Abstract
Stem cell fate in the Arabidopsis thaliana shoot apical meristem (SAM) is controlled by WUSCHEL (WUS) and CLAVATA. Here, we examine BARD1 (for BRCA1-associated RING domain 1), which had previously been implicated in DNA repair functions; we find that it also regulates WUS expression. We observed severe SAM defects in the knockout mutant bard1-3. WUS transcripts accumulated >238-fold in bard1-3 compared with the wild type and were located mainly in the outermost cell layers instead of the usual organizing center. A specific WUS promoter region was recognized by nuclear protein extracts obtained from wild-type plants, and this protein-DNA complex was recognized by antibodies against BARD1. The double mutant (wus-1 bard1-3) showed prematurely terminated SAM structures identical to those of wus-1, indicating that BARD1 functions through regulation of WUS. BARD1 overexpression resulted in reduced WUS transcript levels, giving a wus-1–like phenotype. Either full-length BARD1 or a clone that encoded the C-terminal domain (BARD1:C-ter;bard1-3) was sufficient to complement the bard1-3 phenotype, indicating that BARD1 functions through its C-terminal domain. Our data suggest that BARD1 regulates SAM organization and maintenance by limiting WUS expression to the organizing center.
INTRODUCTION
The ability of flowering plants to continuously produce new organs depends on the activity of stem cell pools, which are located close to the tip of the meristem (Mayer et al., 1998). WUSCHEL (WUS) is a key gene involved in positioning the stem cells and is essential for organization and maintenance of the shoot apical meristem (SAM) (Laux et al., 1996; Schoof et al., 2000; Muller et al., 2006). In Arabidopsis thaliana, activation of the CLAVATA3 (CLV3)–dependent signaling pathway reduces the rate of stem cell proliferation and enhances organ initiation via a feedback loop that inhibits WUS expression (Clark et al., 1997; Brand et al., 2000; Schoof et al., 2000; Muller et al., 2006). Ectopic expression of a WUS transgene in Arabidopsis induces shoot stem cell activity in root and floral meristems on the mature stem surface (Gallois et al., 2004; Xu et al., 2005). Transgenic Arabidopsis plants expressing a cauliflower mosaic virus 35S promoter (CaMV35S)∷WUS construct showed severe growth inhibition and substantially reduced cotyledon expansion and greening (Brand et al., 2002; Lenhard et al., 2002; Kieffer et al., 2006). A similar interaction between WUS and AGAMOUS operates during Arabidopsis flower development (Lenhard et al., 2001; Lohmann et al., 2001). Recently, WUSCHEL-RELATED HOMEOBOX5 (WOX5), a homolog of WUS that is expressed specifically in the quiescent center of the root, was found to serve as the root stem cell organizer (Sarkar et al., 2007).
WUS might function as a repressor of transcription in concert with the groucho-type corepressor protein TOPLESS (TPL), which functions by recruiting gene silencing machinery such as histone deacetylase 19 (Long et al., 2006). The C-terminal domain of both Arabidopsis WUS and its Antirrhinum majus ortholog ROSULATA binds the TPL protein (Kieffer et al., 2006). In the Arabidopsis tpl-1 mutant, the embryonic shoot apex is transformed into a second root pole, and WUS is expressed normally until globular-stage embryos but is totally absent in the transition stage that produces two root axes (Long et al., 2006). In the Arabidopsis mutant I28, in which WUS and CLV3 expression is abolished as the result of a mutation in the transcription factor APETALA2, the shoot meristem was prematurely terminated (Wurschum et al., 2006). Expression of HANABA TARANU, which encodes a GATA-3–like transcription factor, in vascular tissues and cells separating the meristem from organ primordia controls the number and the correct positioning of WUS-expressing cells (Zhao et al., 2004; Tucker and Laux, 2007).
Upon execution of a large-scale promoter scanning experiment, Baurle and Laux (2005) found that a 57-bp cis-element including an HD-ZIP consensus binding site–like motif located in the distal (−530 to −586) promoter region of the WUS gene provides all the spatial and temporal information necessary for WUS transcription in the stem cell niche. WUS transcription is also modulated through direct binding of SPLAYED (SYD), a SNF2 chromatin-remodeling ATPase, to its proximal (−435 to −70) promoter region. In a chromatin immunoprecipitation assay that used polyclonal antibodies raised against the N-terminal domain of SYD, this proximal region was highly represented, but a distal region (−1664 to −1348) and the transcribed region were not (Kwon et al., 2005).
BARD1 (At1g04020) encodes a protein containing two tandem BRCA1 C-Terminal (BRCT) domains, which function in phosphorylation-dependent protein–protein interactions (Glover et al., 2004; Narod and Foulkes, 2004; Williams et al., 2004), and a RING domain, and it is located on Arabidopsis chromosome 1; BARD1 reportedly is involved in DNA repair (Reidt et al., 2006). A similar gene on chromosome 4 (At4g21070) with almost identical BRCT and RING domain structures was named At BRCA1 after the original human breast cancer–associated gene 1 (BRCA1) (Lafarge and Montane, 2003). Human BRCA1 and BARD1, the BRCA1-associated ring domain 1 protein, can form heterodimers through their common N-terminal RING domains and together function as tumor suppressors. These proteins have been implicated in many processes, including cell cycle control, DNA repair, recombination, and transcriptional regulation (Wu et al., 1996; Baer and Ludwig, 2002; Irminger-Finger and Jefford, 2006). BARD1 is indispensable for cell viability in mammals because loss-of-function mutations result in early embryonic lethality (Irminger-Finger and Jefford, 2006). To elucidate the mode of BARD1 function in Arabidopsis, we obtained all three T-DNA insertion lines that disrupted its expression from the SALK collections. Here, we provide molecular and genetic evidence to show that BARD1 mutations cause severe SAM defects in Arabidopsis by releasing WUS expression from its normal confinement to the organizing center (OC), allowing its expression to spread to the outermost cell layers in the SAM.
RESULTS
Identification of Homozygous BARD1 Knockout Mutant Lines
Three Arabidopsis mutant lines with disrupted BARD1 were obtained from the SALK collection; T-DNA insertions were found in the first intron and in the third exon (SALK_097601 and SALK_031862 lines or bard1-1 and bard1-2, respectively) and also in the last exon (SALK_003498 line or bard1-3) (see Supplemental Figure 1A online). We identified plants homozygous for bard1-3, with a single T-DNA insertion that completely blocked BARD1 expression (see Supplemental Figures 1B to 1D online). Confirming that the phenotype is specific to the BARD1 locus, RT-PCR analysis did not reveal a significant shift in the expression pattern of 10 putative open reading frames (ORFs) located adjacent to BARD1 on the chromosome, either upstream or downstream of the bard1-3 mutation (see Supplemental Figure 1E online). Because the bard1-1 and bard1-2 alleles had previously been implicated in DNA repair (Reidt et al., 2006), we tested bard1-3 for the phenotype. The UV-C recovery assay (see Supplemental Figure 2A online) and terminal transferase dUTP nick end labeling (TUNEL)–based in situ cell death analysis (see Supplemental Figure 2B online) revealed obvious defects in DNA repair in bard1-3 seedlings.
Phenotypic Characterization of the bard1-3 Mutant
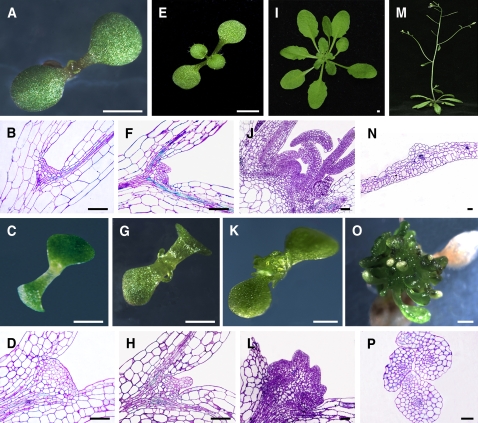
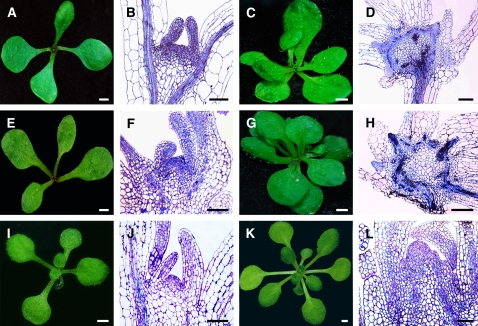
Severe developmental defects in plant architecture, especially in SAM organization, were observed in homozygous bard1-3 seedlings (Figure 1). At 5 d after germination (DAG), SAMs of the wild type and mutant were not fundamentally different, except that the apical tissue of bard1-3 was somewhat enlarged (Figures 1A to 1D). Significant differences were observed in seedlings at 7 DAG or older. Instead of the well-organized tunica-corpus structure observed in wild-type sections (Figures 1F and 1J), one or more irregular primordia with compact and presumably actively dividing cell populations was observed in mutant apical tissue, as indicated by condensed toluidine blue staining (Figures 1H and 1L). Also, in contrast with wild-type leaves (Figure 1N), a large number of tubular and finger-like structures was produced on the mutant (Figures 1O and 1P), indicating that proper BARD1 function is required for the development of leaf dorsoventrality in Arabidopsis.
Figure 1.
Phenotypic Characterization of the bard1-3 Arabidopsis Mutant.
Wild-type Columbia (Col-0) plants are shown in the top two rows, and bard1-3 plants are shown in the bottom two rows. (A), (E), (I), and (M) were photographed at the same growth period with panels (C), (G), (K), and (O), respectively. White bars = 1 mm; black bars = 50 μm.
(A) to (D) Seedlings 5 DAG.
(E) to (H) One-week-old seedlings.
(I) to (L) Three-week-old plants.
(M) and (O) Five-week-old plants.
(A), (E), (I), and (M) Photographs of wild-type Col-0 plants.
(C), (G), (K), and (O) Photographs of the bard1-3 mutant.
(B), (F), and (J) Median longitudinal semithin sections stained with toluidine blue showing wild-type SAMs at corresponding growth stages.
(D), (H), and (I) Sections of bard1-3 mutant SAMs at growth stages described above.
(N) Cross section of a mature wild-type leaf.
(P) Cross section of the leaf-like structures from the 5-week-old bard1-3 mutant.
In the bard1-3 mutant, the elongation growth of primary roots, but not root hairs, was also significantly inhibited (see Supplemental Figure 3C online and the wild type in Supplemental Figure 3A online). Similar to what was observed in the SAM of the mutant, its root apical meristem and all the lateral root meristems were significantly enlarged, with huge cell masses (see Supplemental Figures 3E and 3G online). Lugol's staining (Sarkar et al., 2007) revealed that no starch granules and no columella cells were present in the root tips of the mutant (see Supplemental Figures 3B and 3D online), while 4′,6-diamidino-2-phenylindole (DAPI) staining showed the existence of large quantities of undifferentiated cells in the meristems (see Supplemental Figures 3F and 3H online). These results indicated that the bard1-3 mutation affected the development of both the SAM and the root apical meristem during the embryo stage.
Identification of WUS as a Molecular Target of BARD1
To determine the molecular target of BARD1, we searched the Genevestigator database (Zimmermann et al., 2004) and looked for genes that were specifically expressed in the same proliferating cell types as BARD1. A total of 25 genes showed expression ratios >10 (comparing transcript level in dividing tissues, either shoot apex, embryo, or radicles, to ground tissues, either leaves, stems, or roots). Extensive RT-PCR analyses showed that, of the whole set of candidate genes, WUS was the most significantly upregulated in the bard1-3 background (see Supplemental Figure 4A online). STM, CLV3, WOX2, WOX5, and CUC1 expression was also significantly, but not as dramatically, upregulated in the mutant (see Supplemental Figure 4A online). Quantitative RT-PCR (QRT-PCR) data, represented as fold changes relative to the wild-type values, are reported beneath the respective RT-PCR data for those candidate genes that showed significantly different transcript levels in the mutant by visual inspection (see Supplemental Figure 4A online). Wild-type levels of BARD1 transcripts were found in the wus-1 mutant (see Supplemental Figure 4B online), indicating that WUS does not regulate BARD1 expression levels in Arabidopsis.
In Situ and QRT-PCR Analyses of WUS and BARD1 Expression in Wild-Type and bard1-3 Mutants
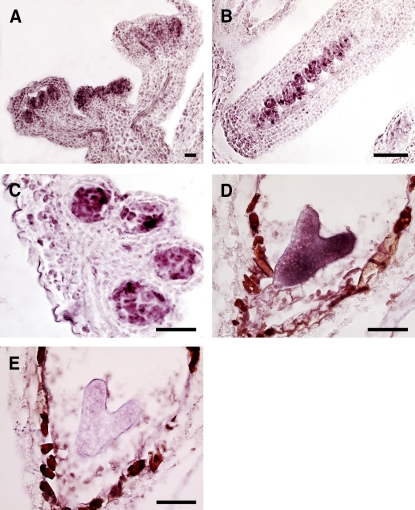
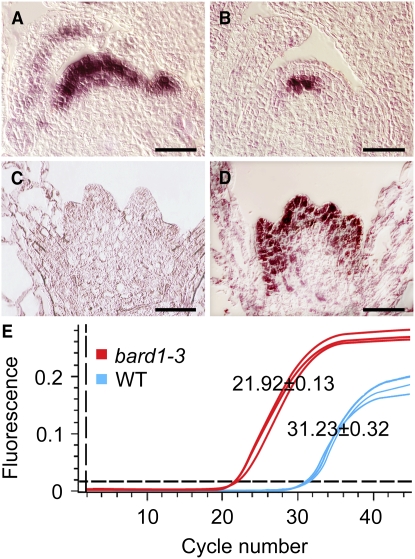
A series of in situ RNA hybridization experiments indicated that BARD1 is expressed specifically in the apical domains of Arabidopsis inflorescence (Figure 2A), ovules (Figure 2B), anthers (Figure 2C), and embryos (Figure 2D). Hybridization results using a sense BARD1 probe showed that the heavy staining in the surrounding seed coat of the developing embryo was not due to BARD1 mRNA (Figure 2E). When very young seedlings were analyzed, BARD1 transcripts localized mainly in the outermost three to four cell layers of the main shoot apex in wild-type seedlings and in developing leaf primordia and young leaves (Figure 3A), whereas WUS was localized specifically in the OC (Figure 3B). In bard1-3 mutant seedlings, in which BARD1 mRNA was not detectable (Figure 3C), a very strong WUS signal was observed in the outermost cell layers of multiple apical primordia–like structures (Figure 3D). QRT-PCR analysis using RNA samples prepared from whole young shoots indicated that WUS transcripts were 238.0 ± 12.3-fold higher (a difference of 9.31 ± 0.35 cycles) in the mutant relative to the wild type (Figure 3E).
Figure 2.
Tissue-Specific Expression of BARD1 in Wild-Type Arabidopsis Plants.
In situ hybridization experiments using an antisense BARD1 probe were performed on Arabidopsis inflorescence (A), carpels (B), anthers (C), and the heart-shaped-stage embryo (D). The same stage developing embryo shown in (D) was probed with a sense BARD1 sequence (E) to demonstrate the specificity of our hybridization experiments. Bars = 50 μm.
Figure 3.
WUS Transcripts Are Detected in the Outermost Cell Layers, Instead of the Usual OC, in bard1-3 SAMs.
(A) and (B) In situ hybridization of BARD1 and WUS, respectively, in wild-type Arabidopsis.
(C) and (D) In situ hybridization of BARD1 and WUS, respectively, in bard1-3. Bars = 20 μm.
(E) QRT-PCR analysis of WUS mRNA levels in the wild type and the bard1-3 mutant. Mean (±se) comparative threshold (CT) values were calculated from triplicate QRT-PCR experiments using independent RNA samples prepared from different batches of Arabidopsis plants.
The BARD1 expression pattern was further studied by producing transgenic Arabidopsis plants that expressed a β-glucuronidase (GUS) construct driven by a 1.91-kb 5′ BARD1 upstream promoter fragment (see Supplemental Figure 5 online). When cleared whole seedlings were assayed, strong GUS activity was observed in the shoot apex, including the developing leaf primordia (see Supplemental Figure 5A online). When longitudinal sections of Arabidopsis shoot apices were stained, GUS activity was localized mainly in the apical layers of the SAM (see Supplemental Figure 5B online), similar to the in situ hybridization results reported in Figure 3A. Strong GUS activity was also observed mainly in the meristem zone of primary roots (see Supplemental Figure 5C online) and in the initiation site of lateral roots (see Supplemental Figure 5D online). We also constructed a GUS construct driven by a fragment of the WUS promoter, but we were unable to produce WUS:GUS transgenic Arabidopsis on a bard1-3 genetic background because the homozygous mutant was seedling lethal, and no WUS:GUS bard1-3 seedlings were obtained even by a genetic cross using heterozygous BARD1/bard1-3 plants.
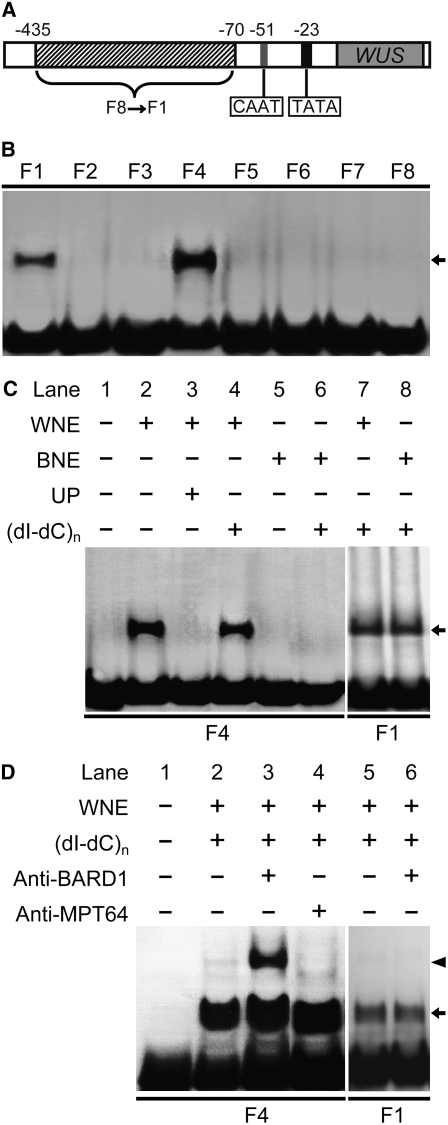
BARD1 Is Involved in the Formation of a Protein-DNA Complex with Specific Regions on the WUS Promoter
Because BARD1 activity seemed to repress WUS expression, we used a gel shift assay to study possible molecular interactions between BARD1 and WUS using part of the WUS promoter (Kwon et al., 2005) as depicted in Figure 4A. Significant mobility shifts were observed when fragments 1 or 4 (F1 or F4) were incubated with wild-type nuclear extract (Figure 4B). The DNA-bound protein complex specifically appeared after the addition of nuclear extract prepared from wild-type Arabidopsis seedlings but not from bard1-3 mutants (Figure 4C, lanes 2 and 4). Formation of the DNA-protein complex was blocked upon addition of specific unlabeled competitor probes but not when the nonspecific competitor poly(dI-dC) was added (Figure 4C, lanes 3 and 4), demonstrating sequence-specific binding. The protein-bound complex on F1 was most likely not related to BARD1 because similar binding was observed using wild-type or mutant nuclear extracts (Figure 4C, lanes 7 and 8). When a polyclonal antiserum to BARD1 was included in the binding reaction together with wild-type nuclear extract, another band with a higher molecular mass appeared (Figure 4D, lane 3), suggesting that BARD1 is part of the DNA-protein complex. This supershifted band was not observed when antiserum against a nonspecific bacterial protein was included (Figure 4D, lane 4). The complex on F1 did not react with anti-BARD1 (Figure 4D, lanes 5 and 6).
Figure 4.
BARD1 Forms a Specific Complex on Fragment 4 of the WUS Promoter.
(A) Schematic diagram showing the WUS promoter region used for gel shift assays. Fragments: F1, −70 ∼ −125; F2, −116 ∼ −170; F3, −161 ∼ −215; F4, −206 ∼ −260; F5, −251 ∼ −305; F6, −296 ∼ −350; F7, −341 ∼ −395; F8, −386 ∼ −435 bp.
(B) Nuclear extracts prepared from wild-type plants form binding complexes with F1 and F4. The arrows in (B) to (D) indicate the DNA-protein complex.
(C) The DNA-bound protein complex on F4 only appears after adding wild-type nuclear extracts but does not form after adding bard1-3 extract. The protein complex on F1 is not specific. WNE, 10 μg wild-type nuclear extract; BNE, 10 μg bard1-3 mutant nuclear extract; UP, 50-fold unlabeled probe; (dI-dC)n, 0.5 μg poly(dI-dC).
(D) The protein-bound DNA complex on F4 is recognized by BARD1 antiserum, whereas that on F1 is not. Anti-BARD1, 20 nmol of polyclonal antibody against the C-terminal region of BARD1; anti-MPT64, 20 nmol of polyclonal antibody against MPT64 (Cai et al., 2005). The arrowhead indicates the DNA-protein-antibody complex.
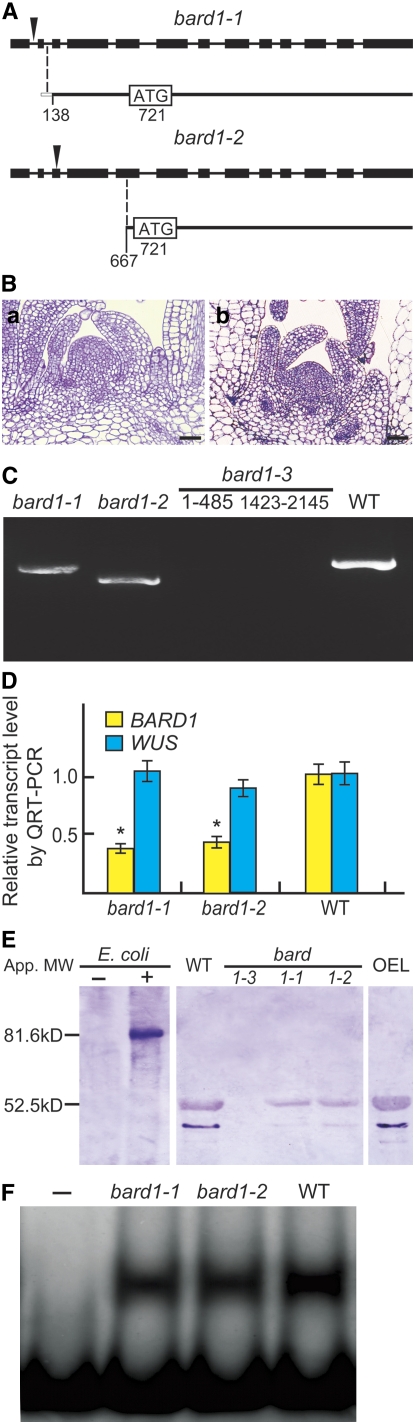
Truncated mRNAs Encoding Potentially Functional Polypeptides Are Found in Both bard1-1 and bard1-2 Mutants
To examine the differences between bard1-1/bard1-2 and bard1-3, we performed detailed sequence analyses using genomic DNA from wild-type Arabidopsis plants. Putative ORFs were found in bard1-1 and bard1-2, downstream of the T-DNA insertions (Figure 5A). We further sequenced the longest transcripts after repeated 5′ rapid amplification of cDNA ends (RACE) experiments using RNA templates from either bard1-1 or bard1-2 plants to find new potential transcription start sites (Figure 5A). The longest RNA from bard1-1 included 19 nucleotides of the second intron at its 5′ end with all other intron sequences removed in the mature mRNA. The longest RNA from bard1-2 started from the middle of the original exon 5 with no intron sequences attached (Figure 5A). Semithin sections of shoot meristems of bard1-1 and bard1-2 are shown in Figure 5B. Apart from the fact that both mutants were smaller and developed slower than the wild type, no other defects were observed. Full-length (2145 nucleotides) mRNA from the wild type and truncated mRNAs from bard1-1 (2027 nucleotides) and bard1-2 (1479 nucleotides) were successfully amplified, whereas no product was found in bard1-3 using primer pairs derived either from the 5′ (1 to 485 nucleotides) or 3′ (1423 to 2145 nucleotides) regions for up to 35 PCR cycles (Figure 5C). QRT-PCR indicated that bard1-1 and bard1-2 mRNA levels were 39 and 46%, respectively, of the wild-type level (Figure 5D). No significant increase in WUS expression was detected in these two mutants, indicating that the C-terminal part of BARD1 is sufficient to repress WUS expression (Figure 5D).
Figure 5.
Molecular Analyses of Different Mutant Lines.
(A) Analysis of bard1-1 and bard1-2 transcripts. The 5′ RACE experiments were performed on RNA templates derived from bard1-1 or bard1-2. Inverted triangles indicate the sites of the T-DNA insertions, black boxes denote exons, and the thin lines denote introns.
(B) Median longitudinal semithin sections stained with toluidine blue showing the SAMs of 3-week-old bard1-1 (left) and bard1-2 (right). Bars = 20 μm.
(C) RT-PCR analysis of BARD1 RNAs. Full-length RNA from wild-type and truncated RNAs from bard1-1 and bard1-2 are shown. Two primer pairs derived from 5′ (1 to 485 nucleotides) and 3′ (1423 to 2145 nucleotides) regions were used to amplify bard1-3.
(D) Comparison of WUS and BARD1 mRNA levels in wild-type, bard1-1, 1-2, and 1-3 plants by QRT-PCR. Relative transcript levels (mean ± se) were calculated from triplicate QRT-PCR reactions of independent RNA samples prepared from different batches of 3-week-old Arabidopsis plants. Both WUS and BARD1 mRNA levels in the wild type were arbitrarily set to 1. *, P < 0.05 compared with the wild type.
(E) Protein gel blotting showing the presence or absence of anti-BARD1-reactive proteins in various Arabidopsis lines, as well an E. coli strain expressing a full-length ORF of BARD1. Lanes were loaded with total protein (20 μg) extracted from 3-week-old Arabidopsis lines and 10 μg total E. coli extract before (–) and after (+) IPTG induction. OEL, a protein sample was prepared from BARD1 overexpressing line (CaMV35S∷BARD1).
(F) Nuclear extracts prepared from bard1-1, bard1-2 and wild-type plants retained similar ability to form a protein-DNA complex with F4 from the WUS promoter. Nuclear extract (10 μg) obtained from 2-week-old bard1-1, bard1-2, or wild-type plants was incubated with the DNA fragment and subjected to nondenaturing gel electrophoresis as described in Methods. –, no nuclear extract was added.
Protein gel blotting experiments showed that our polyclonal antibody recognized BARD1 specifically because a single band with an apparent molecular mass similar to that calculated from the vector (79.7 kD for BARD1 + 2.6 kD for the tag) was observed in the lane containing total Escherichia coli protein after (+) but not before (–) isopropylthio-β-d-galactoside (IPTG) inductions (Figure 5E). One major band (apparent molecular mass of 52.5 kD) along with a weaker band (a presumptive degradation product of 43.4 kD) was observed for total protein prepared from the wild type and bard1-1 and bard1-2 but not from that of bard1-3 (Figure 5E). Full-length BARD1 protein was not detected even from the BARD1+overexpressing line (wild-type Arabidopsis plants that expressed a CaMV35S∷BARD1 construct) (Figure 5E, labeled OEL). Similar protein bands were also detected using antibodies produced from a synthesized peptide. The ∼80 kD protein band produced in E. coli only after IPTG induction was purified and subjected to matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) identification. As shown in Supplemental Table 2 online, we obtained a total of 26 peptides corresponding to various parts of BARD1. Our results indicate that the antibody recognized BARD1 specifically. Additionally, gel shift experiments showed that the proteins produced in bard1-1 and bard1-2, like the wild-type protein, retained the ability to form the BARD1-dependent protein-DNA complex (Figure 5F).
Phenotypes Similar to the bard1-3 Mutant Are Produced in Multiple RNA Insertion Lines
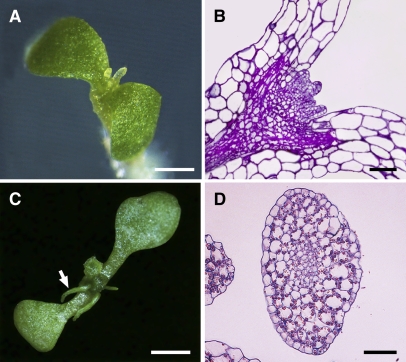
Because no independent knockout lines except bard1-3 showed severe defects in SAM organization, we generated multiple RNA interference (RNAi) lines in which the interfering RNA was targeted specifically against the last BRCT domain coding region of BARD1. A large number of these RNAi plants showed phenotypes similar to bard1-3 during the early seedling stages. We photographed one such seedling (Figure 6A) and a longitudinal semithin section showing its SAM organization (Figure 6B). The phenotype was less extreme at later stages because older seedlings produced significantly expanded, although still centric, leaf-like structures (Figures 6C and 6D). Similar to the bard1-3 mutant plants, RNAi plants typically grew very slowly and did not go through the reproductive stage. In general, 15 to 20% of the plant lines showed phenotypes similar to those of bard1-3, and seedlings having the most severe growth defects were the ones that produced the lowest amounts of BARD1 mRNA, although no quantitative analysis was performed due to limited RNA sources.
Figure 6.
The Phenotype of RNAi Seedlings Expressing an RNA Directed against the Last BRCT Domain Coding Region of BARD1 Is Similar to that of bard1-3 Mutants.
(A) Close-up photo of a 14-d-old RNAi Arabidopsis seedling.
(B) Semithin longitudinal section of an RNAi seedling that was harvested at the same growth stage as in (A). Note that the SAM structure is similar to that of bard1-3 mutants.
(C) Close-up photo of a 21-d-old RNAi Arabidopsis seedling.
(D) Cross section of a tubular leaf-like structure observed from (C) (denoted by a white arrow) indicating the complete loss of leaf dorsoventrality.
White bars = 1 mm; black bars = 50 μm.
The bard1-3 Phenotype Is Suppressed in the wus-1 Genetic Background
The wus-1 loss-of-function mutant was genetically crossed to bard1-3 to obtain a homozygous wus-1 bard1-3 double mutant Arabidopsis line. We used standard genotyping procedures with previously designed derived cleaved amplified polymorphic sequence primers (Wurschum et al., 2006) specific for the wus-1 mutation and also selected for the T-DNA in bard1-3. The absence of functional WUS mRNA in this homozygous double mutant line was confirmed using a reverse primer (see Supplemental Table 1 online) derived from the mis-spliced exon 2. We sequenced the wus-1 locus to determine the existence of the original point mutation in the homozygous double mutant. No amplification of BARD1 mRNA was observed in the double mutant using primer pairs derived either from 5′ (1 to 485 nucleotides) or 3′ (1423 to 2145 nucleotides) regions. Loss of WUS function completely suppressed the bard1-3 phenotype; SAM structures were identical to wus-1 (Figures 7A to 7D). However, during the reproductive growth stage, defects were more severe in the double mutant than in wus-1 mutants because these plants could hardly bolt and usually produced no flowers. Surprisingly, BARD1-overexpressing lines (CaMV35S∷BARD1) also showed a phenotype similar to the double mutant throughout the vegetative growth stages (Figures 7E to 7H). The bard1-3 phenotype could be complemented by genetic transformation with a genomic clone that contained the entire BARD1 gene that included the 1.91-kb 5′ upstream and 0.7-kb downstream sequences (BARD1;bard1-3) or with a truncated version that contained the predicted smallest ORF in bard1-1 and bard1-2 plants (amino acids 241 to 714, BARD1:C-ter;bard1-3) but included the same flanking sequences (Figures 7I to 7L). Transgenic plants expressing the BARD1:C-ter;bard1-3 construct showed obvious defects in DNA repair when compared with the wild type (see Supplemental Figure 2A online). However, much less DNA breakage (observed as TUNEL signals) was found in BARD1:C-ter;bard1-3 seedlings than in bard1-3 (see Supplemental Figure 2B online), indicating that the BARD1 C-terminal polypeptide may be necessary for efficient DNA repair. We were unable to produce transgenic plants that expressed the BARD1 N-terminal domain (residues 1 to 240, BARD1:N-ter;bard1-3).
Figure 7.
Comparison of Shoot Meristem Phenotypes.
(A), (E), and (I) Photographs of 14-d-old wus-1 bard1-3 double mutant, CaMV35S∷BARD1 (in wild-type background), and BARD1:C-ter;bard1-3 seedlings, respectively.
(B), (F), and (J) Median longitudinal sections of the same lines.
(C), (G), and (K) Photographs of 25-d-old wus-1 bard1-3, CaMV35S∷BARD1, and BARD1:C-ter;bard1-3 plants, respectively.
(D), (H), and (L) Median longitudinal sections of the same lines. White bars = 1 mm; black bars = 50 μm.
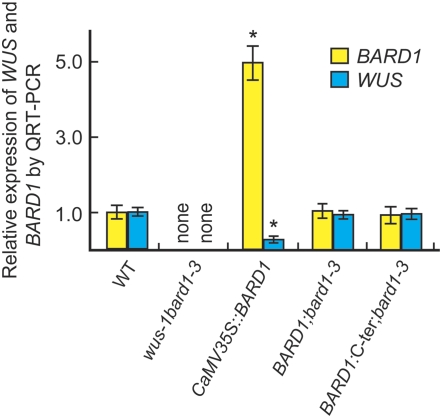
WUS Transcript Levels Are Significantly Reduced in CaMV35S∷BARD1 Plants
Transcript analyses revealed that neither WUS nor BARD1 mRNA was present (<0.003% of wild-type levels) in wus-1 bard1-3 double mutants (Figure 8). Increased BARD1 expression in CaMV35S∷BARD1 plants reduced WUS transcripts to ∼25% of wild-type levels (Figure 8). These plants showed wus-1 or wus-1 bard1-3 double mutant–like structures. Wild-type levels of WUS and BARD1 transcripts were found in BARD1;bard1-3 and also in BARD1:C-ter;bard1-3 plants (Figure 8), suggesting that BARD1 functions primarily through its C-terminal domain.
Figure 8.
QRT-PCR Analysis of BARD1 and WUS Expression in Different Arabidopsis Lines.
Quantitative analysis of WUS and BARD1 transcript levels in the wus-1 bard1-3 double mutant, CaMV35∷BARD1, BARD1;bard1-3, and BARD1:C-ter;bard1-3 seedlings, respectively. The aboveground parts of 3-week-old seedlings of the various genotypes were used for total RNA extraction, reverse transcription, and QRT-PCR analysis. Relative expression levels (mean ± se) were calculated from triplicate independent RNA samples prepared from different batches of Arabidopsis plants. Levels were compared with the wild type, which was arbitrarily set to 1. “None” indicates <0.003% of wild-type WUS and BARD1 transcript levels. *, P < 0.01 compared with the wild type.
WUS May Be Regulated at the Chromosomal Structure Level
Because BARD1 can form a protein-DNA complex with roughly the same WUS promoter region as that bound by SYD (Kwon et al., 2005), we performed coimmunoprecipitation (co-IP) experiments to investigate a potential physical interaction between BARD1 and SYD. In vitro–expressed BARD1 was found to interact with the ATPase domain of SYD (residues 666 to 916, SYD-2-c-Myc) (see Supplemental Figure 6A online, left panel). Both BARD1-HA and SYD-2-c-Myc were coimmunoprecipitated by either anti-HA or anti-c-Myc, indicating the existence of a physical interaction between these two polypeptides (see Supplemental Figure 6A online, middle panel). However, no interaction between BARD1-HA and SYD-1-c-Myc (74.9 kD) was observed after co-IP: anti-HA or anti-c-Myc did not pull down the other protein present in the reaction (see Supplemental Figure 6A online, right panel). Theoretical nucleosome positioning analysis revealed that the immediate WUS promoter region is predicted with high probability to pack into nucleosomes (see Supplemental Figure 6B online). Furthermore, the WUS CAAT and TATA boxes are predicted to locate inside nucleosomes rather than being positioned in internucleosome regions, as is the case for other more ubiquitously expressed genes (see Supplemental Figures 6B and 6C online). These data indicate that substantial chromatin remodeling at or near the WUS promoter may be a prerequisite for WUS expression.
DISCUSSION
BARD1 Regulates Arabidopsis SAM Organization and Maintenance by Repressing WUS Expression
WUS activity is required for the maintenance of stem cell identity, whereas the CLV gene family acts to limit the size of this stem cell pool by promoting differentiation and organ primordia formation via a feedback loop that inhibits WUS transcription (Clark et al., 1997; Brand et al., 2000; Schoof et al., 2000; Muller et al., 2006). When the domain of WUS transcription was no longer confined to the OC and WUS transcript accumulated in the outermost cell layers as in bard1-3, cell division was promoted and organ differentiation was inhibited significantly (Figures 1L and 1P), which resulted in a multiple primordia phenotype (Figures 1K and 1O). Similarly, when WUS was expressed in cells that were programmed to form organ primordia under control of the ANT promoter, these plants could not go through seedling stages due to the accumulation of large amounts of meristem cells that failed to differentiate into organs (Schoof et al., 2000; Groß-Hardt et al., 2002). WUS transcripts localized outside of the OC and massive accumulation of nondifferentiated stem cells was also observed by overexpressing POLTERGEIST LIKE1 (encoding a protein phosphatase acting downstream of the CLV signaling pathway) in a clv mutant background (Song et al., 2006). These results indicate that WUS transcription outside of the OC may be responsible for the observed bard1-3 mutant phenotype.
Formation of the BARD1-Dependent Protein-DNA Complex May Inhibit the Chromatin Remodeling Effects of SYD
BARD1 functions as a heterodimer with BRCA1 in Arabidopsis (Reidt et al., 2006). Here, in vitro gel shift experiments indicated that a BARD1-dependent protein-DNA complex was formed specifically with the F4 fragment upstream of the WUS promoter (Figures 4C and 4D). A co-IP assay showed that SYD, a SWI2-SNF2 ATPase subunit of the chromatin remodeling complex, interacted with BARD1 (see Supplemental Figure 6A online), implying that WUS expression may be regulated at the chromosome structure level. Indeed, when chromosomal DNA sequences of the WUS promoter were loaded into a computational model (Segal et al., 2006), the CAAT and TATA boxes had a high probability of being buried inside nucleosomes (see Supplemental Figures 6B and 6C online); however, this was not the case when we analyzed sequences for more readily expressed genes, such as RBCMT, ADH1, Histone H1, and RPL23A (see Supplemental Figure 6C online). Thus, our data suggest that BARD1 may repress WUS transcription by inhibiting the chromatin remodeling process that is, in theory, essential for WUS expression.
BARD1 May Repress WUS Expression through Its C-Terminal Domain
Analysis of Arabidopsis bard1-1 and bard1-2 mutant lines suggested that BARD1 is involved in DNA repair (Reidt et al., 2006). However, neither bard1-1 nor bard1-2 displayed the same phenotype as bard1-3; to address this unexpected finding, we performed 5′ RACE, which revealed that bard1-1 and bard1-2 plants produced 5′-truncated RNAs of 2027 nucleotides (starting from exon 3 with 19 nucleotides of intron 2 attached) and 1479 bp (starting from the middle of exon 5), respectively (Figure 5). In-frame translation start sites were found in both truncated mRNAs, indicating that these mutants might produce BARD1 C-terminal polypeptides containing functional BRCT domains important for phosphorylation-dependent protein–protein interactions (Glover et al., 2004; Narod and Foulkes, 2004; Williams et al., 2004). These truncated mRNAs in bard1-1 and bard1-2 were ∼39 and 46%, respectively, of the wild-type mRNA level, whereas no part of the BARD1 mRNA could be amplified from bard1-3. The existence of polypeptides encoded by mRNAs from bard1-1 and bard1-2 was confirmed by protein gel blotting experiments (Figure 5E). Our results seem to suggest that only a 52.5-kD polypeptide, but not the full-length BARD1 protein, is present in Arabidopsis plants. However, our data do not indicate whether the 52.5-kD form is the sole polypeptide encoded by BARD1 or the putative full-length BARD1 is cleaved in plant cells by an efficient and plant-specific protein cleavage process to produce two separate polypeptides. Also, in contrast with bard1-3, WUS mRNA in both bard1-1 and bard1-2 lines remained at the wild-type level, indicating that the C-terminal part of BARD1 produced in these plants was sufficient to repress WUS transcription, which is necessary for maintaining normal SAM organization. Indeed, homozygous transgenic Arabidopsis plants expressing the two C-terminal BRCT domains in the bard1-3 background produced wild-type SAM structures (Figures 7I to 7L). These data support the hypothesis that BARD1 is essential for inhibiting WUS transcription outside the OC and that BARD1 functions mainly through the C-terminal BRCT domains. However, the fact that the wus-1 bard1-3 double mutant had a more severe phenotype than wus-1 apart from identical SAM structure suggests that BARD1 has additional effects via pathways not involving WUS, as reported previously (Reidt et al., 2006).
BARD1 Is Required for Proper Development of Leaf Dorsoventrality
Leaf dorsoventral polarity requires lateral information from meristem cells. Centric organs, instead of leaves with proper dorsoventrality, can be produced if the apical meristem is isolated by cutting across the apex extending from the edge of P1 to the edge of P2 (the two youngest leaf primordia) (Sussex, 1951). However, if the apex is only isolated from lateral information flow arising in or near P1 and P2, a dorsoventral leaf is produced (Sussex, 1951; Reinhardt et al., 2005). In this study, tubular-like structures, similar to those obtained by physically separating leaf primordia from developing apices, were observed on the bard1-3 mutant (Figure 1P), indicating that BARD1 activity or a BARD1-dependent signal(s) is required for proper leaf development. Similar morphology was observed in the Arabidopsis phabulosa-1d (phb-1d) mutant and in the loss of PHANTASTICA (PHAN) Antirrhinum mutant (Waites and Hudson, 1995; McConnell and Barton, 1998; Hudson, 2000). Further studies are warranted to determine whether BARD1 affects the expression pattern or the function of PHB-1D in Arabidopsis and PHAN in Antirrhinum.
In summary, our data indicate that BARD1 is essential for confining WUS expression to the OC and for maintaining its relative transcription level, which is vital for proper Arabidopsis growth and development. The identification of putative new factors involved in regulation of WUS expression will help elucidate the molecular mechanisms that fine-tune this master regulator.
METHODS
Plant Lines and Growth Conditions
The Arabidopsis thaliana plants used were mainly derived from Col-0 accessions. Three Arabidopsis mutant lines with disrupted BARD1 were obtained from SALK collections (ABRC; http://signal.salk.edu): bard1-1, SALK_097601; bard1-2, SALK_031862; and bard1-3, SALK_003498. Seeds were surface-sterilized with 0.1% HgCl2, germinated on Murashige and Skoog medium for 2 weeks, transferred to soil, and grown in fully automated growth chambers (Conviron) with a 16/8 h light/dark cycle at 23°C in 70% humidity (Wang et al., 2003). The wus-1 mutant originated from Thomas Laux and was a gift from K. Chong.
Histological Analysis
To prepare semithin sections, seedlings harvested at different stages were fixed overnight in 2% paraformaldehyde and 2.5% glutaraldehyde in PBS, pH 7.2, on ice. Specimens were dehydrated in an ethanol series (30, 50, 70, 80, 90, 95, and 100%) and embedded in Spurr's resin (Spi-Chem) according to the manufacturer's instructions. The tissue was sectioned at a thickness of 4 μm on a Leica RM 2265 microtome (Leica). After staining with 0.05% toluidine blue, sections were observed under bright-field optics using a Leica DMR microscope.
RNA Extraction and QRT-PCR
Rosette leaves, stems, roots, and shoot apices were harvested from 3-week-old wild-type Arabidopsis seedlings. Radicles were prepared from Arabidopsis seeds 30 h after germination, flowers were collected on the day before full opening, and embryos were obtained from Arabidopsis siliques 1 to 4 d after anthesis. RNA extraction and QRT-PCR were performed as reported (Gong et al., 2004), 5 μg of RNAs were used for each reaction, and the housekeeping UBQ gene was used as internal standard. We used triplet replicates of independent plant samples. All primer sequences are shown in Supplemental Table 1 online. All Arabidopsis lines used in QRT-PCR experiments, including wus-1 bard1-3, BARD1;bard1-3, and BARD1 overexpressers (CaMV35S∷BARD1), were genotyped as reported (Wurschum et al., 2006) before being used for RNA extraction and analyses. We used the CT values method to quantify the relative amounts of target gene transcripts as reported (Xu et al., 2007).
In Situ Hybridization
In situ hybridization experiments were done as described (Mayer et al., 1998). A 696-bp fragment from the 3′ end of the BARD1 coding region was used to generate probes using the gene-specific primers described in Supplemental Table 1 online. The full-length WUS coding region was used to generate the riboprobes as reported (Mayer et al., 1998). The PCR fragments were cloned into pGEM-T-easy vector (Promega) and linearized using NcoI or SalI before being used for synthesis of 11-digoxigenin-UTP–labeled sense or antisense probes, respectively. The probes were synthesized in accordance with the manufacturer's instructions (Roche Diagnostics).
Lugol and DAPI Staining of Root Cells
For Lugol staining, 7-d-old roots were dipped in Lugol's staining solution (Sigma-Aldrich) for 3 min, washed with distilled water, and observed under a differential interference contrast microscope (Leica). For DAPI staining, Arabidopsis roots were fixed in 3% glutaraldehyde (Sigma-Aldrich) overnight and stained with 1% DAPI (Sigma-Aldrich) for 4 h. The roots with stained nuclei were visualized on the microscope with a UV fluorescence filter set and photographed using the Leica DFC 480 camera.
Gel Shift Assays
Oligonucleotides complementary to different motifs of the WUS promoter from −435 to −70 bp, as depicted in Figure 3A, were synthesized by Sunbiotech. The gel shift assay was performed in a total volume of 20 μL DNA binding buffer (25 mM HEPES-KOH, pH 7.5, 10% glycerol, 4 mM NaCl, 1 mM DTT, 40 mM KCl, and 0.5 mM EDTA). Nuclear extract (10 μg) prepared from wild-type or bard1-3 mutant seedlings and 2 pmol of 32P-end–labeled oligonucleotides synthesized from different DNA fragments were added to the reaction. After incubating for 2 h at room temperature, the reaction was loaded onto an 8% nondenaturing PAGE gel in 0.5× TBE (45 mM Tris-borate and 1 mM EDTA). DNA binding activity was quantified using the Typhoon 9200 PhosphorImager (GE Healthcare) as reported (Wei et al., 2005). For supershift assays, ∼20 nmol of anti-BARD1 antiserum was added to the reaction after an initial 30-min incubation of the DNA fragments with nuclear extracts at room temperature. The reactions were further incubated for a total of 2 h before gel loading.
Preparation of Antiserum to BARD1
A 723-bp C-terminal part of BARD1 was cloned into the pET-28a expression vector for protein purification using primers reported in Supplemental Table 1 online. Approximately 3 mg of proteins purified using a His-tag affinity column (Novagen) was used to immunize mice for polyclonal antibody production (performed commercially in the Antibody Center, National Institute of Biological Sciences of China). We further affinity purified the antibody using the polypeptide ELGAESSNNVNDQR (residues 639 to 652), identified by Peptide-antigen Finder (Chinese Peptide), as the affinity column tag and confirmed the quality of the antibody by an enzyme-linked immunosorbent assay before protein gel blotting analyses. A second antibody was produced from rabbit using this synthesized polypeptide as the antigen at the Antibody Center (Institute of Molecular Immunology, Peking University).
Protein Gel Blotting Analysis and MALDI-TOF Identification of BARD1
Shoots of various Arabidopsis plant lines were harvested, stored in liquid nitrogen, and subsequently ground into a fine powder using a mortar and pestle. Samples were homogenized in extraction buffer containing 50 mM Tris-acetate, pH 7.9, 100 mM potassium acetate, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, and 20% glycerol. Cell debris was removed by centrifugation at 10,000g for 15 min. Immunoblotting was performed after 20 μg protein (quantified by a protein assay kit [Bio-Rad]) was subjected to SDS-PAGE (10% gel) and then transferred to a polyvinylidene difluoride membrane; purified anti-BARD1 was diluted 1:1000 in 1× PBS buffer containing 0.05% Tween 20 and 5% fat-free milk. In parallel experiments, preimmune serum was used as the negative control for antibody specificity. We also used samples extracted from one Escherichia coli strain carrying an expression cassette for a full-length BARD1 before and after IPTG induction to demonstrate the specificity of our antibody. This protein band was further purified and sent for MALDI-TOF identification following a previously reported procedure (Wang et al., 2006).
UV-C Treatment and Detection of in Situ Cell Death by TUNEL Assay
Wild-type and bard1-3 mutant apical Arabidopsis tissues were fixed in formalin/acetic acid fixation solution, dehydrated in a series of graded ethanol, and embedded in paraffin. Sections were cut at a thickness of 10 μm using a rotary microtome (Leitz 1512). Samples were digested with 10 μg/mL proteinase K at 37°C for 30 min. UV lamps (254-nm radiation at a fluency rate of 1.0 J/m2/s) were used to treat the plants at the indicated dosages. TUNEL assay was performed according to the manufacturer's instructions (Boehringer Mannheim) with details as reported (Li et al., 2004).
Co-IP Assay
We cloned the full-length BARD1 downstream of the HA-tag in pGADT7 and cloned two fragments covering the N-terminal part of the SYD coding region (1 to 665 and 666 to 916, designated as SYD-1 and SYD-2, respectively; the SYD-2 fragment contained the functional ATPase domain), downstream of the cMyc-tag in pGBKT7. We used TNT-coupled wheat germ extract systems (Promega) and [35S]methionine (Perkin-Elmer) for in vitro translation of the above polypeptides and performed the co-IP assay using the Matchmaker kit (Becton-Dickinson) following the manufacturer's instructions. Gels were dried and scanned using the Typhoon 9200 PhosphorImager (GE Healthcare).
Plant Transformation and Crossing
For genetic complementation of the bard1-3 phenotype, a 6.1-kb genomic DNA that encompassed the whole BARD1 (At1g04020) coding region plus 1.91 kb 5′ upstream and 0.7 kb downstream flanking sequences was cloned into pCAMBIA1305 using primers described in Supplemental Table 1 online. This construct was then transformed into heterozygous bard1-3/+ Arabidopsis plants. The N terminus (residues 1 to 240) or C terminus (residues 241 to 714) of BARD1 was cloned into pCAMBIA3301 that contained the same 1.91-kb BARD1 5′ upstream promoter and 0.7-kb downstream flanking sequences. Transgenic lines were selected by antibiotic resistance, genomic PCR, and also by cosegregation studies that looked for single-copy insertion events into the bard1-3 homozygous background. A BARD1 overexpression line (CaMV35S∷BARD1) was obtained by cloning the full-length BARD1 coding region under the CaMV35S promoter and the NOS terminator in pCAMBIA1305 and then transforming wild-type plants. Pollen collected from wus-1 plants was used to pollinate heterozygous bard1-3 plants to produce the homozygous wus-1 bard1-3 double mutant.
Production of RNAi Lines
To produce transgenic plants expressing a BARD1-RNAi construct, a 553-nucleotide BARD1 C-terminal coding region (1593 to 2145) was amplified using primers described in Supplemental Table 1 online and fused to the same CaMV35S promoter and NOS terminator in pCAMBIA1305, as described in the previous section. The PCR products were cloned in both sense (digested with SacI and KpnI) and antisense (digested with XbaI and BamHI) directions, separated by a 10-nucleotide spacer.
Analysis of GUS Activity
pBARD1:GUS was constructed by amplifying the 1.91-kb BARD1 promoter and inserting it between HindIII and XbaI sites of pBI121. GUS activity was assayed using 10-d-old seedlings, and stained tissues were processed for histological evaluation (Schoof et al., 2000; Kwon et al., 2005).
Predictions of Intrinsic Nucleosome Organization
For intrinsic nucleosome organization prediction, we used genomic sequences obtained from 2.0 kb upstream to 2.0 kb downstream of the putative transcription initiation site for WUS and a few readily expressed housekeeping genes and performed computational analyses using the online algorithm model obtained from yeast (Segal et al., 2006; also see http://132.77.150.113/pubs/nucleosomes06/segal06_prediction.html). For CAAT and TATA boxes analysis, promoter sequences were scanned by searching http://www.dna.affrc.go.jp/PLACE/signalscan.html.
Statistical Analysis
Statistical significance was evaluated by one-way analysis of variance followed by Tukey's test (SigmaStat 3.5; Systat Software).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BARD1 (At1g04020, NM_100283), WUS (AT2G17950, NM_127349), BRCA1 (At4g21070, NM_118225), and BARD1 variant transcript in bard1-1 (EU817406) and in bard1-2 (EU817407).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Characterization of Different bard1 Mutant Alleles.
Supplemental Figure 2. bard1-3 Is Hypersensitive to DNA Damage.
Supplemental Figure 3. Phenotypic Characterization of the bard1-3 Root System.
Supplemental Figure 4. RT- and QRT-PCR Analyses of Potential Target Gene Expression.
Supplemental Figure 5. GUS Expression in Transgenic Arabidopsis Plants Carrying the BARD1:GUS Construct.
Supplemental Figure 6. BARD1 May Repress WUS Expression by Inhibiting the Chromatin Remodeling Process That Is Essential for WUS Promoter Function.
Supplemental Table 1. Primers Used for RT-, QRT-PCR, and Other Analyses.
Supplemental Table 2. MALDI-TOF Analysis of At BARD1 Expressed in E. coli after IPTG Induction.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (Grants 90717009 and 30221120261).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Yu-Xian Zhu (zhuyx@water.pku.edu.cn).
Online version contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Baer, R., and Ludwig, T. (2002). The BRCA/BARD1 heterodimer, a tumor suppressor complex with ubiquitin E3 ligase activity. Curr. Opin. Genet. Dev. 12 86–91. [DOI] [PubMed] [Google Scholar]
- Baurle, I., and Laux, T. (2005). Regulation of WUSCHEL transcription in the stem cell niche of the Arabidopsis shoot meristem. Plant Cell 17 2271–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand, U., Fletcher, J.C., Hobe, M., Meyerowitz, E.M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289 617–619. [DOI] [PubMed] [Google Scholar]
- Brand, U., Grunewald, M., Hobe, M., and Simon, R. (2002). Regulation of CLV3 expression by two homeobox genes in Arabidopsis. Plant Physiol. 129 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, H., Tian, X., Hu, X.D., Li, S.X., Yu, D.H., and Zhu, Y.X. (2005). Combined DNA vaccines formulated either in DDA or in saline protect cattle from Mycobacterium bovis infection. Vaccine 23 3887–3895. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89 575–585. [DOI] [PubMed] [Google Scholar]
- Gallois, J.L., Nora, F.R., Mizukami, Y., and Sablowski, R. (2004). WUSCHEL induces shoot stem cell activity and developmental plasticity in the root meristem. Genes Dev. 18 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover, J.N.M., Williams, R.S., and Lee, M.S. (2004). Interactions between BRCT repeats and phosphoproteins: Tangled up in two. Trends Biochem. Sci. 29 579–585. [DOI] [PubMed] [Google Scholar]
- Gong, W., et al. (2004). Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol. 135 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groß-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, A. (2000). Development of symmetry in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51 349–370. [DOI] [PubMed] [Google Scholar]
- Irminger-Finger, I., and Jefford, C.W. (2006). Is there more to BARD1 than BRCA1? Nat. Rev. Cancer 6 382–391. [DOI] [PubMed] [Google Scholar]
- Kieffer, M., Stern, Y., Cook, H., Clerici, E., Maulbetsch, C., Laux, T., and Davies, B. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18 560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, C.S., Chen, C., and Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19 992–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarge, S., and Montane, M.H. (2003). Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 31 1148–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laux, T., Mayer, K.F.X., Berger, J., and Jurgens, G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814. [DOI] [PubMed] [Google Scholar]
- Lenhard, M., Jurgens, G., and Laux, T. (2002). The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129 3195–3206. [DOI] [PubMed] [Google Scholar]
- Li, J., Wang, D.Y., Li, Q., Xu, Y.J., Cui, K.M., and Zhu, Y.X. (2004). PPF1 inhibits programmed cell death in apical meristems of both G2 pea and transgenic Arabidopsis plants possibly by delaying cytosolic Ca2+ elevation. Cell Calcium 35 71–77. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Hong, R.L., Hobe, M., Busch, M.A., Parcy, F., Simon, R., and Weigel, D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. [DOI] [PubMed] [Google Scholar]
- Long, J.A., Ohno, C., Smith, Z.R., and Meyerowitz, E.M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312 1520–1523. [DOI] [PubMed] [Google Scholar]
- Mayer, K.F.X., Schoof, H., Haecker, A., Lenhard, M., Jurgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125 2935–2942. [DOI] [PubMed] [Google Scholar]
- Muller, R., Borghi, L., Kwiatkowska, D., Laufs, P., and Simon, R. (2006). Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell 18 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narod, S.A., and Foulkes, W.D. (2004). BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer 4 665–676. [DOI] [PubMed] [Google Scholar]
- Reidt, W., Wurz, R., Wanieck, K., Chu, H.H., and Puchta, H. (2006). A homologue of the breast cancer-associated gene BARD1 is involved in DNA repair in plants. EMBO J. 25 4326–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, D., Frenz, M., Mendel, T., and Kuhlemeier, C. (2005). Microsurgical and laser ablation analysis of leaf positioning and dorsoventral patterning in tomato. Development 132 15–26. [DOI] [PubMed] [Google Scholar]
- Sarkar, A.K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., Scheres, B., Heidstra, R., and Laux, T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446 811–814. [DOI] [PubMed] [Google Scholar]
- Schoof, H., Lenhard, M., Haecker, A., Mayer, K.F.X., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100 635–644. [DOI] [PubMed] [Google Scholar]
- Segal, E., Fondufe-Mittendorf, Y., Chen, L., Thastrom, A., Field, Y., Moore, I.K., Wang, J.-P.Z., and Widom, J. (2006). A genomic code for nucleosome positioning. Nature 442 772–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, S.-k., Lee, M.M., and Clark, S.E. (2006). POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133 4691–4698. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1951). Experiments on the cause of dorsiventrality in leaves. Nature 167 651–652. [DOI] [PubMed] [Google Scholar]
- Tucker, M.R., and Laux, T. (2007). Connecting the paths in plant stem cell regulation. Trends Cell Biol. 17 403–410. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). Phantastica: A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Wang, B.C., Wang, H.Y., Feng, J.X., Meng, D.Z., Qu, L.J., and Zhu, Y.X. (2006). Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6 2555–2563. [DOI] [PubMed] [Google Scholar]
- Wang, D.Y., Xu, Y.J., Li, Q., Hao, X.M., Cui, K.M., Sun, F.Z., and Zhu, Y.X. (2003). Transgenic expression of a putative calcium transporter affects the time of Arabidopsis flowering. Plant J. 33 285–292. [DOI] [PubMed] [Google Scholar]
- Wei, G., Pan, Y., Lei, J., and Zhu, Y.X. (2005). Molecular cloning, phylogenetic analysis, expressional profiling and in vitro studies of TINY2 from Arabidopsis thaliana. J. Biochem. Mol. Biol. 38 440–446. [DOI] [PubMed] [Google Scholar]
- Williams, R.S., Lee, M.S., Hau, D.D., and Glover, J.N.M. (2004). Structural basis of phosphopeptide recognition by the BRCT domain of BRCA1. Nat. Struct. Mol. Biol. 11 519–525. [DOI] [PubMed] [Google Scholar]
- Wu, L.C., Wang, Z.W., Tsan, J.T., Spillman, M.A., Phung, A., Xu, X.L., Yang, M.-C.W., Hwang, L.-Y., Bowcock, A.M., and Baer, R. (1996). Identification of a RING protein that can interact in vivo with the BRCA gene product. Nat. Genet. 14 430–440. [DOI] [PubMed] [Google Scholar]
- Wurschum, T., Groß-Hardt, R., and Laux, T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Wang, B.C., and Zhu, Y.X. (2007). Identification of proteins expressed at extremely low level in Arabidopsis leaves. Biochem. Biophys. Res. Commun. 358 808–812. [DOI] [PubMed] [Google Scholar]
- Xu, Y.Y., Wang, X.M., Li, J., Li, J.H., Wu, J.S., Walker, J.C., Xu, Z.H., and Zhong, K. (2005). Activation of the WUS gene induces ectopic initiation of floral meristems on mature stem surface in Arabidopsis thaliana. Plant Mol. Biol. 57 773–784. [DOI] [PubMed] [Google Scholar]
- Zhao, Y., Medrano, L., Ohashi, K., Fletcher, J.C., Yu, H., Sakai, H., and Meyerowitz, E.M. (2004). HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 16 2586–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR, Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.