Abstract
Following meiosis, plant gametophytes develop through two or three rounds of mitosis. Although the ontogeny of gametophyte development has been defined in Arabidopsis thaliana, the molecular mechanisms regulating mitotic cell cycle progression are not well understood. Here, we report that RING-H2 group F 1a (RHF1a) and RHF2a, two RING-finger E3 ligases, play an important role in Arabidopsis gametogenesis. The rhf1a rhf2a double mutants are defective in the formation of male and female gametophytes due to interphase arrest of the mitotic cell cycle at the microspore stage of pollen development and at female gametophyte stage 1 of embryo sac development. We demonstrate that RHF1a directly interacts with and targets a cyclin-dependent kinase inhibitor ICK4/KRP6 (for Interactors of Cdc2 Kinase 4/Kip-related protein 6) for proteasome-mediated degradation. Inactivation of the two redundant RHF genes leads to the accumulation of ICK4/KRP6, and reduction of ICK4/KRP6 expression largely rescues the gametophytic defects in rhf1a rhf2a double mutants, indicating that ICK4/KRP6 is a substrate of the RHF E3 ligases. Interestingly, in situ hybridization showed that ICK4/KRP6 was predominantly expressed in sporophytes during meiosis. Our findings indicate that RHF1a/2a-mediated degradation of the meiosis-accumulated ICK4/KRP6 is essential to ensure the progression of subsequent mitoses to form gametophytes in Arabidopsis.
INTRODUCTION
The plant life cycle alternates between a diploid sporophytic phase and a haploid gametophytic phase. In angiosperms, the diploid male and female sporophytes produce haploid microspores and megaspores, respectively; these give rise to gametophytes after meiosis and a subsequent two or three rounds of mitosis that are not seen in animals or fungi (McCormick, 1993, 2004; Yadegari and Drews, 2004). During megagametogenesis, meiosis of the megaspore mother cell gives rise to four megaspores, but only one survives. This cell then undergoes three rounds of mitosis to form a seven-celled mature embryo sac (female gametophyte) at female gametophyte stage 6 (FG6) (Grossniklaus and Schneitz, 1998). During microgametogenesis, meiosis of the microspore mother cell produces a tetrad of microspores. After release from the tetrad, each microspore goes through an asymmetric cell division, pollen mitosis I (PM I), to produce a bicellular pollen grain containing a generative cell and a much larger vegetative cell. Only the smaller generative cell undergoes a second round of cell division, pollen mitosis II (PM II), to give two sperm cells (McCormick, 1993, 2004). Therefore, the development of both gametophytes requires that mitosis be strictly regulated according to the genetic programs of mega- and microgametogenesis. However, the molecular basis of this mitotic regulatory mechanism is not well understood.
In eukaryotic organisms, cyclin-dependent kinases (CDKs) play an essential role in regulating cell cycle progression and cell division. CDKs are normally inhibited by a family of proteins called CDK inhibitors, and degradation of CDK inhibitors is a key regulatory step in the cell cycle (Huntley and Murray, 1999; Mironov et al., 1999; Barroco et al., 2003; Zhou et al., 2003a). In mammals, the CDK inhibitor p27Kip1 regulates correct cell cycle progression and integrates developmental signals with the core cell cycle machinery (Wang et al., 2000; De Veylder et al., 2001). Human p27Kip1 is regulated by ubiquitin-mediated proteolysis, which is mediated by either the SCF (for SKP1-CUL1-F-Box) ubiquitin E3 ligase complex in nucleus or the RING-type ubiquitin ligase KPC (for Kip1 ubiquitination-Promoting Complex) complex in cytoplasm (Verma et al., 1997; Vlach et al., 1997; Tomoda et al., 1999; Hengst, 2004; Kamura et al., 2004). On the basis of sequence homology to p27Kip1, seven putative CDK inhibitors have been identified in Arabidopsis thaliana, designated as Interactors of Cdc2 Kinase (ICKs) or Kip-related proteins (KRPs). ICK1/KRP1 and ICK2/KRP2 in Arabidopsis were shown to be regulated through the action of the 26S proteasome (Zhou et al., 2003b; Weinl et al., 2005; Jakoby et al., 2006). Although it was recently reported that KRP1 degradation was dependent both on SCFSKP2b and the RING protein RKP (Ren et al., 2008), the physiological roles of these ICK/KRPs and the ubiquitin E3 ligases responsible for their degradation are largely unknown.
Ubiquitin E3 ligases add ubiquitin chains to specific proteins to target them for degradation. E3 ligases are classified into two main groups: single and multiple subunit E3 ligases (Gagne et al., 2002). The RING-type E3 ligases belong to the single subunit group. In Arabidopsis, ∼470 putative RING E3 ligases have been found (Stone et al., 2005), but only a few of them, including SINAT5 (Xie et al., 2002), COP1 (Hardtke et al., 2000; Osterlund et al., 2000), KEG (Stone et al., 2006), AIP2 (Zhang et al., 2005) and HOS1 (Dong et al., 2006), have been functionally characterized and their target proteins defined. Here, we report the molecular analysis of two closely related RING E3 ligases, RING-H2 group F 1a (RHF1a) and RHF2a, the identification of their target protein ICK4/KRP6, a CDK inhibitor, and the elucidation of their redundant roles in Arabidopsis gametogenesis. Our data demonstrate that RHF1a/2a-mediated degradation of the ICK4/KRP6 that accumulated during meiosis is critical for the progression of the subsequent mitotic cell cycles during gametophyte development.
RESULTS
rhf1a rhf2a Double Mutants Have Significantly Reduced Fertility
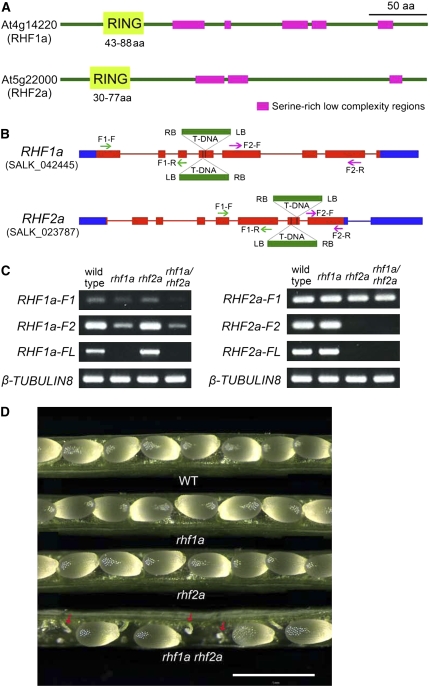
RHF1a and RHF2a are two putative RING-finger type E3 ligases that are clustered into the same clade (Stone et al., 2005). The two proteins are homologous and appear to be plant specific, sharing 30% overall amino acid sequence identity (see Supplemental Figure 1 online). RHF1a has been shown to have E3 ubiquitin ligase activities in vitro (Stone et al., 2005). Both RHF1a and RHF2a contain a RING-H2 domain at their N terminus and several Ser-rich, low-complexity regions toward the C terminus (Figure 1A).
Figure 1.
SALK T-DNA Insertion Mutants of RHF1a and RHF2a.
(A) Protein domains of two Arabidopsis RING-finger proteins, RHF1a and RHF2a, showing a RING-finger domain at the N terminus (light green) and several Ser-rich low-complexity regions (magenta). aa, amino acids.
(B) T-DNA insertion sites in RHF1a and RHF2a genes. Blue box, untranslated regions; red box, exons; red line, introns. Two neighboring inverted T-DNAs were found inserted in the fourth exon of RHF1a in rhf1a (SALK_042445) and in the seventh exon of RHF2a in rhf2a (SALK_023787) through sequencing the amplified flanking sequence. The green and purple arrows show the positions of primers used to amplify the truncated transcripts in (C). The green arrows indicate the primers used to amplify the F1 fragment. The purple arrows indicate the primers used to amplify the F2 fragment.
(C) RHF1a and RHF2a full-length and truncated transcript level in wild-type plants, single insertion mutants, and rhf1a rhf2a double mutants. RNA was extracted from 5-week-old plants. F1, truncated transcript before the T-DNA insertion site; F2, truncated transcript after the T-DNA insertion site; FL, full-length transcript. β-Tubulin8 was used as an internal control.
(D) Seed development in the wild-type plant, rhf1a single mutant, rhf2a single mutant, and rhf1a rhf2a double mutant. Aborted ovules (indicated by red arrows) were evident in rhf1a rhf2a mutants. Bar = 1 mm.
To define the physiological roles of RHF1a and RHF2a, we isolated T-DNA insertion mutants for the two genes (Figure 1B). By RT-PCR analyses we did not detect full-length transcripts for either gene, although truncated transcripts were detected (Figure 1C). Inactivation of either RHF1a or RHF2a did not cause any obvious developmental defects (Figure 1D). However, the rhf1a rhf2a double mutant displayed a low fertility phenotype with siliques significantly shorter than those of the wild type. We categorized the phenotypes as weak, medium, or severe because the number of aborted ovules ranged from 4.4 to 63%, whereas the number of total ovules per silique was not much affected (Figure 1D, Table 1). Reciprocal crosses to wild-type plants with RHF1a/rhf1a rhf2a or rhf1a RHF2a/rhf2a plants revealed that the transmission of the rhf1a and rhf2a alleles occurred at lower frequencies in both male and female gametophytes, suggesting that these ovule abortion phenotypes were mainly due to gametophytic defects (Table 2). These findings indicate that RHF1a and RHF2a have overlapping functions in both male and female gametogenesis.
Table 1.
Statistical Analysis of rhf1a, rhf2a, and rhf1a rhf2a Siliques Compared with Wild-Type Plants
| Developed Seeds | Aborted Ovules | Percentage of Aborted Ovules | Silique Length | Developed Seeds per Silique | Total Ovules per Silique | Seed Set | ||
|---|---|---|---|---|---|---|---|---|
| Wild Type | 1353 | 9 | 0.7% | 1.49 ± 0.02 | 52 ± 1.19 | 52 ± 1.19 | 1.00 | |
| rhf1a | 1663 | 12 | 0.7% | 1.51 ± 0.04 | 55 ± 1.27 | 55 ± 1.27 | 1.00 | |
| rhf2a | 1035 | 8 | 0.8% | 1.52 ± 0.02 | 54 ± 1.94 | 54 ± 1.94 | 1.00 | |
| rhf1a rhf2a | Weak (32%) | 1722 | 80 | 4.4% | 1.32 ± 0.08 | 43 ± 3.07 | 45 ± 2.89 | 0.96 |
| Medium (16%) | 627 | 285 | 31.3% | 1.19 ± 0.12 | 31 ± 4.56 | 46 ± 2.46 | 0.67 | |
| Severe (52%) | 1217 | 2074 | 63.0% | 1.00 ± 0.05 | 19 ± 0.72 | 50 ± 1.43 | 0.38 |
All the measurements were performed with the first five siliques on the 6-week-old plants. For the wild type and rhf1a and rhf2a single mutant plants, the samples were from five independent plants. For rhf1a rhf2a double mutants, the samples were from 25 independent plants. Mean ± sd is shown. Differences between values for the wild type and other single or double mutants were significant at the 0.05 level. Seed set: normally developed seeds per silique/total ovules per silique.
Table 2.
Transmission Efficiency Test through Reciprocal Crosses
| Parental Genotypes (Female × Male) | Progeny
|
||||||
|---|---|---|---|---|---|---|---|
| Cross | rhf1a/RHF1a | RHF1a/RHF1a | rhf2a/RHF2a | RHF2a/RHF2a | Total | TEF | TEM |
| RHF1a/rhf1a rhf2a/rhf2a × wild type | 104 | 143 | 247 | 247 | 72.7% | NA | |
| Wild type × RHF1a/rhf1a rhf2a/rhf2a | 117 | 136 | 253 | 253 | NA | 86.0% | |
| rhf1a/rhf1a RHF2a/rhf2a × wild type | 262 | 121 | 141 | 262 | 85.8% | NA | |
| Wild type × rhf1a/rhf1a RHF2a/rhf2a | 257 | 110 | 147 | 257 | NA | 74.8% | |
Reciprocal crosses among wild-type plants, RHF1a/rhf1a rhf2a, and rhf1a RHF2a/rhf2a were used to determine the transmission efficiency of gametes from RHF1a/rhf1a rhf2a and rhf1a RHF2a/rhf2a. Because of the loss of the kanamycin selection marker for the SALK mutants, we used PCR to determine the transmission efficiencies (TE). TE is calculated according to the following: TE = number of progenies with T-DNA insertion/number of progenies without T-DNA insertion × 100%. 1:1 for the reciprocal crosses is the expected value for the normal gamete transmission. TEF, female transmission efficiency; TEM, male transmission efficiency; NA, not applicable.
The rhf1a rhf2a Double Mutants Have Defects in Female Gametophyte Development at the One-Nucleate Stage
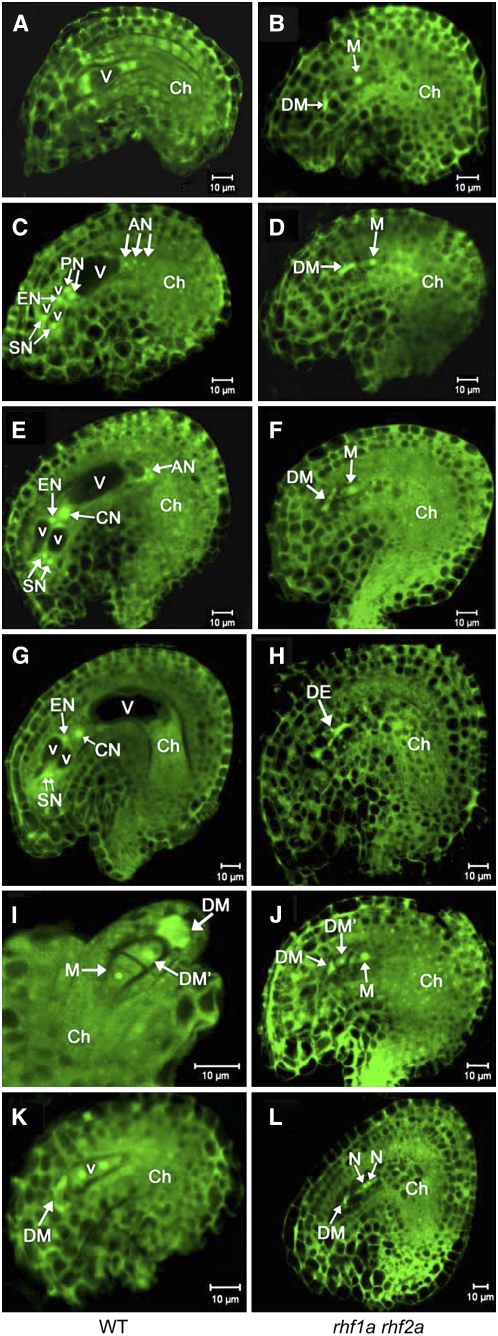
To further characterize the ovule abortion phenotype of the rhf1a rhf2a double mutants, we analyzed the development of the female gametophyte (megagametogenesis) in rhf1a rhf2a plants with a severe phenotype. Megagametogenesis of wild-type plants proceeds normally from the one-nucleate stage FG1 (female gametophyte stage 1) to FG7 (for later stages, see Figures 2A, 2C, 2E, and 2G). Ovule development in the same wild-type pistil was primarily synchronous, spanning at most three neighboring stages (Table 3). However, in the rhf1a rhf2a double mutants, the developmental synchronicity of embryo sacs was impaired (Table 3), and in ∼60% of the ovules, megagametogenesis had not proceeded beyond FG1 before the female gametophyte was completely degenerated (Figures 2B, 2D, 2F, and 2H). In the wild type, the three megaspores at the micropylar pole in FG1 ovules had generally degenerated, concomitant with completion of meiosis (Figure 2I, Table 3). By contrast, one of the three megaspores destined to degenerate was often visible in rhf1a rhf2a until stages corresponding to FG6-FG7 in wild-type ovules, probably due to slower degradation (Figure 2J, Table 3). Occasionally, the rhf1a rhf2a ovules were arrested at the two-nucleate stage (FG2) (Figure 2K), with an integument at later stages, relative to wild-type ovules at the same stage (Figure 2L). Despite these developmental defects of ovules in rhf1a rhf2a mutants, ∼30% of the rhf1a rhf2a embryo sacs developed up to FG7 and formed fertile seeds upon fertilization.
Figure 2.
Ovule Development in Wild-Type Plants and the rhf1a rhf2a Double Mutants Revealed by Confocal Laser Scanning Microscopy Analysis.
Ovule cells are visualized by autofluorescence. AN, antipodal nuclei; Ch, chalazal end; CN, central nucleus; DE, degenerated embryo sac; DM, degenerated megaspore; DM', nondegenerated megaspore other than the functional megaspore; EN, egg nucleus; N, nucellus; PN, polar nuclei; SN, synergid nuclei; V, large vacuole; v, small vacuole. Bars = 10 μm.
(A) and (B) Wild-type ovule at early FG4, containing a four-nucleate embryo sac (A) compared with the abnormal female gametophytes from the rhf1a rhf2a double mutant pistils at the same stage (B).
(C) and (D) Wild-type ovule at FG5 with an eight-nucleate embryo sac (C) compared with the rhf1a rhf2a double mutant ovule at the same stage (D). At this stage in the wild type, cellularization took place and cell differentiation was completed with the formation of two synergid nuclei (SN), an egg nucleus (EN), three antipodal nuclei (AN), and two prominent unfused polar nuclei (PN).
(E) and (F) Wild-type ovule with a mature seven-celled embryo sac at FG6 (E) compared with the rhf1a rhf2a double mutant ovule at the same stage (F). During this stage, the two polar nuclei fused to form a diploid central nucleus (CN) in the wild type.
(G) and (H) Mature wild-type ovule with a four-celled embryo sac at FG7 (G) and an rhf1a rhf2a double mutant ovule (H). Note that the three antipodal cells have degenerated completely in the wild type. Note also that, in the rhf1a rhf2a double mutant, female gametophytes were arrested at the one-nucleate stage FG1 until the embryo sac (DE) degenerated completely, visualized by its strong autofluorescence (B), (D), (F), and (H).
(I) Wild-type ovule at FG1 with one functional megaspore (M) and three degenerating megaspores, of which two micropylar megaspores have degenerated completely (DM) and the other has not degenerated yet (DM'). Note that the outer and inner integuments do not completely enclose the nucellus (N).
(J) Ovule from the rhf1a rhf2a double mutant showing a nondegraded megaspore (DM') and a functional megaspore with outer and inner integuments enclosing them.
(K) Wild-type ovule at FG2 with two-nucleate embryo sac.
(L) Ovule from the rhf1a rhf2a double mutant arresting at the two-nucleate stage. Note that the developmental extent of the ovule in (L) with mature integuments enclosing is much higher than the ovule in (K).
Table 3.
Synchrony of Female Gametophyte Development in Wild Type and rhf1a rhf2a Double Mutants
| Number of Female Gametophytes at Developmental Stages
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Pistil Number | Completely Degenerated | FG1 with the Third Undegraded Nonfunctional Megaspore | FG1 | FG2 | FG3 | FG4 | FG5 | FG6 | FG7 | FG8 | |
| Wild type | sili1 | 6 | 22 | 1 | |||||||
| sili2 | 7 | 15 | 6 | 3 | |||||||
| sili3 | 4 | 13 | 8 | ||||||||
| sili4 | 7 | 12 | |||||||||
| sili5 | 2 | 17 | 4 | ||||||||
| sili6 | 4 | 13 | 7 | ||||||||
| sili7 | 2 | 17 | 9 | ||||||||
| sili8 | 2 | 24 | |||||||||
| rhf1a rhf2a | sili1 | 4 | 13 | ||||||||
| sili2 | 1 | 8 | 4 | ||||||||
| sili3 | 11 | 4 | 7 | ||||||||
| sili4 | 2 | 14 | 5 | 3 | 1 | ||||||
| sili5 | 1 | 10 | 1 | 4 | |||||||
| sili6 | 1 | 11 | 8 | 2 | |||||||
| sili7 | 4 | 12 | 3 | 3 | 3 | ||||||
| sili8 | 2 | 1 | 17 | 1 | 1 | 5 | 4 | ||||
| sili9 | 3 | 11 | 1 | 2 | 2 | ||||||
| sili10 | 6 | 13 | 1 | 1 | 2 | 8 | |||||
FG stages defined according to Christensen et al. (1998)
Arrest at Interphase of the Mitotic Cell Cycle Occurs in Pollen Development in the rhf1a rhf2a Double Mutants
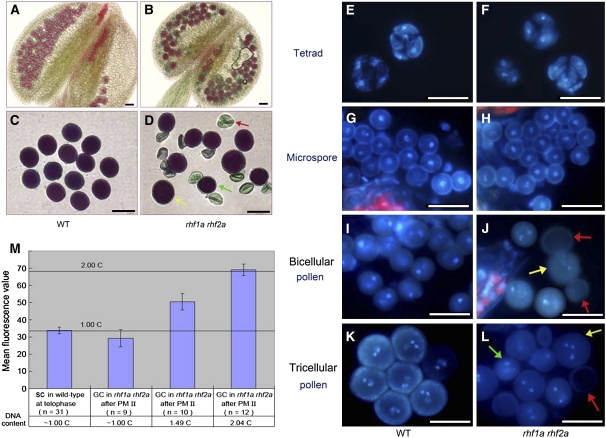
Because the transmission efficiency of rhf1a and rhf2a alleles was reduced in the rhf1a rhf2a double mutant through both male and female gametophytes (Table 2), we investigated whether any defects occurred during male gametogenesis in the rhf1a rhf2a double mutant, using Alexander's staining, which distinguishes viable pollen from nonviable pollen (Alexander, 1969). The rhf1a rhf2a plants had smaller anthers with 50 to 70% nonviable pollen grains that were small and shrunken, whereas the wild type produced <0.5% of abnormal pollen (Figures 3A to 3D, Table 4).
Figure 3.
Pollen Development in Wild-Type Plants and the rhf1a rhf2a Double Mutants.
(A) Alexander's staining of wild-type anthers.
(B) Alexander's staining of double mutant anthers containing degenerated pollen.
(C) Alexander's staining of wild-type mature pollen grains of uniform size.
(D) Alexander's staining of the rhf1a rhf2a double mutant mature pollen grains from the same anther. Note that the double mutant mature pollen is of variable size. Yellow arrow: much larger than normal pollen grain. Red arrow: shrunken pollen. Green arrow: smaller than normal pollen grain.
(E) and (F) DAPI staining of tetrads in wild-type (E) and the rhf1a rhf2a double mutant anthers (F). No visible difference was seen between wild-type and double mutant tetrads.
(G) and (H) DAPI staining of microspores in wild-type (G) and the rhf1a rhf2a double mutant anthers (H). Note no visible difference between wild-type and double mutant microspores.
(I) and (J) DAPI staining of binucleate stage pollen in wild-type (I) and the rhf1a rhf2a double mutant anthers (J). Note that the binucleate pollen are of uniform size in the wild type and of variable size in the double mutant. The yellow arrow indicates larger pollen, and the red arrow indicates abnormal pollen with no DAPI staining.
(K) and (L) DAPI staining of mature pollen grains in wild-type (K) and the rhf1a rhf2a double mutant anthers (L). Note that mature pollen grains from the double mutant contain shrunken pollen walls (red arrow), larger (yellow arrow), and smaller pollen grains than normal (green arrow). Bars = 25 μm.
(M) Relative DNA content (DAPI fluorescence values) in generative cells (GC) and sperm cells (SC) in wild-type and the rhf1a rhf2a mutant pollen. The mean C values calculated relative to the 1C content of telophase sperm cell nuclei (1C) after the PM II stage are shown. Error bars = se.
Table 4.
Pollen Abnormalities Observed and Statistical Analysis
| Normal Pollen with Staining | Shrunken Pollen without Staining | Total | Percentage of Shrunken Pollen | ||
|---|---|---|---|---|---|
| Wild-type plants | Wild type-1 | 1128 | 5 | 1133 | 0.4% |
| Wild type-2 | 959 | 3 | 962 | 0.3% | |
| Wild type-3 | 1206 | 4 | 1212 | 0.3% | |
| rhf1a rhf2a double mutants | Line-7 | 365 | 609 | 974 | 62.5% |
| Line-8 | 595 | 747 | 1342 | 55.7% | |
| Line-13 | 557 | 785 | 1342 | 58.5% | |
| Line-15 | 423 | 490 | 913 | 53.7% | |
| Line-23 | 411 | 609 | 1020 | 59.7% | |
| Line-25 | 418 | 658 | 1076 | 61.2% | |
| Line-26 | 613 | 694 | 1307 | 53.1% | |
| Line-28 | 363 | 693 | 1056 | 65.6% |
Alexander's staining analysis was performed to assess pollen grain development of wild-type plants and rhf1a rhf2a double mutants.
We further examined the microspores released from rhf1a rhf2a anthers at four sequential developmental stages. In the rhf1a rhf2a double mutants, although no obvious defects were observed at stages prior to PM I, which corresponds to the tetrad and microspore stages (Figures 3E to 3H), striking differences became apparent from the early bicellular stage, where 30 to 40% of the rhf1a rhf2a microspores failed to go through PM I, exhibited no fluorescence, and then quickly became shrunken (Figures 3I and 3J). In addition, this aberrant microspore phenotype was also found at the early tricellular stage in those rhf1a rhf2a grains that had proceeded normally through PM I. Approximately 20 to 30% of the surviving microspores failed to go through PM II and later were shrunken (Figures 3K and 3L).
To define the exact phase at which rhf1a rhf2a pollen development was arrested, we examined rhf1a rhf2a pollen at the stage when pollen grains in wild-type anthers are about to dehisce. At this stage, the generative cell usually has divided into two sperm cells in wild-type pollen. However, we found that undivided generative cells were still present in pollen of the rhf1a rhf2a double mutants (Figure 3L). These undivided generative cells in rhf1a rhf2a mature pollen had different intensity fluorescences after 4′,6-diamidino-2-phenylindole (DAPI) staining, suggesting that they were at different phases of the cell cycle (Figure 3L).
We conducted DNA content analysis on the generative cells of rhf1a rhf2a at this stage. It was previously demonstrated that wild-type generative cells at early interphase contain ∼1C of the mean DNA content, which increases to almost 2C prior to PM II (Friedman, 1999). In the rhf1a rhf2a double mutant, we found either 1C, 1.49C, or 2.04C in those generative cells that failed to go through PM II (Figure 3M), suggesting that the generative cells in rhf1a rhf2a could be arrested at either G1, S, or G2 phase.
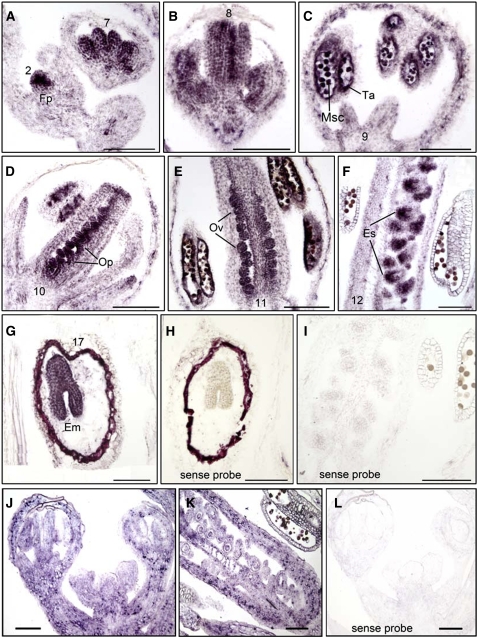
In Inflorescences, RHF1a Is Primarily Expressed in Developing Gametophytes and Embryos, whereas RHF2a Is Expressed Ubiquitously
To investigate the expression patterns of RHF1a and RHF2a during ovule and pollen development, we performed RNA in situ hybridization on wild-type inflorescences. The mRNA of the RHF1a gene was detected in the newly emerging floral primordia from a very early stage (Figure 4A). As flower development proceeded, the RHF1a transcript was mainly detected in the stamens and carpels. The RHF1a transcript in stamens was first found in the stamen primordia, then accumulated in pollen mother cells and tapetal cells, and later in microspores, until flower stage 11 (Figures 4A to 4E). However, no transcript was detected in anthers of stage 12 flowers (Figure 4F). By contrast, carpels expressed RHF1a throughout all developmental stages, from primordia to the mature embryo sac stage (Figures 4A, 4B, and 4D to 4F). After fertilization, RHF1a mRNA was mainly present in the developing embryos (Figure 4G). RHF2a was expressed throughout the inflorescence, including all four whorls of the floral organs (Figures 4J and 4K). This ubiquitous expression pattern may explain why the expression level of RHF2a is eightfold higher than that of RHF1a in inflorescences, as indicated by quantitative real-time PCR (see Supplemental Figure 2 online). The spatial pattern of RHF1a and RHF2a correlated well with the gametophytic defects found in the rhf1a rhf2a mutant, although RHF1a might be more specific and important to gametogenesis than RHF2a.
Figure 4.
RNA in Situ Expression of RHF1a and RHF2a in Wild-Type Inflorescences and Siliques.
Longitudinal sections ([A] to [G]) of inflorescences were hybridized with RHF1a antisense probe.
(A) to (F) Expression of RHF1a in wild-type inflorescence from an early stage to stage 12.
(G) Expression of RHF1a in wild-type developing seeds.
(H) and (I) Hybridized with RHF1a sense probe.
(J) Expression of RHF2a in inflorescence.
(K) Expression of RHF2a in developing carpels.
(L) Hybridized with RHF2a sense probe.
Em, embryos; Es, embryo sac; Fp, floral primordia; Msc, microsporocyte; Op, ovule primordia; Ov, ovule; Ta, tapetum. Bars = 100 μm.
RHF1a Primarily Interacts with and Degrades the CDK Inhibitor ICK4/KRP6 in a 26S Proteasome-Dependent Manner
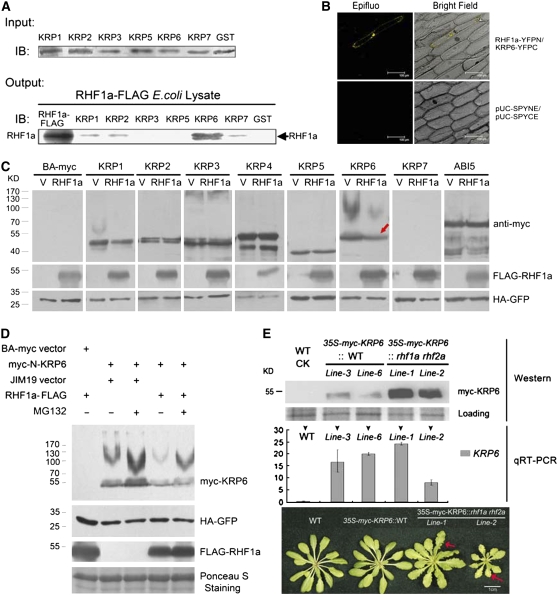
The generative cells in rhf1a rhf2a mutants were arrested at either the G1, S, or G2 phases of the cell cycle, and RHF1a posseses E3 ligase activity in ubiquitin-dependent proteolysis (Stone et al., 2005). These findings together suggested that RHF1a and RHF2a are likely involved in degrading a negative regulator that might function in the interphase of the mitotic cell cycle in gametogenesis. Dozens of proteins are known to negatively regulate cell cycle progression in Arabidopsis, including Retinoblastoma-Related Protein 1, WEE1, PASTICCINO2, SIAMESE, Cell Division Cycle 20, and ICKs/KRPs (Inzé and Veylder, 2006; De Veylder et al., 2007). Among these negative regulators, ICKs/KRPs, a type of CDK inhibitor, were primarily considered to have wide-spectrum effects in interphase (Nakayama, 1998). Therefore, we used an in vitro glutathione S-transferase (GST) pull-down assay to investigate whether RHF1a interacts with these negative regulators. One member of the Arabidopsis ICK/KRP family, ICK4/KRP6, was found to strongly interact with RHF1a-FLAG, while ICK1/KRP1, ICK2/KRP2, and ICK5/KRP7 had weak interactions with RHF1a-FLAG (Figure 5A). The interaction between RHF1a and KRP6 was further confirmed in vivo (Figure 5B) using bimolecular fluorescence complementation (BiFC) assays (Walter et al., 2004).
Figure 5.
RHF1a Primarily Interacts with ICK4/KRP6 and Targets It for Degradation.
(A) Pull-down assays of RHF1a and ICKs/KRPs (except for KRP4). Input, protein gel blot using anti-GST antibody, shows comparable protein loading in the GST pull-down assays. Output, Escherichia coli lysates containing RHF1a-FLAG were used in GST pull-down assays with GST-ICK/KRP fusions. RHF1a-FLAG represents lysate from E. coli expressing RHF1a-FLAG. IB, immunoblot detected with anti-FLAG antibody.
(B) BiFC assays in vivo. RHF1a-YFPN is able to interact with KRP6-YFPC to generate yellow fluorescent protein (YFP) fluorescence in onion epidermal cells. The expression of pUC-SPYNE and pUC-SPYCE was used as the control.
(C) Effects of RHF1a on the degradation of ICKs/KRPs and ABI5 in vivo. N. benthamiana leaves were coinfiltrated with indicated binary vectors. For each experiment, two constructs were infiltrated, one expressing a specific KRP protein (top row of labels) and the other expressing either RHF1a or an empty vector (bottom row of labels). 35S-HA-GFP (green fluorescent protein) and 35S-p19 were also simultaneously coinfiltrated to serve as an internal control and to suppress gene silencing, respectively. Total proteins were extracted and resolved by SDS-PAGE analysis followed by immunoblotting with different antibodies. Anti-myc was used to detect the protein level of ICKs/KRPs or ABI5 (top panel), anti-FLAG was used for detection of FLAG-RHF1a (middle panel), and anti-HA was used to detect the internal control HA-GFP (bottom panel). The red arrow indicates the reduced protein level of ICK4/KRP6.
(D) ICK4/KRP6 degradation in a 26S proteasome-dependent manner. Immunoblots were used to detect ICK4/KRP6 levels in the absence or presence of RHF1a and with or without the proteasome inhibitor, MG132 (40 μM), infiltrated into N. benthamiana leaves 12 h before sample collection. Anti-FLAG was used for detection of FLAG-RHF1a, and anti-HA was used to detect HA-GFP as internal controls. Ponceau S staining of the transferred membrane is displayed as a loading control.
(E) ICK4/KRP6 protein accumulated much more in the rhf1a rhf2a double mutant plants than in wild-type plants. 35S-KRP6 construct was transformed into wild-type and rhf1a rhf2a double mutant plants, named as 35S-myc-KRP6∷WT and 35S-myc-KRP6∷rhf1a rhf2a, respectively. The analysis with two of the 35S-myc-KRP6∷WT independent lines and two of the 35S-myc-KRP6∷rhf1a rhf2a independent lines is shown. Proteins from seedlings ∼20 d after germination (DAG) were processed for protein gel blots with anti-myc antibody to detect the ICK4/KRP6 protein level in vivo (top panel). Ponceau S staining of the transferred membrane is displayed as a loading control. Total RNAs from the same transgenic plants were used for quantitative RT-PCR analysis (middle panel). Leaf phenotypes (bottom panel) of wild-type plants, one of the two 35S-myc-KRP6∷WT lines, and two of the 35S-myc-KRP6∷rhf1a rhf2a lines were shown (bottom panel). Photos were taken from 3-week-old plants. Red arrows indicate serrated leaves.
To determine the target specificity of the degradation, we then examined the protein level of all the ICKs/KRPs and ABI5 (as a control) in the absence or presence of RHF1a in vivo, using a transient expression assay in tobacco (Nicotiana benthamiana) leaves. The results showed that the protein level of ICK4/KRP6 decreased in the presence of FLAG-RHF1a and that the level of ICK1/KRP1 was slightly reduced, whereas the protein levels of other ICKs/KRPs and ABI5 exhibited no significant differences in the presence or absence of FLAG-RHF1a (Figure 5C). These results were largely consistent with the in vitro interaction results, suggesting that RHF1a mainly targets ICK4/KRP6 for degradation. We further investigated the relationship between ICK4/KRP6 and the 26S proteasome-dependent proteolysis pathway using MG132, an inhibitor of such proteolysis. The results showed that more ICK4/KRP6 protein accumulated after MG132 treatment, and the accumulation level of ICK4/KRP6 was reduced in the presence of RHF1a (Figure 5D), indicating that the ICK4/KRP6 protein level was regulated by 26S proteasome-dependent proteolysis.
RHF1a/RHF2a Function Correlates with the Accumulation of ICK4/KRP6
To further confirm the regulation of ICK4/KRP6 by RHF1a/RHF2a in Arabidopsis, we transformed an ICK4/KRP6-myc fusion construct driven by the cauliflower mosaic virus (CaMV) 35S promoter (pBA002-KRP6-myc) into rhf1a rhf2a and wild-type plants, respectively. The protein level of ICK4/KRP6-myc was determined by protein gel blots, using a commercially available myc antibody. As shown in Figure 5E, the protein level of ICK4/KRP6-myc was rather low in the two independent transgenic lines in the wild-type background, suggesting that ICK4/KRP6 is rapidly degraded under normal growth conditions. However, in the rhf1a rhf2a background, although the transcription level was similar or even lower, ICK4/KRP6-myc was significantly accumulated, in the two independent transgenic lines (Figure 5E). Moreover, the accumulation of ICK4/KRP6-myc in the rhf1a rhf2a background led to serrated leaves (Figure 5E), a previously reported phenotype that was observed in ICK4/KRP6-overexpressing plants (Zhou et al., 2002; Butaye et al., 2004). These results, together with the in vivo and in vitro results (Figures 5A to 5D), demonstrate that inactivation of these two RHF E3 ligases led to the accumulation of the ICK4/KRP6-myc protein and that RHF1a targets ICK4/KRP6 to the 26S proteasome for proteolysis.
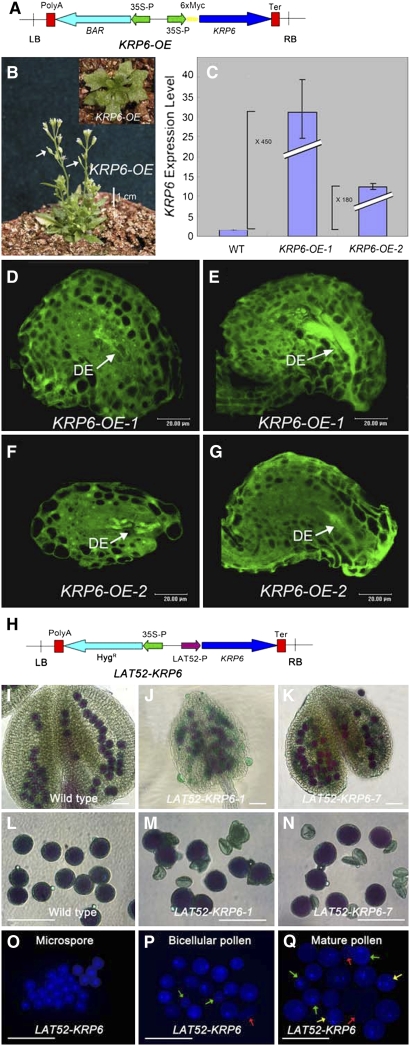
ICK4/KRP6 Overexpression Lines Exhibited Gametogenesis Defects Similar to Those Observed in the rhf1a rhf2a Double Mutants
Because RHF1a and RHF2a appear to be required for ICK4/KRP6 degradation as shown above, we tested whether ICK4/KRP6 overexpression can phenocopy the rhf1a rhf2a defects. We analyzed >50 independent transgenic plants overexpressing ICK4/KRP6 from the CaMV 35S promoter (Figure 6A). Approximately 20% of them exhibited pleiotropic phenotypes, including serrated leaves, small stature, abnormal flowers, and low fertility (Figure 6B), similar to a previous report (Zhou et al., 2002). ICK4/KRP6 was highly expressed in these lines, two of which, ICK4/KRP6-OE-1 and ICK4/KRP6-OE-2, were chosen for further analysis of pollen and ovule development (Figure 6C). Confocal laser scanning microscopy of these ICK4/KRP6-OE lines showed that female gametophyte development was inhibited; 40 to 50% of the female gametophytes completely degenerated at the terminal stage, their embryo sac cavities filling with autofluorescent material. The degeneration of the embryo sacs occurred during a series of stages in megagametogenesis, including the one-, two-, and four-nucleate stages (Figures 6D to 6G). These phenotypes were very similar to, although seemingly less severe than, those of the rhf1a rhf2a double mutants.
Figure 6.
Elevated Expression of ICK4/KRP6 Phenocopies the Gametophytic Defects of rhf1a rhf2a Mutant.
(A) Construct of the KRP6-OE plasmid. LB, T-DNA left border; polyA, CaMV 35S poly(A); BAR, resistance gene; 35S-P, CaMV 35S promoter; 6×myc, six myc tags fusion; KRP6, open reading frame of KRP6; Ter, nopaline synthase terminator; RB, T-DNA right border.
(B) ICK4/KRP6 overexpression transgenic plants ∼50 DAG. The fertility of the transgenic plants was low as shown by white arrows indicating the extremely short siliques.
(C) Real-time quantitative RT-PCR analysis of ICK4/KRP6 expression level in two independent ICK4/KRP6 OE lines. Expression is normalized to a wild-type level of 1. Error bars indicate sd of three real-time PCR experiments of each biological sample.
(D) to (G) Confocal laser scanning microscopy analysis of embryo sac development in ovules from the KRP6 OE-1 line ([D] and [E]) and the KRP6 OE-2 line ([F] and [G]) independently. The completely degenerated embryo sacs (DE) were indicated by high autofluorescence regions.
(H) Construction of the LAT52-KRP6 plasmid. LAT52P, LAT52 promoter; HygR, hygromycin resistance gene; LB, polyA, 35S-P, Ter, and RB are as in (A).
(I) Alexander's staining of wild-type anthers.
(J) and (K) Alexander's staining of LAT52-KRP6-1 (J) and LAT52-KRP6-7 (K) transgenic plant anthers.
(L) Alexander's staining of wild-type pollen grains with even size.
(M) and (N) Alexander's staining of LAT52-KRP6-1 (M) and LAT52-KRP6-7 (N) transgenic plant pollen grains with shrunken pollen walls.
(O) DAPI staining of LAT52-KRP6 microspores.
(P) DAPI staining of LAT52-KRP6 pollen at bicellular stage with some aberrantly stained pollen (green arrows) and shrunken pollen walls (red arrows).
(Q) DAPI staining of LAT52-KRP6 pollen at tricellular stage with microspores (green arrows), bicellular pollen (yellow arrows), and shrunken pollen walls (red arrows) other than the normally stained tricellular pollen grains.
To overcome the reported low expression problem of the CaMV 35S promoter in pollen, we transformed wild-type Arabidopsis with the ICK4/KRP6 gene driven by the LAT52 promoter, which is expressed starting from PM I (Twell et al., 1990; Eady et al., 1995), to investigate whether the phenotype in the male gametophyte could be recapitulated (Figure 6H). Alexander's staining showed that LAT52-ICK4/KRP6 pollen grains displayed similar phenotypes to those found in rhf1a rhf2a (Figures 6I to 6N). DAPI staining results showed that the developmental defects in the LAT52-ICK4/KRP6 pollen only emerged after the microspore stage (Figure 6O) (i.e., during PM I and PM II; Figures 6P and 6Q), which is consistent with the male gametophyte defects in rhf1a rhf2a. These results indicate that elevated expression of ICK4/KRP6 in pollen recapitulates the male gametophyte defects seen in the rhf1a rhf2a double mutant.
To confirm that ICK4/KRP6 is indeed the functionally relevant target of RHF1a/2a, we lowered the expression of ICK4/KRP6 in the rhf1a rhf2a double mutants by RNA interference (RNAi). Statistical analysis showed that 48% of ICK4/KRP6-RNAi/rhf1a rhf2a plants displayed a wild-type phenotype for the ovule development and fertility, and many fewer plants with severe phenotypes were found (Table 5). Compared with the ovule development of rhf1a rhf2a double mutants (Table 1), these genetic data demonstrate that knocking down the ICK4/KRP6 expression largely rescues the ovule development defects in the rhf1a rhf2a double mutants and clearly confirm that ICK4/KRP6 is the functionally relevant substrate of RHF1a/2a.
Table 5.
Statistical Analysis of ICK4/KRP6-RNAi/rhf1a rhf2a Siliques Compared with Wild-Type Plants
| Developed Seeds | Aborted Ovules | Percentage of Aborted Ovules | Silique Length | Developed Seeds per Silique | Total Ovules per Silique | Seed Set | ||
|---|---|---|---|---|---|---|---|---|
| Wild type | 1314 | 11 | 0.8% | 1.49 ± 0.04 | 53 ± 1.77 | 53 ± 1.77 | 1.00 | |
| ICK4/KRP6-RNAi/rhf1a rhf2a | Wild type-like (48%) | 3214 | 26 | 0.8% | 1.51 ± 0.03 | 54 ± 1.56 | 54 ± 1.56 | 1.00 |
| Weak (28%) | 1645 | 70 | 4.1% | 1.37 ± 0.06 | 45 ± 2.23 | 47 ± 3.05 | 0.96 | |
| Medium (16%) | 651 | 309 | 32.2% | 1.18 ± 0.15 | 32 ± 3.26 | 47 ± 1.27 | 0.68 | |
| Severe (8%) | 191 | 296 | 60.8% | 1.00 ± 0.11 | 19 ± 1.12 | 48 ± 1.23 | 0.39 |
All the measurements were performed with the first five siliques on the 6-week-old plants. For wild-type plants, the samples were from five independent plants. For ICK4/KRP6-RNAi/rhf1a rhf2a double mutants, the samples were from 25 transgenic plants. Mean ± sd is shown. Seed set: normally developed seeds per silique/total ovules per silique.
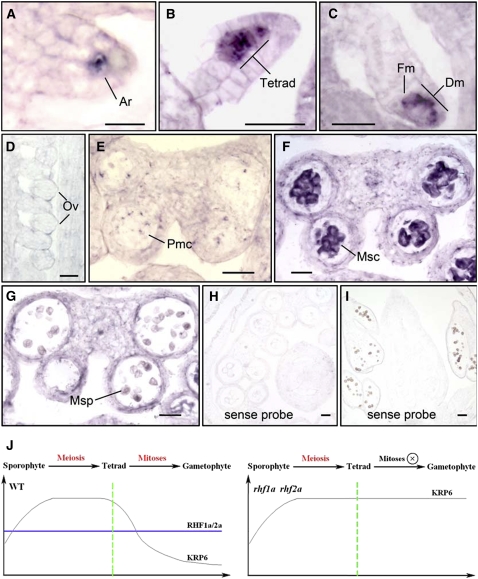
ICK4/KRP6 Transcription Is Specifically Switched on during Meiosis but Turned off before Gametophytic Mitosis
To further clarify how ICK4/KRP6, a negative regulator of mitotic cell cycle progression, was involved in gametophyte-specific defects in the rhf1a rhf2a mutant, we used RNA in situ hybridization to investigate the expression pattern of ICK4/KRP6 in wild-type inflorescences. During ovule development, ICK4/KRP6 RNA was first detectable in female archesporial cells (Figure 7A). The signal became stronger in female tetrads (Figure 7B) and then in functional megaspores and degenerating megaspores (Figure 7C). However, the ICK4/KRP6 RNA signal was no longer detectable after the functional megaspore was distinguished and established (Figure 7D). In anther development, the ICK4/KRP6 transcripts were weakly detectable in pollen mother cells in anthers at stage 4 (Figure 7E). Then, the hybridization signal was predominantly observed in microsporocytes in stage 5 and 6 anthers (Figure 7F). In anthers after stage 6, the ICK4/KRP6 RNA level was nearly undetectable (Figure 7G). These expression patterns of ICK4/KRP6 suggest that the transcription of ICK4/KRP6 is tightly regulated during gametogenesis and that it is specifically switched on during meiosis but switched off before mitosis. Taken together, our data demonstrate that RHF1a/2a-mediated degradation of the ICK4/KRP6 that was accumulated during meiosis is critical for the progression of the subsequent mitotic cell cycles during gametophyte development (Figure 7J).
Figure 7.
The Transcription of ICK4/KRP6 Was Tightly Regulated and Only Detected in Stages before Gametophytic Mitosis by in Situ Hybridization.
(A) KRP6 RNA expression in ovule primordia at early stages, showing the KRP6 RNA signal in the female archesporial cell (Ar).
(B) and (C) KRP6 RNA expression in ovule primordia at late stages, showing the stronger KRP6 RNA signal in the female tetrad, shown as four neighboring megaspores (B), and later in functional megaspore (Fm) and three other degenerating megaspores (Dm) (C).
(D) KRP6 RNA expression in the ovule, showing no or very low KRP6 RNA signal in developing ovules (Ov).
(E) Weak KRP6 RNA expression in pollen mother cells (Pmc) in an anther at stage 4.
(F) KRP6 expression in anthers at stages 5 to 6, showing that KRP6 RNA signal was predominantly in microsporocyte (Msc).
(G) KRP6 expression was not detected in microspores (Msp) in anthers after stage 6.
(H) and (I) Control sections hybridized with the sense KRP6 RNA probe, showing no signal detected.
(J) A possible working model for RHF1a/2a and ICK4/KRP6 during gametophyte formation in Arabidopsis. The transcript of ICK4/KRP6 was accumulated during meiosis and turned off right before the subsequent gametophytic mitosis. RHF1a/RHF2a-mediated 26S proteasome-dependent proteolysis may serve as a watchdog in Arabidopsis, to keep the level of meiosis-accumulated ICK4/KRP6 below a certain threshold, so that the subsequent gametophytic mitoses can proceed smoothly.
Bars = 10 μm in (A) and 20 μm in all other panels.
DISCUSSION
We demonstrated that two functionally overlapping RING-type E3 ligases, RHF1a and RHF2a, regulate mitotic cell cycle progression during Arabidopsis gametogenesis by activating the degradation of the CDK inhibitor ICK4/KRP6. Our findings show that plants use the 26S proteasome-mediated proteolysis to keep the protein level of ICK4/KRP6, a negative regulator of CDKs and a preferentially expressed protein during meiosis, below a certain threshold, to allow the mitotic divisions that produce the gametophytes.
Although gametogenesis is an essential process for plant reproduction and has been studied extensively, the detailed regulatory mechanisms controlling this process are not well defined. Many mutants with defects in forming gametophytes in Arabidopsis have been identified. Among the various Arabidopsis mutants, those mutants whose gametophyte development is arrested at early stages (e.g., at FG1 stage of megagametogenesis and/or PM I stage of microgametogenesis) are particularly interesting because molecular characterization of these mutants will facilitate our understanding of the determining factors for gametophyte development. Although mutants defective at FG1 and/or PM I of gametophyte development have been identified in Arabidopsis (Feldmann et al., 1997; Christensen et al., 1998; Drews et al., 1998; Park et al., 1998; Yang and Sundaresan, 2000; Twell et al., 2002), molecular analysis of these mutants has proven difficult. For example, female gametophyte development is known to be arrested at FG1 in the Gf, fem2, fem3, gfa4, and gfa5 mutants (Redei, 1965; Christensen et al., 1998; Durbarry et al., 2005), but the molecular lesions responsible for these phenotypes have not been identified. The identification of the rhf1a rhf2a double mutant, which is blocked at FG1 and PM I of gametogenesis, and the biochemical characterization of these two RING E3 genes provide a genetic and molecular basis for further defining the mechanisms of initiation and regulation of gametogenesis. Interestingly, the male and female gametophyte defects in the rhf1a rhf2a double mutant were incompletely penetrant and showed variable expressivity (Table 1). We have examined additional mutant lines from SALK (i.e., SALK_079844 for RHF1a and SALK_138317 for RHF2a) but unfortunately the transcription of either gene was not affected. One possible explanation for the genetic complexity is that the truncated proteins of RHF1a and RHF2a in the mutants we studied might have partial E3 ligase activities because we detected truncated transcripts of RHF1a and RHF2a. In both mutants, the two T-DNA insertion sites were located at the positions after the RING-finger domain, which is responsible for the E3 ligase activity. It is also possible that the variable silencing of gene transcription caused by the combination of the two 35S driven mutants might contribute to the phenotype variability in the double mutants (Daxinger et al., 2008).
In plants, meiosis is a conserved cell division that is essential for eukaryotic sexual reproduction, but the subsequent gametophytic mitoses are specific to plants. In general, mitotic cell cycle progression is achieved by regulating CDK activities with a suite of regulatory mechanisms, including controlled destruction of cyclins, phosphorylation and dephosphorylation of the CDKs, and degradation of inhibitory proteins (CDK inhibitors). Although some cell cycle–related proteins have been shown to play important roles in either male or female gametogenesis, for example, CDKA;1 (Iwakawa et al., 2006; Nowack et al., 2006) and NOMEGA (Kwee and Sundaresan, 2003), how the general regulatory machinery of the cell cycle specifically regulates gametogenesis is still far from clear. In this study, although the transcripts of RHF1a and RHF2a were detected in most developmental stages during gametogenesis, we found that the transcription of ICK4/KRP6 was tightly controlled and primarily concentrated in the sporophytes, right before the onset of mega- and microspore mitosis. Taken together with the evidence that degradation of ICK4/KRP6 by RHF1a/RHF2a is essential for gametophytic mitosis, these results indicate that ICK4/KRP6 is regulated not only at the protein level but also at the transcriptional level to define its spatial and temporal expression.
Because overexpression of ICK4/KRP6 leads to the arrest of embryo sac development at FG1, FG2, or FG4 and to pollen defects at PM I and PM II (Figure 6), it is likely that one of the targets of ICK4/KRP6 is CDKA;1 because mutations in CDKA;1 were shown to have defects in PM II. However, a mutation in CDKA;1 only led to defective male gametophytes arrested at PM II but did not affect the female gametophyte (Iwakawa et al., 2006; Nowack et al., 2006). It is likely that ICK4/KRP6 may have additional CDK targets. This hypothesis is consistent with the observation that all of the Arabidopsis ICK/KRP proteins can bind to active cyclin D2 (CYCD2)/CDKA or CYCD2/CDKB complexes with similar affinities and can inhibit their kinase activities (Nakai et al., 2006). Moreover, the meiosis-specific expression pattern of ICK4/KRP6 suggests that ICK4/KRP6 is somehow required during meiosis and that there might also be other CDKs involved in meiosis.
In mammals, the CDK inhibitor p27Kip1 was reported to regulate correct cell cycle progression and the integration of developmental signals with the core cell cycle machinery (Kato et al., 1994; Nourse et al., 1994; Polyak et al., 1994). Thus, p27Kip1 nullizygous mice are significantly larger than control mice because of an increase in the number of cells (Fero et al., 1996; Nakayama et al., 1996). In Arabidopsis, seven putative CDK inhibitors homologous to human p27Kip1 have been identified, the transcript accumulation of which are spatially diversified (De Veylder et al., 2001), suggesting that each of the ICK/KRP members might function in a tissue-specific manner. That the transcription of ICK4/KRP6 was turned off right before the subsequent gametophytic mitosis supports this hypothesis and implies that transcription of ICK4/KRP6 is not needed for the subsequent plant-specific mitoses. Taking into consideration that RHF1a and RHF2a are expressed throughout meiosis and mitosis during gametogenesis, it is reasonable to speculate that RHF1a/RHF2a-mediated 26S proteasome-dependent proteolysis may serve as a watchdog in Arabidopsis, to keep the level of meiosis-accumulated ICK4/KRP6 below a certain threshold, so that the subsequent mitoses can proceed smoothly. The variable amount of ICK4/KRP6 left over from meiosis, which may affect the subsequent mitoses to different extents, may also contribute to the incomplete penetrance and variable expressivity found in the rhf1a rhf2a double mutants. Why ICK4/KRP6 is predominantly expressed during meiosis and how the plant-specific RING E3 ligases (RHF1a/2a) coevolve with the targets ICKs/KRPs (ICK4/KRP6) to guarantee the formation of a plant-specific organ (gametophyte) are open questions and need to be further investigated.
In conclusion, our findings in this study establish that regulated degradation of the CDK inhibitor ICK4/KRP6 by two RING-type E3 ligases is essential for gametophytic mitosis in Arabidopsis.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia glabra served as the wild type. All the transgenic lines used in this study were in the Columbia ecotype. Plants were grown as previously reported (Qin et al., 2005). The rhf1a/rhf1a mutant (SALK_042445) and the rhf2a/rhf2a mutant (SALK_023787) were obtained from the ABRC. The insertion sites of the two SALK mutants were confirmed by sequencing.
Genotype Determination of the SALK T-DNA Insertion Mutants by PCR
The primers AbS1 and RHF1a-042445-R (see Supplemental Table 1 online for all primer sequences) were used to amplify an 1162-bp fragment containing the RHF1a wild-type allele. The rhf1a mutant allele was detected using the T-DNA–specific primer LBb1 and primers Abs2 or RHF1a-042445-R. The primers RHF2a-023787-F and RHF2a-023787-R were used to amplify an 856-bp fragment containing the wild-type RHF2a allele. The rhf2a mutant allele was detected using the T-DNA–specific primer LBb1 and primers RHF2a-023787-F or RHF2a-023787-R. Because two neighboring inverted T-DNAs were inserted in both of the rhf1a and rhf2a mutants, either combination of LBb1 and the forward or reverse primers could be used in the genotyping assays.
Vector Constructions and Arabidopsis Transformation
The cDNA of RHF1a was amplified from wild-type Arabidopsis by RT-PCR with the primers AbS1 and AbAS1 and cloned into the EcoRV site of pBluescript SK+, designated as pBS-RHF1a. 35S-RHF1a was constructed by the ligation of the XbaI/SalI-digested fragment from pBS-RHF1a and XbaI/XhoI-digested vector pJim19.
The cDNA of KRP6 was amplified from wild-type Arabidopsis by RT-PCR and cloned into pBluescript SK+. For the 35S construct, KRP6-NT-MYC-F2 and KRP6-NT-MYC-R2 were used to amplify and clone KRP6 into pBA002 plasmid with 6xmyc at the N terminus. For the LAT52 promoter construct, KRP6-LAT52-F and KRP6-LAT52-R were used to amplify and clone KRP6, together with LAT52 fragment digested from pBS-LAT52, into a pJim19 vector.
For the RNAi construct of KRP6, a 460-bp KRP6-specific fragment in the coding region, amplified with the primers KRP6-ISH-F and KRP6-ISH-R, was used in both sense and antisense orientations, which were then cloned into the binary vector pJim19 with a β-glucuronidase (GUS) fragment in the middle.
Arabidopsis transformation and transgenic plant screening were performed as previously reported (Qin et al., 2005).
Total RNA Isolation and Quantitative RT-PCR Analyses
To check the expression pattern of KRP6 in 35S-KRP6 independent transgenic plants through RT-PCR, total RNA was prepared from 20-DAG seedlings using a TRIZOL total RNA isolation reagent (Invitrogen). The cDNA synthesis and quantitative RT-PCR with the primers KRP6-RT-F1 and KRP6-RT-R1 were conducted as previously reported (Guo et al., 2006). Three biological repeats were examined and showed highly reproducible results.
Detection of Transcripts of RHF1a and RHF2a in the Mutants
The primer pairs RHF1a-salk-FN1/RHF1a-salk-RN1 and RHF1a-Salk-F2/RHF1a-Salk-R2 were used to amplify the fragments before and after the T-DNA insertion site in SALK_042445 mutant, respectively. The primers R1-F and R1-R were used to amplify the full-length fragment of RHF1a. The primer pairs RHF2a-Salk-F1/RHF2a-Salk-R1 and RHF2a-SALK-F2/RHF2a-SALK-R2 were used to amplify the fragments before and after the T-DNA insertion site in the SALK_023787 mutant, respectively. The primers R2-F and R2-R were used to amplify the full-length fragment of RHF2a. β-TUBULIN8 as a control was amplified with TUB8-5′ and TUB8-3′.
Confocal Laser Scanning Microscopy Analysis
Confocal observation of ovules was performed as previously described (Shi et al., 2005).
In Situ Hybridization
In situ hybridization experiments were performed as previously described with some modifications (Qu et al., 2003). Arabidopsis inflorescence tissues were collected from 40-d-old wild-type plants. The complete coding sequence of RHF1a was used as the probe. A 589-bp fragment in the coding region specific to RHF2a, amplified with the primers RHF2a-ISH-F and RHF2a-ISH-R, was used as the RHF2a probes. A 460-bp KRP6-specific fragment in the coding region, amplified with the primers KRP6-ISH-F and KRP6-ISH-R, was used as the KRP6 probes.
Pollen Observations Using Alexander's Staining, DAPI Staining, and DNA Content Analyses
For pollen analysis, pollen grains were mounted with Alexander's stain (Alexander, 1969). To visualize nuclei in pollen grains, pollen was processed as described by Park et al. (1998). Detached stamens were collected into a DAPI staining solution (0.1 M sodium phosphate, pH 7.0, 1 mM EDTA, 0.1% Triton X-100, and 0.5 μg/mL DAPI). The Alexander-stained slides and DAPI fluorescence were examined under a microscope (Olympus BX51) equipped with a CCD camera (Eastman Kodak).
Relative nuclear DNA content was determined from DAPI fluorescence. Relative fluorescence values were recorded with a fixed exposure and area of interest using IMAGEQUANT software (Molecular Dynamics). A net value for each nucleus was measured after subtraction of background taken from the corresponding cytoplasm. Fluorescence was normalized by comparison with sperm nuclei at telophase that contain 1C DNA.
In Vitro GST Pull-Down Assays and Protein Gel Blotting
RHF1a complete coding sequence was fused with 3×FLAG in pBluescript SK+, designated as pBS-RHF1a-FLAG. The related primers used were AbS1, RHF1a-R-FLAG, Flag-1, and Flag-2. The protein-expressing construct was made through ligation of two fragments, the XbaI/SacI-digested RHF1a-3×FLAG fused fragment from pBS-RHF1a-FLAG and the XbaI/SacI-digested vector pET23a. The FLAG fusion protein and empty GST (pGEX-5x-2) were expressed in Escherichia coli strain BL21 (DE3). The GST-fused KRP proteins (pDEST15-KRP1-3 and pDEST15-KRP5-7) were expressed in E. coli strain BL21-AI. For GST pull-down assays, the protocol previously described was adopted (Feng et al., 2008), except that anti-GST beads were used.
BiFC Assays
BiFC assays were performed as described previously (Walter et al., 2004). For generation of the BiFC vectors, the coding region of RHF1a was cloned via SmaI-KpnI into pUC-SPYCE, resulting in RHF1a-YFPC. The coding region of KRP6 was cloned via SmaI-KpnI into pUC-SPYNE, resulting in KRP6-YFPN. The constructs for fusion protein production were verified by sequencing. The onion cells were observed with a Leica TCS SPE confocal laser scanning microscope with a 514-nm laser.
Transient Expression Assay in Nicotiana benthamiana for Detection of Protein Degradation in Vivo
RHF1a and one of the KRP proteins or ABI5 were transiently expressed in N. benthamiana leaf cells. Agrobacterium tumefaciens cells containing the constructs with either one of the KRP members or ABI5 coding sequence fused with myc at the N terminus driven by a CaMV 35S promoter and the pJim vector constructed with RHF1a with FLAG at the C terminus were coinfiltrated with pCam-P19 into N. benthamiana leaves as previously described (English et al., 1997; Voinnet et al., 2003). Each sample was coinfiltrated with the 35S-HA-GFP construct as an internal control. After incubation for 3 d, the infiltrated parts of leaves were subjected to protein extraction and protein gel blots. Total protein extracts were obtained from leaf tissues by grinding in liquid nitrogen and then suspension in 2 volumes of extraction buffer (50 mM sodium phosphate, 200 mM NaCl, 10 mM MgCl2, 10% glycerol, and protease inhibitor cocktail tablets) on ice. The total extract was centrifuged at 15,000g at 4°C for 30 min, and aliquots of supernatant were added with SDS-PAGE loading buffer for protein gel analysis. Buffers and the procedure were as described (Zhang et al., 2007). The protein-transferred filter was incubated in blocking solution (5% skim milk in PBS buffer) for 3 h at room temperature and then in 2.5% skim milk PBS blocking solution containing either anti-FLAG (Sigma-Aldrich), anti-HA (Santa Cruz Biotechnology), or anti-MYC (Santa Cruz Biotechnology) overnight at 4°C. After the incubation with the corresponding antibody, the filter was incubated with anti-mouse IgG HRP conjugate (Promega; 1:2500) in a PBS solution of 2.5% skim milk. After three washes, the filter was visualized using chemiluminescence according to the manufacturer's instructions (ECL; Amersham Pharmacia). For MG132 treatment, 40 μM MG132 was injected into the tobacco leaves 12 to 16 h before sample collection.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RHF1a (At4g14220), RHF2a (At5g22000), ICK1/KRP1 (At2g23430), ICK2/KRP2 (At3g50630), ICK6/KRP3 (At5g48820), ICK7/KRP4 (At2g32710), ICK3/KRP5 (At3g24810), ICK4/KRP6 (At3g19150), and ICK5/KRP7 (At1g49620).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Amino Acid Sequence Alignment of RHF1a and RHF2a with Their Homologs in Other Plant Species.
Supplemental Figure 2. Expression Analysis of RHF1a and RHF2a in Wild-Type Inflorescence via Quantitative RT-PCR.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Sheila McCormick (University of California, Berkeley, CA) for revising the manuscript and De Ye (China Agriculture University, Beijing, China), Hong Ma (Penn State University, University Park, PA), Xing-Wang Deng (Yale University, New Haven, CT), and Yunde Zhao (University of California, San Diego, CA) for helpful suggestions and valuable discussions. We also thank Nam-Hai Chua (Rockefeller University, New York, NY) for the vector pBA002-myc. The work was supported by the National Key Basic Research Program of People's Republic of China (973-2007CB108702) and the National Natural Science Foundation of China (NSFC Grant 30625002 to L.-J.Q. and NSFC-JSPS 30711140112 to L.-J.Q. and T.T.). This work was partially supported by the 111 Project, an NSFC grant (30530400 to Q.X.), the MOESCST of Japan (Grant 19370019 to M.U.), the Heiwa Nakajima Foundation (International Research Grant 2006 to T.T. and L.-J.Q.), the Kyoto University Foundation (Fellowship Grant 2007 to T.T. and L.-J.Q.), and the Program for Promotion of Basic Research Activities for Innovative Biosciences to M.U.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Li-Jia Qu (qulj@pku.edu.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. [DOI] [PubMed] [Google Scholar]
- Barroco, R.M., De Veylder, L., Magyar, Z., Engler, G., Inzé, D., and Mironov, V. (2003). Novel complexes of cyclin-dependent kinases and a cyclin-like protein from Arabidopsis thaliana with a function unrelated to cell division. Cell. Mol. Life Sci. 60 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye, K.M., Goderis, I.J., Wouters, P.F., Pues, J.M., Delaure, S.L., Broekaert, W.F., Depicker, A., Cammue, B.P., and De Bolle, M.F. (2004). Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. Plant J. 39 440–449. [DOI] [PubMed] [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202 136–151. [DOI] [PubMed] [Google Scholar]
- Daxinger, L., Hunter, B., Sheikh, M., Jauvion, V., Gasciolli, V., Vaucheret, H., Matzke, M., and Furner, I. (2008). Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 13 4–6. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T., Krols, L., Terras, F., Landrieu, I., van der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13 1653–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., and Inzé, D. (2007). The ins and outs of the plant cell cycle. Nat. Rev. Mol. Cell Biol. 8 655–665. [DOI] [PubMed] [Google Scholar]
- Dong, C.H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J.K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. USA 103 8281–8286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbarry, A., Vizir, I., and Twell, D. (2005). Male germ line development in Arabidopsis. duo pollen mutants reveal gametophytic regulators of generative cell cycle progression. Plant Physiol. 137 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady, C., Lindsey, K., and Twell, D. (1995). The significance of microspore division and division symmetry for vegetative cell-specific transcription and generative cell differentiation. Plant Cell 7 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J.J., Davenport, G.F., Elmayan, T., Vaucheret, D., and Baulcombe, C. (1997). Requirement of sense transcription for homology-dependent virus resistance and trans-inactivation. Plant J. 12 597–603. [Google Scholar]
- Feldmann, K.A., Coury, D.A., and Christianson, M.L. (1997). Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fero, M.L., Rivkin, M., Tasch, M., Porter, P., Carow, C.E., Firpo, E., Polyak, K., Tsai, L.H., Broudy, V., Perlmutter, R.M., Kaushansky, K., and Roberts, J.M. (1996). A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27Kip1_deficient mice. Cell 85 733–744. [DOI] [PubMed] [Google Scholar]
- Friedman, W.E. (1999). Expression of the cell cycle in sperm of Arabidopsis: Implications for understanding patterns of gametogenesis and fertilization in plants and other eukaryotes. Development 126 1065–1075. [DOI] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., and Schneitz, K. (1998). The molecular and genetic basis of ovule and megagametophyte development. Semin. Cell Dev. Biol. 9 227–238. [DOI] [PubMed] [Google Scholar]
- Guo, L., Wang, Z.Y., Lin, H., Cui, W.E., Chen, J., Liu, M., Chen, Z.L., Qu, L.J., and Gu, H. (2006). Expression and functional analysis of the rice plasma-membrane intrinsic protein gene family. Cell Res. 16 277–286. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. (2000). HY5 stability and activity in arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst, L. (2004). A second RING to destroy p27Kip1. Nat. Cell Biol. 6 1153–1155. [DOI] [PubMed] [Google Scholar]
- Huntley, R.P., and Murray, J.A. (1999). The plant cell cycle. Curr. Opin. Plant Biol. 2 440–446. [DOI] [PubMed] [Google Scholar]
- Inzé, D., and Veylder, L.D. (2006). Cell cycle regulation in plant development. Annu Rev Genet. 40 77–105. [DOI] [PubMed]
- Iwakawa, H., Shinmyo, A., and Sekine, M. (2006). Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 45 819–831. [DOI] [PubMed] [Google Scholar]
- Jakoby, M.J., Weinl, C., Pusch, S., Kuijt, S.J., Merkle, T., Dissmeyer, N., and Schnittger, A. (2006). Analysis of the subcellular localization, function, and proteolytic control of the Arabidopsis cyclin-dependent kinase inhibitor ICK1/KRP1. Plant Physiol. 141 1293–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T., Hara, T., Matsumoto, M., Ishida, N., Okumura, F., Hatakeyama, S., Yoshida, M., Nakayama, K., and Nakayama, K.I. (2004). Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27Kip1 at G1 phase. Nat. Cell Biol. 6 1229–1235. [DOI] [PubMed] [Google Scholar]
- Kato, J.Y., Matsuoka, M., Polyak, K., Massague, J., and Sherr, C.J. (1994). Cyclic AMP-induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell 79 487–496. [DOI] [PubMed] [Google Scholar]
- Kwee, H.S., and Sundaresan, V. (2003). The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the Anaphase Promoting Complex in Arabidopsis. Plant J. 36 853–866. [DOI] [PubMed] [Google Scholar]
- McCormick, S. (1993). Male gametophyte development. Plant Cell 5 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick, S. (2004). Control of male gametophyte development. Plant Cell 16(Suppl): S142–S153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, V.V., De Veylder, L., Van Montagu, M., and Inzé, D. (1999). Cyclin-dependent kinases and cell division in plants - The nexus. Plant Cell 11 509–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, T., Kato, K., Shinmyo, A., and Sekine, M. (2006). Arabidopsis KRPs have distinct inhibitory activity toward cyclin D2-associated kinases, including plant-specific B-type cyclin-dependent kinase. FEBS Lett. 580 336–340. [DOI] [PubMed] [Google Scholar]
- Nakayama, K. (1998). Cip/Kip cyclin-dependent kinase inhibitors: Brakes of the cell cycle engine during development. Bioessays 20 1020–1029. [DOI] [PubMed] [Google Scholar]
- Nakayama, K., Ishida, N., Shirane, M., Inomata, A., Inoue, T., Shishido, N., Horii, I., and Loh, D.Y. (1996). Mice lacking p27Kip1 display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85 707–720. [DOI] [PubMed] [Google Scholar]
- Nourse, J., Firpo, E., Flanagan, W.M., Coats, S., Polyak, K., Lee, M.H., Massague, J., Crabtree, G.R., and Roberts, J.M. (1994). Interleukin-2-mediated elimination of the p27Kip1 cyclin-dependent kinase inhibitor prevented by rapamycin. Nature 372 570–573. [DOI] [PubMed] [Google Scholar]
- Nowack, M.K., Grini, P.E., Jakoby, M.J., Lafos, M., Koncz, C., and Schnittger, A. (2006). A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat. Genet. 38 63–67. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405 462–466. [DOI] [PubMed] [Google Scholar]
- Park, S.K., Howden, R., and Twell, D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125 3789–3799. [DOI] [PubMed] [Google Scholar]
- Polyak, K., Lee, M.H., Erdjument-Bromage, H., Koff, A., Roberts, J.M., Tempst, P., and Massague, J. (1994). Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell 78 59–66. [DOI] [PubMed] [Google Scholar]
- Qin, G., Gu, H., Zhao, Y., Ma, Z., Shi, G., Yang, Y., Pichersky, E., Chen, H., Liu, M., Chen, Z., and Qu, L.J. (2005). An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell 17 2693–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.J., et al. (2003). Molecular cloning and functional analysis of a novel type of Bowman-Birk inhibitor gene family in rice. Plant Physiol. 133 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redei, G.P. (1965). Non-Mendelian megagametogenesis in Arabidopsis. Genetics 51 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, H., Santner, A., Del Pozo, J.C., Murray, J.A., and Estelle, M. (2008). Degradation of the cyclin-dependent kinase inhibitor KRP1 is regulated by two different ubiquitin E3 ligases. Plant J. 53 705–716. [DOI] [PubMed] [Google Scholar]
- Shi, D.Q., Liu, J., Xiang, Y.H., Ye, D., Sundaresan, V., and Yang, W.C. (2005). SLOW WALKER1, essential for gametogenesis in Arabidopsis, encodes a WD40 protein involved in 18S ribosomal RNA biogenesis. Plant Cell 17 2340–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Hauksdottir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137 13–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Williams, L.A., Farmer, L.M., Vierstra, R.D., and Callis, J. (2006). KEEP ON GOING, a RING E3 Ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda, K., Kubota, Y., and Kato, J. (1999). Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 398 160–165. [DOI] [PubMed] [Google Scholar]
- Twell, D., Park, S.K., Hawkins, T.J., Schubert, D., Schmidt, R., Smertenko, A., and Hussey, P.J. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4 711–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., and McCormick, S. (1990). Pollen-specific gene expression in transgenic plants: coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109 705–713. [DOI] [PubMed] [Google Scholar]
- Verma, R., Annan, R.S., Huddleston, M.J., Carr, S.A., Reynard, G., and Deshaies, R.J. (1997). Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science 278 455–460. [DOI] [PubMed] [Google Scholar]
- Vlach, J., Hennecke, S., and Amati, B. (1997). Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 16 5334–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33 949–956. [DOI] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Wang, H., Zhou, Y., Gilmer, S., Whitwill, S., and Fowke, L.C. (2000). Expression of the plant cyclin-dependent kinase inhibitor ICK1 affects cell division, plant growth and morphology. Plant J. 24 613–623. [DOI] [PubMed] [Google Scholar]
- Weinl, C., Marquardt, S., Kuijt, S.J., Nowack, M.K., Jakoby, M.J., Hulskamp, M., and Schnittger, A. (2005). Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. Plant Cell 17 1704–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.H. (2002). SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419 167–170. [DOI] [PubMed] [Google Scholar]
- Yadegari, R., and Drews, G.N. (2004). Female gametophyte development. Plant Cell 16(Suppl): S133–S141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W.C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3 53–57. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Garreton, V., and Chua, N.H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Yang, C., Li, Y., Zheng, N., Chen, H., Zhao, Q., Gao, T., Guo, H., and Xie, Q. (2007). SDIR1 is a RING Finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19 1912–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., Fowke, L.C., and Wang, H. (2002). Plant CDK inhibitors: Studies of interactions with cell cycle regulators in the yeast two-hybrid system and functional comparisons in transgenic Arabidopsis plants. Plant Cell Rep. 20 967–975. [Google Scholar]
- Zhou, Y., Li, G., Brandizzi, F., Fowke, L.C., and Wang, H. (2003. b). The plant cyclin-dependent kinase inhibitor ICK1 has distinct functional domains for in vivo kinase inhibition, protein instability and nuclear localization. Plant J. 35 476–489. [DOI] [PubMed] [Google Scholar]
- Zhou, Y., Wang, H., Gilmer, S., Whitwill, S., and Fowke, L.C. (2003. a). Effects of co-expressing the plant CDK inhibitor ICK1 and D-type cyclin genes on plant growth, cell size and ploidy in Arabidopsis thaliana. Planta 216 604–613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.