early and late night correlated with significant reduction of Hd3a expression in Spin1-OX in SD when compared with NPB (Figures 8C, 8D, and 8F) suggesting that Spin1 represses Hd3a expression via Hd1. By contrast, the expression levels of SOC1 did not vary significantly between Spin1-OX and NPB under SD (Figure 8C). Under LD, Hd1 transcript levels either did not vary significantly between NPB and Spin1-OX or the variations did not correlate with the delayed flowering phenotype (Figures 8C and 8E). By contrast, significant differences in Hd3a expression between NPB and Spin1-OX at midmorning and, especially, at midnight correlated with the delayed flowering in Spin1-OX plants (Figures 8C and 8G). The enhanced downregulation of Hd3a transcript levels at midnight correlated with significant reduction of SOC1 expression at this time point in Spin1-OX lines compared with NPB, but not at the other time points (Figure 8C). These results indicated that Spin1 represses flowering independently of daylength via a mechanism that primarily targets the flowering promoter Hd3a, especially in SD.
DISCUSSION
We have shown that SPL11 interacts with and ubiquitinates SPIN1, a nuclear RNA/DNA binding protein of the STAR family. In addition, we have shown that Spl11 negatively regulates Spin1 expression during light periods in both SD and LD. The spl11 mutant has a delayed flowering phenotype in LD, while overexpression of Spin1 causes delayed flowering in both SD and LD. Our data are in agreement with SPL11 acting as a positive regulator of flowering via negative regulation of the SPIN1 flowering repressor. Our results reveal that transcriptional regulation of a gene encoding an RNA/DNA binding protein, possibly via monoubiquitination, is the mechanism of flowering time control by this E3 ligase in rice.
Putative paralogs and orthologs of SPIN1 were identified in rice and other plant species, respectively. The high level of conservation at the amino acid level between monocot and dicot SPIN1-like sequences suggests that their functions might be conserved across distantly related species. In total, seven paralogs including SPIN1 were found in rice. It is likely that these genes have redundant functions in rice, which might explain our observation that silencing of Spin1 did not significantly affect flowering time. Functional redundancy was confirmed for the SPA family and the AGL15 and AGL18 floral repressors in Arabidopsis, in which double or triple, but not single, loss-of-function mutations cause early flowering (Laubinger et al., 2006; Adamczyk et al., 2007). We have checked the expression of the closest rice paralog of Spin1, Os 07g0227400 (encoding Os NP_001059216; Figure 2B), in our transgenic plants and found a slight to nonsignificant decrease in its transcript levels depending on the RNAi line tested (see Supplemental Figure 6 online). Os 07g0227400's coding sequence shows 79% identity with Spin1 at the nucleotide level and 90% similarity in the amino acid sequence (Figure 2B). Therefore, the lack of phenotype in our RNAi lines is most likely due to genetic redundancy. Silencing Spin1 and one or a few paralogous members simultaneously will be central to testing such functional redundancy.
Spl11 was initially characterized as a gene that negatively regulates PCD and broad-spectrum disease resistance (Zeng et al., 2004). Our results in this study add a role for SPL11 in the control of flowering time. In plants, examples have been reported of an E3 ligase that plays different roles depending on the biological processes. For example, the F-box protein ORE9 regulates both senescence and shoot branching in Arabidopsis (Woo et al., 2001; Stirnberg et al., 2007). The Spin1-RNAi transgenic plants did not show disease resistance or cell death–related phenotypes, suggesting that SPIN1 might not be involved in these pathways. Due to possible functional redundancy in the Spin1 gene family (see above), we cannot rule out a role of SPIN1 in PCD and pathogen resistance. However, Spin1-OX lines were not affected in either defense or cell death. Moreover, Spin1 is upregulated in leaves at booting stage and in panicles, which is in agreement with its involvement in flowering control. We have identified other interactors of SPL11 in the yeast two-hybrid screen (see Supplemental Table 1 online). Whether other SPL11-interacting partners play a role in PCD and defense is under investigation.
Our results showed that SPIN1 is ubiquitinated in vitro by SPL11 (Figure 6). Polyubiquitination through Lys-48 ubiquitin residues usually serves as a signal for degradation by the 26S proteasome (Thrower et al., 2000). However, polyubiquitination through Lys residues other than Lys-48 as well as monoubiquitination have been implicated in nonproteolytic cellular processes (Terrell et al., 1998; Schnell and Hicke, 2003; Sigismund et al., 2004; Mukhopadhyay and Riezman, 2007). Our initial attempts to test whether SPL11 regulates SPIN1 protein stability using a cycloheximide-chase experiment indicated that SPIN1 is not degraded when SPL11 is overexpressed in rice protoplast. However, the experiment was inconclusive due to our failure to detect the ubiquitinated form of SPIN1 in protoplasts. Nevertheless, it is likely that SPIN1 is not degraded by SPL11 in vivo, based on the results indicating that SPIN1 is monoubiquitinated by SPL11 and by a time-course analysis of the TAP-SPIN1 protein accumulation under SD and LD over a 24-h period (see Supplemental Figure 7 online): no significant variations in protein levels were observed throughout the day, which would not be expected if SPIN1 was a substrate for proteolysis. Although the TAP-SPIN1 protein does not represent the endogenous levels of SPIN1 accumulation, in the case of the flowering repressor CDF1 in Arabidopsis, which is targeted for degradation by FKF1-mediated ubiquitination, the levels of CDF1 protein in plants overexpressing an HA-CDF1 construct were significantly reduced in the dark in a proteasome-dependent manner (Imaizumi et al., 2005). Several examples of U-box–type E3 ligases targeting their substrates for nonproteolytical ubiquitination have been reported. In animals, the U-box cochaperone CHIP ubiquitinates, but does not degrade, Hsc70 (Jiang et al., 2001). Similarly, the Arabidopsis homolog of CHIP monoubiquitinates the A subunit of phosphatase 2A (PP2A); since overexpression of At CHIP increased the activity of PP2A, monoubiquitination of PP2A does not promote its degradation (Luo et al., 2006).
Overexpression of Spin1 under both SD and LD delays flowering (Figure 8B). However, the spl11 mutant shows no significant difference in flowering time under SD, but it flowers late in LD (Figure 1). These results suggest that Spin1 acts as a flowering repressor and Spl11 acts as a positive regulator of flowering time in rice. Consistent with this notion, the expression levels of the flowering promoter gene Hd3a are significantly reduced in plants overexpressing Spin1, especially in SD, and in the spl11 mutant in LD. In Arabidopsis, the Hd3a ortholog FT activates some of the floral meristem identity genes involved in flowering transition at the shoot apical meristem (Simpson and Dean, 2002). Recently, both FT and Hd3a proteins have been shown to move from the leaf to the shoot apex, where floral organ identity genes are activated (Corbesier et al., 2007; Tamaki et al., 2007). Hence, it is not surprising that many floral repressor genes function by targeting, either directly or indirectly, FT/Hd3a accumulation. The MADS domain proteins AGL15 and AGL18 directly downregulate FT expression (Adamczyk et al., 2007). Many genes indirectly target FT by negatively regulating CO at transcript or protein levels. For example, CDF1 binds to the CO promoter to repress its expression (Imaizumi et al., 2005), while SPA1 interacts with and negatively controls CO protein levels (Laubinger et al., 2006). COP1 was recently shown to negatively regulate flowering by ubiquitinating CO and promoting its degradation in the dark (Liu et al., 2008). It is unclear whether similar negative regulation is conserved in rice. Knowledge of the regulation of Hd1 is limited compared with its Arabidopsis ortholog CO. However, it has been shown that under LD conditions, Hd1 acts as a repressor of Hd3a expression (Izawa et al., 2002; Hayama et al., 2003; Hayama and Coupland, 2004), which is in contrast with the function of CO in Arabidopsis.
Based on our results, it appears that Spin1 negatively regulates flowering via Hd1-dependent and -independent mechanisms in SD and LD, respectively. In flowering-promoting SD conditions, downregulation of Hd1 at midmorning and early and late evening correlates with decreased Hd3a levels in the Spin1-OX line compared with wild-type NPB plants (Figures 8C, 8D, and 8F). By contrast, under flowering-restrictive LD conditions, Hd1 mRNA levels do not vary significantly between NPB and Spin1-OX. Hd3a levels are very low in both Spin1-OX and NPB in LD, except at time point 16 h (around midnight), when Hd3a transcript accumulation is significantly higher in NPB (Figures 8C and 8G). It is unlikely, however, that the decrease in Hd3a mRNA level at midnight is solely responsible for the delayed flowering phenotype of Spin1-OX plants in LD. Similar to what we observed in Spin1-OX plants, Hd3a expression was strongly repressed in LD conditions in rice plants overexpressing Os GI (Hayama et al., 2003). Moreover, the Os MADS51 mutant shows no significant difference in flowering time compared with the wild type in LD, although Hd3a expression is greatly reduced in the mutant (Kim et al., 2007). Therefore, additional factors regulated by SPIN1 in LD might also contribute to the delay in flowering compared with the wild type.
Spl11 and Spin1 have opposite expression patterns in immature panicles but are both upregulated in leaves at booting stage (Figure 5B), and their mRNA abundances are diurnally regulated in SD (Figure 5C). These results support the notion that both of these genes are related to the plant flowering process. We know that Spl11 negatively regulates the accumulation of Spin1 mRNA in the light and that in the absence of Spl11, Spin1 transcript accumulates with an opposite phase compared with the wild type (Figures 5C and 5D). The only times that this change in phase correlates with changes in flowering time is when Spin1 is upregulated in the morning and during extended light hours, corresponding to early evening in LD in the spl11 mutant (Figure 5D, middle). Thus, high expression of Spin1 in the morning and early evening in the light period is required to repress flowering. This explains why there is no difference in flowering time between spl11 and the wild type in SD, since high Spin1 expression does not coincide with the light at these time points (Figure 5C, left and middle). However, high Spin1 expression during LD in the wild type does not coincide with the light either (Figure 5D, left), and yet flowering is delayed compared with SD. Delayed flowering in wild-type plants under LD can thus be attributed to other flowering-repressing mechanisms, most likely via Hd1, a strong floral repressor in LD (Izawa et al., 2002; Hayama et al., 2003; Hayama and Coupland, 2004). This is in agreement with the observation that in the spl11 mutant, flowering is further delayed in LD compared with the wild type, which can be explained by the additive contributions of Hd1 and SPIN1. In Spin1-OX lines, high accumulation of Spin1 throughout the day, regardless of light and dark phases, causes delayed flowering in both SD and LD (Figure 8B). This is reminiscent of Hd1 regulation in SD versus LD, in which high Hd1 accumulation in the dark promotes flowering in SD, while the coincidence of high Hd1 levels occurring in the light inhibits flowering in LD (Hayama and Coupland, 2004). However, a major difference is that the phase of Hd1 accumulation is not altered by changes in daylength, as is the case for Spin1.
Interestingly, overexpression of Spin1 affects Spl11 diurnal regulation. As shown in Supplemental Figure 8 online, Spl11 shows diurnal regulation in LD but not in SD in Spin1-OX, which is the opposite of what we observed in wild-type IR64 plants (Figures 5C and 5D, right). These results suggest that Spin1 regulates Spl11 expression. In LD, loss of Spl11 diurnal regulation may be a mechanism to keep SPL11 levels high enough to inhibit Spin1 expression in the light to lead to a delay in flowering time. Thus, the restoration of Spl11 diurnal expression pattern in LD when Spin1 is overexpressed would serve to decrease SPL11 levels during the light phase to inhibit flowering. However, the role, if any, of the loss of Spl11 diurnal pattern in SD when Spin1 is overexpressed is less clear. The fact that Spl11 loses diurnal regulation in LD in the wild type while Spin1 retains its diurnal regulation regardless of photoperiod argues that clock-related components act differentially in the control of oscillation patterns for the two genes. Supporting the notion of circadian control of flowering regulators, it has been widely documented that loss-of-function mutants in clock feedback loop components cause changes in CO protein and mRNA levels that result in early or late flowering depending on the mutant (Simpson and Dean, 2002; Putterill et al., 2004; Izawa, 2007). Taken together, we propose the following model of flowering time control (Figure 9). Under SD, both Spin1 and Spl11 are diurnally regulated with opposing phases; negative regulation of Hd1 and Hd3a by SPIN1 is inhibited by SPL11 on Spin1 mRNA, protein, or both, and flowering is promoted. In the absence of Spl11, the phase of Spin1 expression changes, but since high expression levels do not coincide with the light during morning and early evening, flowering is not affected. By contrast, in Spin1-OX lines, high accumulation of SPIN1 throughout the day is enough to repress flowering and to cause a loss of diurnal Spl11 expression by an unknown mechanism. Under LD, SPIN1 and Hd1 may act additively to repress Hd3a and possibly other components. Loss of Spl11 diurnal regulation might be required for the negative control of Spin1 transcript and/or protein levels by keeping high levels of SPL11 throughout the day to alleviate flowering repression; in the absence of Spl11, high levels of Spin1 coincide with the light in the morning and early evening, which triggers increased inhibition of flowering time. Similarly, overexpression of Spin1 maintains high SPIN1 levels throughout the day, which both represses Hd3a and other factors and restores the diurnal regulation of Spl11, which ultimately decreases SPL11 overall levels during the light and delays flowering.
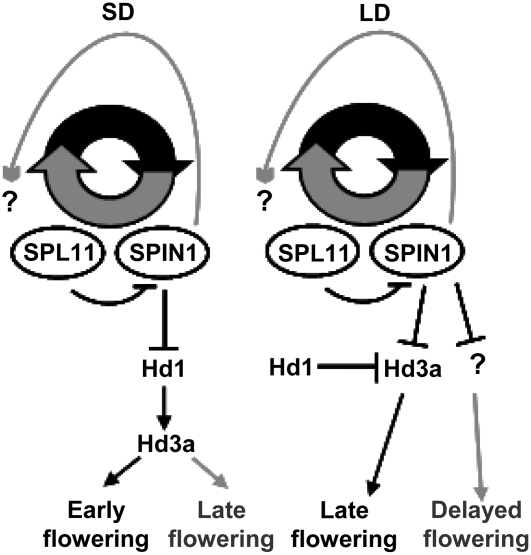
Figure 9.
Proposed Model for the Involvement of SPIN1 and SPL11 in Flowering Time Control under SD and LD.
In SD, SPL11 negatively regulates Spin1 expression to allow early flowering. Overexpression of Spin1 affects Spl11 expression by unknown mechanisms and causes repression of Hd3a via Hd1, which ultimately leads to delayed flowering. In LD, Hd1 represses Hd3a, causing late flowering; however, since Spl11 accumulates at high levels throughout the day, SPIN1 is repressed, which attenuates the late-flowering effect. Overexpression of Spin1 affects Spl11 expression by unknown mechanisms and causes an increase in late flowering by targeting unknown factors and Hd3a. Black lines indicate regulation in the wild type, and gray lines indicate regulation in Spin1-OX lines. Arrows represent positive regulation, and blunt ends represent negative regulation. Question marks indicate unknown additional components.
Our results indicate that SPL11 negatively regulates Spin1 expression and that SPIN1 affects the Spl11 expression pattern as well. The mechanisms underlying the transcriptional regulation of Spin1 by SPL11 and vice versa are unknown at this point. As a nuclear RNA binding protein, SPIN1 might bind directly to the Spl11 pre-mRNA or to the pre-mRNA of putative Spl11 circadian clock regulators. In addition, SPIN1 most likely represses flowering by binding to other targets in the cell. We speculate that it is at this level that the SPL11-mediated ubiquitination of SPIN1 affects flowering time by negatively regulating these interactions. Negative regulation by non-proteasome-mediated ubiquitination was recently shown in the case of endocytic proteins such as Sts2, in which monoubiquitination causes intramolecular interactions that impair Sts2 binding to other proteins, abolishing its regulatory function in receptor trafficking (Hoeller et al., 2006). Similarly, monoubiquitination of SPIN1 might affect its RNA binding activity or its interaction with other proteins, which are questions to be addressed in future studies.
METHODS
Plant Material
Seeds of wild-type, mutant, and transgenic rice (Oryza sativa) were first sterilized by treatment in 75% ethanol for 1 min followed by immersion in 2% sodium hypochlorite for 15 to 20 min. After thoroughly washing with sterile distilled water, seeds were germinated in half-strength Murashige and Skoog medium for 7 to 10 d and then transferred to soil. Growth chamber conditions were 10 h of light, 26°C, 80% RH, followed by 14 h of dark, 20°C, 60% RH, unless otherwise specified.
Flowering Time Measurements
Plants were grown as described above with the following modifications: for SD, plants were grown either in 12 h of light/12 h of dark or 10 h of light/14 h of dark; for LD, plants were grown in 16 h of light/8 h of dark for 120 d, then 14 h of light/10 h of dark for the rest of the experiment. Heading date or flowering time was measured in days from sowing until emergence of the first panicle.
Plasmids
In general, constructs were generated by PCR amplification of the target gene using primers containing appropriate restriction enzyme sites and Spin1 or Spl11 cDNA template, followed by ligation into a desired vector. For a list of primers and constructs, see Supplemental Table 2 online. For the BiFC method Spl11, Spin1, and GUS constructs in pA7-NYFP and pA7-CYFP vectors are described elsewhere (Chen et al., 2006). Overexpression and RNAi constructs were made into the Gateway (Invitrogen)-compatible vectors Ubix.nc1300.ntap.gck (Chern et al., 2005) and pANDA (Miki and Shimamoto, 2004), respectively, following Gateway cloning protocols.
Yeast Two-Hybrid Screen
The ProQuest system (Invitrogen) was used to screen for SPL11-interacting proteins following the manufacturer's protocol. Briefly, a construct that contains the SPL11 ARM repeat domain (amino acids 379 to 653) was cloned by PCR into the NcoI and NheI sites of Proquest's pDBleu vector (see Supplemental Table 2 online); a rice cDNA library was built into the expression vector pPC86 of the ProQuest system using the restriction enzymes NotI and SalI using mRNA isolated from 3-week-old seedlings of rice line 75-1-127. Total RNA was isolated using Trizol reagent (Invitrogen), and mRNA was then purified using the Absolutely mRNA purification kit (Stratagene) according to the protocol provided by the manufacturer.
GST Pull Down
GST pull down was performed as described (Liu et al., 2002) with the following modifications: a Spin1 construct in pET28-a (Novagen) driven by a T7 promoter was used for in vitro transcription/translation using the TNT rabbit reticulocyte lysate translation system (Promega) in the presence of biotinylated Lys to label the protein (Transcend nonradioactive translation detection system; Promega). Detection of bound SPIN1 was performed by streptavidin–horseradish peroxidase chemiluminescent detection using the Transcend nonradioactive detection system (Promega).
BiFC and Fluorescence Microscopy Analyses
Fluorescence microscopy analyses were done by transfection of rice protoplasts with various constructs as described (Chen et al., 2006).
Gene Expression Analyses
RNA was extracted with Trizol reagent (Invitrogen) and treated with DNase I before being used for reverse transcription using the Promega reverse transcription system. Two to 3 μL of first-strand cDNA was used for the PCR. Details of the collection of tissue for RNA samples of rice developmental stages are as described (Nobuta et al., 2007). The exponential range of amplification was determined for each gene in a pilot experiment using incremental PCR cycles (15 to 30 cycles); 24 cycles were optimal for Spl11 and Ubiquitin (Ubq), 25 for Spin1, Hd1, and SOC1, and 26 for Hd3a Spin1. RT-PCR cycling conditions were denaturing at 94°C for 1 min, annealing at 55°C for 1 min, extension at 72°C for 1 min, for 25 cycles. Ubiquitin was used as an internal control, with denaturing at 94°C for 30 s, annealing at 50°C for 30 s, extension at 72°C, for 24 cycles. Spl11 cycling conditions were as described (Chen et al., 2006). For flowering marker genes, primers were as described (Hayama et al., 2003; Tadege et al., 2003). For quantification of gene expression, band intensity from SYBR Gold–stained agarose gels was determined using ImageJ software. Gene expression values at each time point were divided by their corresponding ubiquitin value to correct for loading, and the highest number was given the arbitrary quantity of 100. The rest of the values were normalized relative to 100 to correct for variations in band intensity between replicate gels. For flowering marker gene expression, RT-PCR followed by DNA gel blotting was done as described (Yanovsky and Kay, 2002) . Briefly, RT-PCR was done as described above, and 8 μL of the reaction was loaded on an agarose gel. DNA was blotted onto a nylon membrane and hybridized using gene-specific probes labeled with [32P]dCTP. Blot images were scanned with a PhosphorImager (Molecular Dynamics), and band intensities were quantified using the ImageQuant TL image analysis software (GE Healthcare). Gene expression analysis using the data from the PhosphorImager was done following the same protocol described above. RNA was extracted from 55-d-old plants every 4 h starting at time 0 (7 am) and ending at time 20 (3 am the next day). The values of three independent experiments were averaged and plotted for all quantitative RT-PCR assays performed.
In Vitro Ubiquitination Assays
The ubiquitination assay was done as described (Zeng et al., 2004) with the following modifications: in a 30-μL final volume, 100 ng of E1, 100 ng of E2, 200 ng of GST-SPIN1-HA, 600 ng of GST-SPL11 or GST-OsSINAT5, 12 μg of ubiquitin (Boston Biochem), and 3 μL of 10× ubiquitination buffer (500 mM Tris-HCl, pH 7.5, 50 mM ATP, 50 mM MgCl2, 20 mM DTT, 30 mM creatine phosphate, and 0.05 mg/mL creatine phosphokinase) were mixed and incubated at 30°C for 1.5 h. Samples were denatured and loaded on a 10% SDS-polyacrylamide gel, and ubiquitination was detected by protein gel blot analysis with anti-HA (Roche) antibodies. At UBC9 was used in the reaction using GST-SPL11 and At UBC7 with GST-OsSINAT5.
RNA/DNA Binding Assay
Beads containing polyadenylic, polycytidylic, polyuridylic, and polyguanylic ribohomopolymers as well as calf thymus single- and double-stranded DNA were purchased from Sigma-Aldrich. Approximately 30 ng of recombinant GST-SPIN1-HA was incubated with the beads in 500 μL of KHN buffer (150 mM KCl, 20 mM HEPES, pH 7.9, 0.01% Nonidet P-40, and complete protease inhibitors) under rotation for 10 min. Beads were washed in KHN buffer three times, and proteins retained in the beads were identified by protein gel blot analysis using either anti-HA (Roche) or anti-GST (Invitrogen) antibodies.
Protein Extraction and Protein Gel Blot Analysis
Protein extraction from transgenic Spin1-TAP plants was done by grinding leaves in liquid nitrogen and homogenizing the ground tissue in buffer 2 (8 M urea, 4% CHAPS, 40 mM Tris, and 2 mM TBP) from the ReadyPrep sequential extraction kit from Bio-Rad following the kit's protocol. For protein gel blots, membranes were blocked for 1 h in 5% milk in 1× Tris-buffered saline Tween 20 (TBST), incubated for 1 h in 2% milk in 1× TBST containing the appropriate primary antibody, and finally incubated for 1 h in 2% milk in 1× TBST with the conjugated secondary antibody. Proteins on blots were detected by chemiluminescence using the Pierce ECL protein gel blotting substrate or the Amersham ECL Plus protein gel blotting detection reagents (GE Healthcare), following each manufacturer's instructions. TAP-tagged proteins were detected with a peroxidase anti-peroxidase (PAP) antibody (Sigma-Aldrich).
Phylogenetic Analysis
The dendogram was constructed using MEGA3.1 software (Kumar et al., 2004) using the neighbor-joining algorithm. Boostrapping was performed with 1000 replicates. Arabidopsis thaliana FLK, a KH domain–containing protein unrelated to the SPIN1-like sequences, was used as an outgroup. Multiple sequence alignment was done using ClustalX, and conserved residue shading was performed using GeneDoc.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers NP_001059216, NP_001055861, NP_001044630, NP_001045029, NP_001055572, and EAY87341 for rice sequences; At2g38610 and At3g08620 for Arabidopsis; BAD06470 (Nicotiana tabacum); ABE77708 (Medicago truncatula); NP_004621 (Homo sapiens); NP_492143 (Caenorhabditis elegans); and NP_524447 (Drosophila melanogaster).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of the STAR Domain of SPIN1 with Mammalian Protein Members of the Family.
Supplemental Figure 2. Alignment of Plant SPIN1-Like Proteins.
Supplemental Figure 3. RT-PCR Analysis of Spin1 Expression in T2 Transgenic RNAi and TAP (Overexpression) Lines.
Supplemental Figure 4. Rice Blast and Bacterial Blight Inoculation Response of Spin1-RNAi and Spin1-OX Lines.
Supplemental Figure 5. Days to Heading in N-TAP Transgenic Lines Transformed with the Empty TAP Tag Vector Used for the Generation of Spin1-TAP Lines.
Supplemental Figure 6. Expression Analysis of Spin1 and Its Closest Paralog Os 07g0227400 by RT-PCR in T5 Spin1-RNAi and Spin1-TAP Lines.
Supplemental Figure 7. Accumulation of TAP-SPIN1 Protein in SD and LD over a 24-h Period.
Supplemental Figure 8. RT-PCR Analysis of Spl11 Expression in Spin1-TAP (Overexpression) Lines 11 to 15 in SD and LD over a 24-h Period.
Supplemental Table 1. Summary of the Yeast Two-Hybrid Screen Using SPL11-ARM as Bait.
Supplemental Table 2. List of Primers and Constructs Used in This Study.
Supplemental Data Set 1. Text File Corresponding to the Phylogenetic Tree in Figure 2B and the Alignment in Supplemental Figure 2 Online.
Supplementary Material
Acknowledgments
We thank Ko Shimamoto and Michael Goodin for providing the pANDA plasmid and pGD vector series, respectively; Pamela Ronald for providing the Ubix.nc1300.ntap.gck vector and seeds of N-TAP lines in the Kitaake background; Biao Ding for sharing the PRH75-RFP construct and for his assistance with fluorescence microscopy; R.C. Venu for sharing the RNA samples from his Massively Parallel Signature Sequencing libraries; and Maria Bellizzi for excellent technical assistance. This work was supported by grants from the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (Grant 2007-01667 to G.-L.W.), the USAID-International Rice Research Institute Linkage Program (to G.-L.W. and H.L), and the Ohio Agricultural Research and Development Center's Graduate Research Enhancement Program (to L.Z. and M.E.V.-S.). M.E.V.-S. is supported by an Excellence in Plant Molecular Biology/Biotechnology Fellowship.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Guo-Liang Wang (wang.620@osu.edu).
Online version contains Web-only data.
References
- Adamczyk, B.J., Lehti-Shiu, M.D., and Fernandez, D.E. (2007). The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 50 1007–1019. [DOI] [PubMed] [Google Scholar]
- Adinolfi, S., Bagni, C., Castiglione Morelli, M.A., Fraternali, F., Musco, G., and Pastore, A. (1999). Novel RNA-binding motif: The KH module. Biopolymers 51 153–164. [DOI] [PubMed] [Google Scholar]
- Baurle, I., Smith, L., Baulcombe, D.C., and Dean, C. (2007). Widespread role for the flowering-time regulators FCA and FPA in RNA-mediated chromatin silencing. Science 318 109–112. [DOI] [PubMed] [Google Scholar]
- Berglund, J.A., Chua, K., Abovich, N., Reed, R., and Rosbash, M. (1997). The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell 89 781–787. [DOI] [PubMed] [Google Scholar]
- Boss, P.K., Bastow, R.M., Mylne, J.S., and Dean, C. (2004). Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 16 (suppl.): S18–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40 419–427. [DOI] [PubMed] [Google Scholar]
- Chen, S., Tao, L., Zeng, L., Vega-Sanchez, M.E., Umemura, K., and Wang, G.L. (2006). A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol. Plant Pathol. 7 417–427. [DOI] [PubMed] [Google Scholar]
- Chern, M., Canlas, P.E., Fitzgerald, H.A., and Ronald, P.C. (2005). Rice NRR, a negative regulator of disease resistance, interacts with Arabidopsis NPR1 and rice NH1. Plant J. 43 623–635. [DOI] [PubMed] [Google Scholar]
- Ciechanover, A. (1998). The ubiquitin-proteasome pathway: On protein death and cell life. EMBO J. 17 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., Giakountis, A., Farrona, S., Gissot, L., Turnbull, C., and Coupland, G. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030–1033. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., and Coupland, G. (2004). The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 135 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama, R., Izawa, T., and Shimamoto, K. (2002). Isolation of rice genes possibly involved in the photoperiodic control of flowering by a fluorescent differential display method. Plant Cell Physiol. 43 494–504. [DOI] [PubMed] [Google Scholar]
- Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422 719–722. [DOI] [PubMed] [Google Scholar]
- Henderson, I.R., and Dean, C. (2004). Control of Arabidopsis flowering: The chill before the bloom. Development 131 3829–3838. [DOI] [PubMed] [Google Scholar]
- Hoeller, D., Crosetto, N., Blagoev, B., Raiborg, C., Tikkanen, R., Wagner, S., Kowanetz, K., Breitling, R., Mann, M., Stenmark, H., and Dikic, I. (2006). Regulation of ubiquitin-binding proteins by monoubiquitination. Nat. Cell Biol. 8 163–169. [DOI] [PubMed] [Google Scholar]
- Imaizumi, T., Schultz, T.F., Harmon, F.G., Ho, L.A., and Kay, S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309 293–297. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Matsuoka, M., and Steber, C.M. (2003). A role for the ubiquitin-26S-proteasome pathway in gibberellin signaling. Trends Plant Sci. 8 492–497. [DOI] [PubMed] [Google Scholar]
- Izawa, T. (2007). Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 58 3091–3097. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Sugiyama, N., Tanisaka, T., Yano, M., and Shimamoto, K. (2002). Phytochrome mediates the external light signal to repress FT orthologs in photoperiodic flowering of rice. Genes Dev. 16 2006–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Tokutomi, S., Okuno, K., and Shimamoto, K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22 391–399. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Takahashi, Y., and Yano, M. (2003). Comparative biology comes into bloom: Genomic and genetic comparison of flowering pathways in rice and Arabidopsis. Curr. Opin. Plant Biol. 6 113–120. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Ballinger, C.A., Wu, Y., Dai, Q., Cyr, D.M., Hohfeld, J., and Patterson, C. (2001). CHIP is a U-box-dependent E3 ubiquitin ligase: Identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276 42938–42944. [DOI] [PubMed] [Google Scholar]
- Kim, S.L., Lee, S., Kim, H.J., Nam, H.G., and An, G. (2007). OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145 1484–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Araki, T., and Yano, M. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43 1096–1105. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- Laubinger, S., Marchal, V., Le Gourrierec, J., Wenkel, S., Adrian, J., Jang, S., Kulajta, C., Braun, H., Coupland, G., and Hoecker, U. (2006). Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133 3213–3222. [DOI] [PubMed] [Google Scholar]
- Lee, H., Suh, S.S., Park, E., Cho, E., Ahn, J.H., Kim, S.G., Lee, J.S., Kwon, Y.M., and Lee, I. (2000). The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev. 14 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, M.H., Kim, J., Kim, Y.S., Chung, K.S., Seo, Y.H., Lee, I., Kim, J., Hong, C.B., Kim, H.J., and Park, C.M. (2004). A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F., Quesada, V., Crevillen, P., Baurle, I., Swiezewski, S., and Dean, C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28 398–407. [DOI] [PubMed] [Google Scholar]
- Liu, L.J., Zhang, Y.C., Li, Q.H., Sang, Y., Mao, J., Lian, H.L., Wang, L., and Yang, H.Q. (2008). COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Serino, G., Deng, X.W., and Dinesh-Kumar, S.P. (2002). Role of SCF ubiquitin ligase and the COP9 signalosome in the N gene-mediated resistance response to tobacco mosaic virus. Plant Cell 14 1483–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic, Z.J., Herrmann, R.G., and Oelmuller, R. (1997). PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants. Mol. Cell. Biol. 17 2257–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas, J.I., Arnau, V., and Marin, I. (2006). Comparative genomics and protein domain graph analyses link ubiquitination and RNA metabolism. J. Mol. Biol. 357 9–17. [DOI] [PubMed] [Google Scholar]
- Luo, J., Shen, G., Yan, J., He, C., and Zhang, H. (2006). AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 46 649–657. [DOI] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89 737–745. [DOI] [PubMed] [Google Scholar]
- Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45 490–495. [DOI] [PubMed] [Google Scholar]
- Mockler, T.C., Yu, X., Shalitin, D., Parikh, D., Michael, T.P., Liou, J., Huang, J., Smith, Z., Alonso, J.M., Ecker, J.R., Chory, J., and Lin, C. (2004). Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc. Natl. Acad. Sci. USA 101 12759–12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J., Parry, G., and Estelle, M. (2004). The ubiquitin-proteasome pathway and plant development. Plant Cell 16 3181–3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay, D., and Riezman, H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315 201–205. [DOI] [PubMed] [Google Scholar]
- Nalivaeva, N.N., and Turner, A.J. (2001). Post-translational modifications of proteins: Acetylcholinesterase as a model system. Proteomics 1 735–747. [DOI] [PubMed] [Google Scholar]
- Nobuta, K., Venu, R.C., Lu, C., Belo, A., Vemaraju, K., Kulkarni, K., Wang, W., Pillay, M., Green, P.J., Wang, G.L., and Meyers, B.C. (2007). An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 25 473–477. [DOI] [PubMed] [Google Scholar]
- Onouchi, H., Igeno, M.I., Perilleux, C., Graves, K., and Coupland, G. (2000). Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12 885–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill, J., Laurie, R., and Macknight, R. (2004). It's time to flower: The genetic control of flowering time. Bioessays 26 363–373. [DOI] [PubMed] [Google Scholar]
- Ryder, S.P., Frater, L.A., Abramovitz, D.L., Goodwin, E.B., and Williamson, J.R. (2004). RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 11 20–28. [DOI] [PubMed] [Google Scholar]
- Samach, A., Onouchi, H., Gold, S.E., Ditta, G.S., Schwarz-Sommer, Z., Yanofsky, M.F., and Coupland, G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288 1613–1616. [DOI] [PubMed] [Google Scholar]
- Schnell, J.D., and Hicke, L. (2003). Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278 35857–35860. [DOI] [PubMed] [Google Scholar]
- Schomburg, F.M., Patton, D.A., Meinke, D.W., and Amasino, R.M. (2001). FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell 13 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund, S., Polo, S., and Di Fiore, P.P. (2004). Signaling through monoubiquitination. Curr. Top. Microbiol. Immunol. 286 149–185. [DOI] [PubMed] [Google Scholar]
- Simpson, G.G., and Dean, C. (2002). Arabidopsis, the Rosetta stone of flowering time? Science 296 285–289. [DOI] [PubMed] [Google Scholar]
- Stirnberg, P., Furner, I.J., and Ottoline Leyser, H.M. (2007). MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50 80–94. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez, P., Wheatley, K., Robson, F., Onouchi, H., Valverde, F., and Coupland, G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410 1116–1120. [DOI] [PubMed] [Google Scholar]
- Tadege, M., Sheldon, C.C., Helliwell, C.A., Upadhyaya, N.M., Dennis, E.S., and Peacock, W.J. (2003). Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 1 361–369. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., Shomura, A., Sasaki, T., and Yano, M. (1998). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. USA 98 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki, S., Matsuo, S., Wong, H.L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316 1033–1036. [DOI] [PubMed] [Google Scholar]
- Terrell, J., Shih, S., Dunn, R., and Hicke, L. (1998). A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol. Cell 1 193–202. [DOI] [PubMed] [Google Scholar]
- Thrower, J.S., Hoffman, L., Rechsteiner, M., and Pickart, C.M. (2000). Recognition of the polyubiquitin proteolytic signal. EMBO J. 19 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde, F., Mouradov, A., Soppe, W., Ravenscroft, D., Samach, A., and Coupland, G. (2004). Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303 1003–1006. [DOI] [PubMed] [Google Scholar]
- Vernet, C., and Artzt, K. (1997). STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13 479–484. [DOI] [PubMed] [Google Scholar]
- Woo, H.R., Chung, K.M., Park, J.H., Oh, S.A., Ahn, T., Hong, S.H., Jang, S.K., and Nam, H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170. [DOI] [PubMed] [Google Scholar]
- Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., Baba, T., Yamamoto, K., Umehara, Y., Nagamura, Y., and Sasaki, T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12 2473–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky, M.J., and Kay, S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419 308–312. [DOI] [PubMed] [Google Scholar]
- Yin, Z., Chen, J., Zeng, L., Goh, M., Leung, H., Khush, G.S., and Wang, G.L. (2000). Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 13 869–876. [DOI] [PubMed] [Google Scholar]
- Zeng, L.R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., Xie, Q., Nahm, B.H., Leung, H., and Wang, G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16 2795–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L.R., Vega-Sanchez, M.E., Zhu, T., and Wang, G.L. (2006). Ubiquitination-mediated protein degradation and modification: An emerging theme in plant-microbe interactions. Cell Res. 16 413–426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.