Abstract
The phytochrome (phy) family of photoreceptors regulates changes in gene expression in response to red/far-red light signals in part by physically interacting with constitutively nucleus-localized phy-interacting basic helix-loop-helix transcription factors (PIFs). Here, we show that PIF1, the member with the highest affinity for phys, is strongly sensitive to the quality and quantity of light. phyA plays a dominant role in regulating the degradation of PIF1 following initial light exposure, while phyB and phyD and possibly other phys also influence PIF1 degradation after prolonged illumination. PIF1 is rapidly phosphorylated and ubiquitinated under red and far-red light before being degraded with a half-life of ∼1 to 2 min under red light. Although PIF1 interacts with phyB through a conserved active phyB binding motif, it interacts with phyA through a novel active phyA binding motif. phy interaction is necessary but not sufficient for the light-induced phosphorylation and degradation of PIF1. Domain-mapping studies reveal that the phy interaction, light-induced degradation, and transcriptional activation domains are located at the N-terminal 150–amino acid region of PIF1. Unlike PIF3, PIF1 does not interact with the two halves of either phyA or phyB separately. Moreover, overexpression of a light-stable truncated form of PIF1 causes constitutively photomorphogenic phenotypes in the dark. Taken together, these data suggest that removal of the negative regulators (e.g., PIFs) by light-induced proteolytic degradation might be sufficient to promote photomorphogenesis.
INTRODUCTION
Growth and development are highly regulated by environmental light signals at all phases of a plant's life cycle. Plants have evolved several light receptors: the phytochrome (phy) family of photoreceptors to monitor the red (R)/far-red (FR) region; the cryptochromes, phototropins, and ZTL/FKF1 family of F-box proteins to monitor the UV-A/blue region; and an unidentified receptor to monitor the UV-B region of the spectrum (Lin and Shalitin, 2003; Chen et al., 2004; Schaefer and Nagy, 2006). The phy family in Arabidopsis thaliana (PHYA to PHYE) encodes ∼125-kD soluble proteins that can form selective homodimers or heterodimers between the family members (Mathews and Sharrock, 1997; Sharrock and Clack, 2004). Their photosensitivity relies on the acquisition of a covalently attached bilin chromophore that enables the existence of two interconvertible forms of phys: the Pr form (biologically inactive) with maximal absorbance in the R region of the spectrum and the Pfr form (biologically active) with maximal absorbance in the FR region of the spectrum. The Pr form is converted to the biologically active Pfr form under R light, and the Pfr form is converted back to the inactive Pr form under FR light (Rockwell et al., 2006). The array of photoreceptors allows plants to monitor and respond to a number of parameters of ambient light signals for optimum photomorphogenic development (Schaefer and Nagy, 2006; Whitelam and Halliday, 2007).
phys in the Pr form are predominantly in the cytosol, but they are induced to translocate to the nucleus upon light activation (Kircher et al., 2002). Light induces the nuclear import of phys via either a conformation change (in phyB), resulting in the unmasking of a nuclear localization signal present in its C-terminal domain (Chen et al., 2005), or an association (of phyA) with other proteins containing a nuclear localization signal (Zhou et al., 2005; Hiltbrunner et al., 2006; Rösler et al., 2007). Light-induced nuclear translocation is necessary for the majority of the biological functions of phyA and phyB (Huq et al., 2003; Matsushita et al., 2003; Hiltbrunner et al., 2006; Rösler et al., 2007). However, cytosolic phyA regulates negative gravitropism in blue light as well as R light–enhanced phototropism (Rösler et al., 2007). In the nucleus, phys interact with a group of unrelated proteins (Quail, 2007) and initiate signaling cascades that result in changes in the expression of ∼10% of the genome (Rockwell et al., 2006; Jiao et al., 2007; Whitelam and Halliday, 2007). However, the primary biochemical mechanism of signal transfer from photoactivated phys to signaling partners is still unknown.
Among the phy-interacting proteins, the PHYTOCHROME-INTERACTING FACTOR (PIF) family of basic helix-loop-helix (bHLH) transcription factors constitutes the best model for understanding phy-regulated gene expression (Duek and Fankhauser, 2005; Castillon et al., 2007; Quail, 2007). Six closely related genes of the Arabidopsis bHLH superfamily encode PIF1 and PIF3 to PIF7 (Toledo-Ortiz et al., 2003; Castillon et al., 2007; Quail, 2007; Leivar et al., 2008). PIFs interact selectively with the Pfr forms of phys with differential affinities in vitro. For example, PIF1 and PIF3 interact with the Pfr forms of both phyA and phyB, while all other PIFs interact with the Pfr form of phyB only (Ni et al., 1999; Huq and Quail, 2002; Huq et al., 2004; Khanna et al., 2004; Leivar et al., 2008). Interaction of PIFs with other phys has not been detected. Of all the PIFs, PIF1 has the highest affinity for both phyA and phyB (Huq et al., 2004), suggesting that PIF1 plays a critical role in phy signaling.
Genetic and photobiological analyses show that the PIF family members have distinct, as well as overlapping, biological functions (Castillon et al., 2007; Quail, 2007). However, contrary to the initial observation, PIFs act predominantly as negative regulators of phy signaling pathways. PIF3 to PIF5 and PIF7 negatively regulate light-induced suppression of hypocotyl elongation and cotyledon expansion (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Huq et al., 2004; Monte et al., 2004; Oh et al., 2004). Strikingly, this negative regulation under prolonged R light conditions is correlated with elevated levels of phyB, suggesting that these PIFs regulate phyB protein levels posttranslationally under continuous R light (Monte et al., 2004; Al-Sady et al., 2008; Khanna et al., 2008; Leivar et al., 2008). PIF3 also positively regulates chlorophyll biosynthesis and anthocyanin production in light (Kim et al., 2003; Monte et al., 2004; Shin et al., 2007). Recently, PIF3 and PIF4 were shown to interact with DELLA proteins to coordinately modulate cell elongation (de Lucas et al., 2008; Feng et al., 2008). PIF1 plays a major role in negatively regulating light-induced seed germination and chlorophyll biosynthesis as well as plays a minor role in light-induced suppression of hypocotyl elongation and cotyledon expansion. PIF1 regulates gibberellic acid metabolic and signaling genes to suppress seed germination (Oh et al., 2006, 2007). PIF1 also directly and indirectly regulates chlorophyll biosynthetic genes to optimize the greening process in Arabidopsis (Moon et al., 2008). These data suggest that although PIFs have the potential to receive the light signals from the photoactivated phys, they have an antagonistic relationship with phys.
The functional significance of the above antagonistic relationship became apparent when it was shown that PIF3 is degraded under both R and FR light conditions in a phy-dependent manner (Bauer et al., 2004). Moreover, the transcriptional activation of both PIF1 and PIF3 is also reduced under both R and FR light in a phy-dependent manner (Huq et al., 2004; Al-Sady et al., 2008). Subsequently, it was shown that PIF1 and PIF3 to PIF5 are degraded under light through the ubiquitin (ubi)/26S proteasomal pathway (Monte et al., 2004; Park et al., 2004; Shen et al., 2005, 2007; Oh et al., 2006; Lorrain et al., 2007; Nozue et al., 2007). PIF3 to PIF5 are also phosphorylated specifically in response to R light, and the phosphorylated form is presumably degraded under light (Al-Sady et al., 2006; Lorrain et al., 2007; Shen et al., 2007). An N-terminal conserved region of PIFs, called the APB (for active phyB binding) motif, is necessary for the physical interactions between PIFs and the photoactivated phyB (Khanna et al., 2004). Similarly, an APA (for active phyA binding) motif within the N-terminal region of PIF3, distinct from the ABP motif, is necessary for the interaction of PIF3 and phyA (Al-Sady et al., 2006). Both APA and APB motifs are necessary for the light-induced phosphorylation and subsequent degradation of PIF3. However, despite the fact that PIF1 has the strongest affinity among the PIFs for both phyA and phyB, the functional significance of its direct physical interaction with photoactivated phys has not been demonstrated. Moreover, the early events in the FR-induced degradation of PIFs are not yet known. Here, we show that although PIF1 has an APB motif similar to other PIFs, it has a different APA motif than PIF3. PIF1 was rapidly phosphorylated and ubiquitinated under both R and FR light, and direct physical interaction of PIF1 with phyA or phyB was necessary for light-induced phosphorylation and degradation. Moreover, overexpression of a light-stable, truncated form of PIF1 generated constitutively photomorphogenic phenotypes in the dark, suggesting that the removal of the negative regulators (e.g., PIFs) by light-induced proteolytic degradation might be sufficient to promote photomorphogenesis.
RESULTS
PIF1 Stability Is Highly Sensitive to the Quality and Quantity of Light
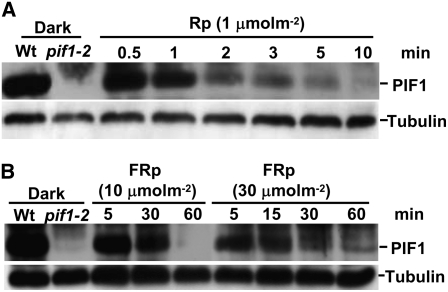
Previously, we and others have shown that PIF1 fusion proteins (LUC-PIF1 and PIF1-HA) are degraded under both R and FR light conditions (Shen et al., 2005; Oh et al., 2006). However, the behavior of native PIF1 is unknown. Antibody specific for native PIF1 was raised and used to investigate the stability of native PIF1 under both R and FR light conditions. Native PIF1 was completely degraded in as little as 100 μmol·m−2 R light within 2 min. Therefore, we used 1 μmol·m−2 R light to test the degradation kinetics, and the results showed that the half-life of PIF1 was ∼1 to 2 min under these conditions (Figure 1A). These results suggested that PIF1 was highly sensitive to R light. Native PIF1 was also degraded under FR light conditions; however, the rate of degradation was much slower under FR light than under R light (Figure 1B). The half-life of native PIF1 was ∼5 to 10 min under 10 μmol·m−2 FR light. These results were largely consistent with our previous data using the luciferase fusion protein LUC-PIF1 (Shen et al., 2005). However, the small difference in half-life between the native PIF1 and LUC-PIF1 fusion proteins might be due to the overexpression of the fusion proteins using the constitutively active 35S promoter and/or to the nature and location of the fusion tags used in previous studies. Overall, these results were consistent with PIF1 having the highest affinity among all of the PIFs for the Pfr forms of both phyA and phyB.
Figure 1.
PIF1 Stability Is Highly Sensitive to the Quality and Quantity of Light.
Native PIF1 is rapidly degraded under Rp (1 μmol·m−2) (A) or FRp (10 or 30 μmol·m−2) (B) light conditions. Four-day-old dark-grown seedlings were exposed to Rp or FRp light and then incubated in the dark for the durations indicated before being harvested for protein extraction. Protein extracts from dark-grown wild-type and pif1 null mutant seedlings are also included in the first and second lanes, respectively. Approximately 30 μg of total protein in each lane was separated on an 8% polyacrylamide gel, transferred to PVDF membranes, and probed with anti-PIF1 antibody. A similar blot was probed with anti-tubulin antibody. The bands corresponding to PIF1 and tubulin are labeled.
phyA Acts Early While phyB and Other phys Induce PIF1 Degradation under Prolonged Light Conditions
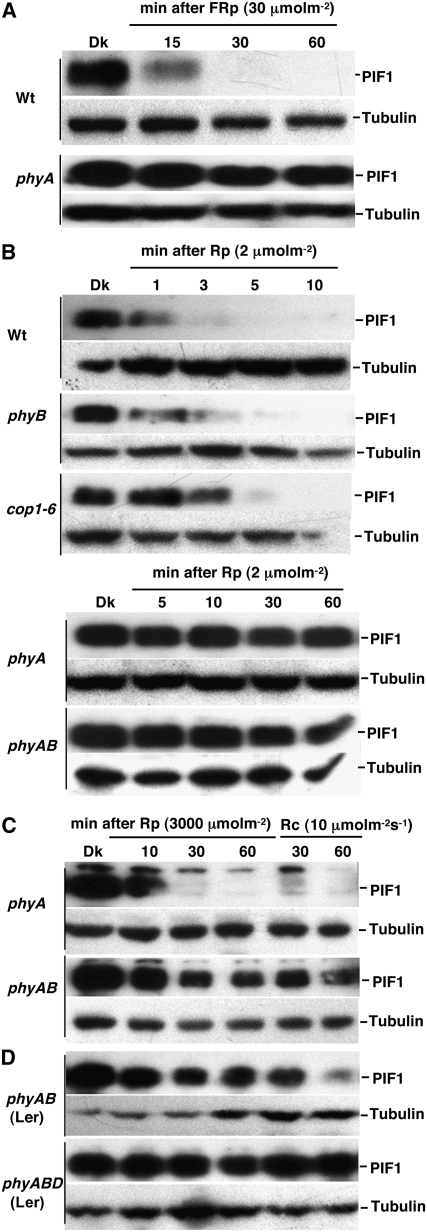
To investigate the relative contributions of different photoreceptors to light-induced degradation of PIF1, we examined the native PIF1 level in various monogenic and higher order photoreceptor mutant lines. PIF1 was stable in the phyA mutant background compared with the wild type after a pulse of FR light (FRp) (Figure 2A), suggesting that phyA was the sole photoreceptor for PIF1 degradation under FR light. PIF1 was also stable in the phyA and phyAB backgrounds for up to 60 min after a pulse of R light (Rp) (Figure 2B). PIF1 degradation was slightly reduced in the phyB background compared with the wild type under these conditions (Figure 2B). However, increased light exposure either by greater fluence rates during the light pulse prior to incubation in the dark or by prolonged illumination at lower fluence rates showed significant degradation of PIF1 in the phyA and phyAB backgrounds (Figure 2C), suggesting that other phys were involved in PIF1 degradation under these conditions. PIF1 was largely stable in the phyABD triple mutant compared with the phyAB double mutant background under prolonged R light exposure (Figure 2D). These data suggested that phyB and phyD and possibly other phys also contributed to the degradation of PIF1 under prolonged light conditions, presumably when phyA levels were reduced. PIF1 degradation was slightly reduced in the constitutive photomorphogenic1 (cop1) mutant background compared with the wild type under R light (Figure 2B), suggesting that COP1 might play a minor role in regulating PIF1 stability under light.
Figure 2.
phyA Plays a Dominant Role during the Initial Light Exposure, While phyB, phyD, and Other phys Regulate PIF1 Stability under Prolonged Light Exposure.
Protein gel blots showing native PIF1 levels in wild-type, phyA, phyB, and phyAB backgrounds. Four-day-old dark-grown seedlings were exposed to FRp (A), Rp (B), or continuous red light (Rc) ([C] and [D]) at the indicated fluences and then incubated in the dark for the times indicated (except Rc) before harvesting for protein extraction. phyAB and phyABD shown in (D) are in the Landsberg erecta (Ler) ecotype; all other mutants are in the Columbia ecotype.
Light Induces Rapid Phosphorylation and Ubiquitination prior to Degradation of PIF1
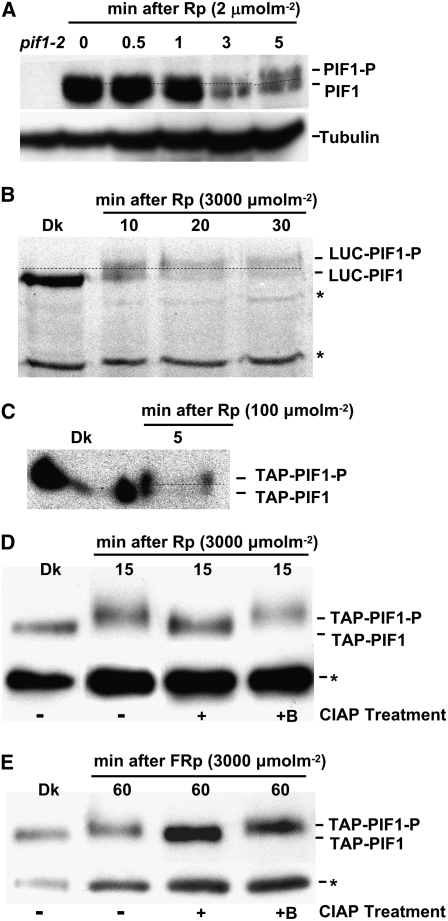
Although we have shown that PIF1 is degraded through the ubi/26S proteasomal system under R and FR light conditions (Figure 1) (Shen et al., 2005), the early events in light-induced degradation of PIF1 have not yet been elucidated. Protein gel blots with anti-PIF1 antibody identified two closely migrating bands under R light conditions (Figure 3A). These closely migrating bands were absent in dark-grown tissues and appeared specifically in light-exposed tissues, suggesting that light induced a rapid posttranslational modification of native PIF1. We investigated whether PIF1 fusion proteins also demonstrated this behavior. Both the LUC-PIF1 and Tandem Affinity Purification (TAP)–PIF1 fusion proteins extracted from the R light–exposed transgenic seedlings migrated as double bands, suggesting that PIF1 fusion proteins were also posttranslationally modified after Rp (Figures 3B and 3C). Both fusion proteins were also degraded following a Rp as expected, while the control proteins (LUC-GFP [for green fluorescent protein] and TAP-GFP) did not migrate as double bands and were not degraded under light (Figure 4B) (Shen et al., 2005), suggesting that the band shift and the degradation under light were specific to PIF1.
Figure 3.
Light Induces Rapid Phosphorylation prior to Degradation of PIF1.
(A) Native PIF1 migrates as two bands (PIF1 and PIF1-P) following Rp (2 μmol·m−2). A blot probed with anti-PIF1 antibody is shown.
(B) LUC-PIF1 also exhibits a slower migrating band (LUC-PIF1-P) after Rp (3000 μmol·m−2). Proteins from plants expressing LUC-PIF1 were probed with anti-LUC antibody.
(C) TAP-PIF1 shows a slower migrating band (TAP-PIF1-P) and is also degraded after Rp (100 μmol·m−2). Proteins from plants expressing TAP-PIF1 were probed with anti-MYC antibody that recognizes the TAP tag.
Dotted lines separate the two forms of PIF1 in (A) to (C).
(D) and (E) The Rp- and FRp-induced slow-migrating band is a phosphorylated form of PIF1. TAP-PIF1 was immunoprecipitated from protein extracts prepared using 4-d-old dark-grown 35S:TAP-PIF1 seedlings kept in the dark or exposed to either Rp (3000 μmol·m−2; [D]) or FRp (3000 μmol·m−2; [E]) followed by dark incubation. The immunoprecipitated pellets from the Rp- or FRp-exposed samples were dissolved in buffer and incubated without (−) or with (+) native CIAP or with boiled CIAP (+B). Samples were then separated on 6.5% SDS-PAGE gels and probed with anti-MYC antibody. Asterisks denote cross-reacting bands.
Figure 4.
Light Induces Rapid Phosphorylation and Ubiquitination prior to Degradation of PIF1.
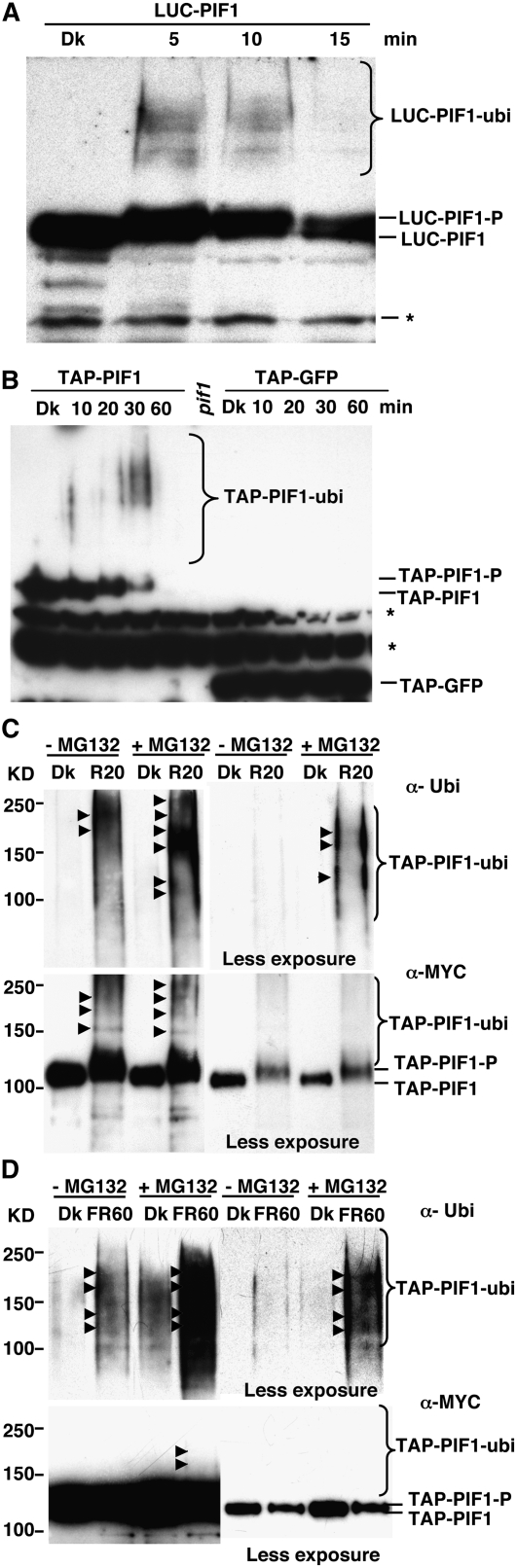
(A) LUC-PIF1 shows high molecular weight bands (LUC-PIF1-ubi) after Rp (3000 μmol·m−2). A blot probed with anti-LUC antibody is shown.
(B) TAP-PIF1 shows high molecular weight bands (TAP-PIF1-ubi) and is also degraded following Rp (100 μmol·m−2), while TAP-GFP is stable under these conditions. A blot probed with anti-MYC antibody that recognizes the TAP tag is shown. Asterisks denote cross-reacting bands.
(C) and (D) The Rp- and FRp-induced slow-migrating bands are ubiquitinated forms of PIF1. TAP-PIF1 was immunoprecipitated from protein extracts prepared using 4-d-old dark-grown seedlings either kept in the dark or exposed briefly to Rp light (3000 μmol·m−2; [C]) or FRp light (3000 μmol·m−2; [D]). The immunoprecipitated samples were then separated on 6.5% SDS-PAGE gels and probed with anti-Ubi or anti-MYC antibody.
To experimentally determine whether the retardation of migration of PIF1 in light-exposed samples was due to a rapid light-induced phosphorylation of PIF1, we immunoprecipitated the TAP-PIF1 fusion protein from dark-incubated or R or FR light–exposed seedling extracts. We then incubated the light-exposed samples with buffer alone or with active or boiled alkaline phosphatase. The slow-migrating PIF1 band in the light-exposed samples was eliminated in the presence of alkaline phosphatase but not in the presence of boiled (inactive) alkaline phosphatase (Figures 3D and 3E). These results demonstrated that PIF1 was indeed phosphorylated under both R and FR light conditions.
Both LUC-PIF1 and TAP-PIF1 extracted from seedlings that were exposed to a pulse of R light also showed several high molecular weight bands on protein gel blots (Figures 4A and 4B) that might have been ubiquitinated forms of PIF1. Protein gel blots of immunoprecipitated TAP-PIF1 using anti-ubiquitin (Ubi) antibody showed that TAP-PIF1 recovered from seedlings under either R or FR light conditions was indeed polyubiquitinated (Figures 4C and 4D). Anti-MYC (recognizing the TAP tag specifically) and anti-Ubi antibodies detected high molecular weight bands from the light-exposed TAP-PIF1 seedling samples but not in samples immunoprecipitated from seedlings incubated in the dark (Figures 4C and 4D). Additionally, the phosphorylated form of tagged PIF1 predominated in samples immunoprecipitated from the light-exposed seedlings (Figures 4C and 4D). These data suggest that PIF1 was phosphorylated and ubiquitinated under both R and FR light conditions before being degraded.
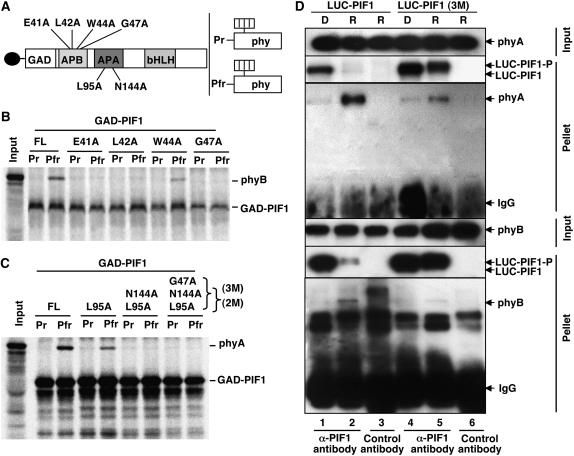
The APB and APA Motifs Present in the N-Terminal 150–Amino Acid Region Are Necessary for the Pfr-Specific Interaction of PIF1 with phyA and phyB Both in Vitro and in Vivo
To understand the functional significance of PIF1–phy interactions, we mapped the phy interaction motifs in PIF1. Recent reports showed that a small motif, the APB, that is present in many phy-interacting bHLH factors is necessary for the physical interaction with the Pfr form of phyB (see Supplemental Figure 1A online) (Khanna et al., 2004; Al-Sady et al., 2006; Shen et al., 2007). Ala scanning by site-directed mutagenesis of conserved amino acids in this region reduced PIF1's interaction with the Pfr form of phyB either severely (E41A, L42A, or G47A) or partially (W44A) (Figures 5A and 5B; see Supplemental Figure 1A online), suggesting that the putative APB motif in PIF1 is also necessary for the interaction with the Pfr form of phyB.
Figure 5.
The APB and APA Motifs Present in the N-Terminal 150–Amino Acid Region of PIF1 Are Necessary for Its Pfr-Specific Interaction with phyA and phyB Both in Vitro and in Vivo.
(A) Schematic representation of the GAD-PIF1 baits (left) and full-length phy (phy) preys (right) used in coimmunoprecipitation assays. Mutations made in GAD-PIF1 for testing phyB binding are shown above the diagram, and those for testing phyA binding are shown below the diagram.
(B) and (C) Autoradiographs show in vitro interactions of wild-type PIF1 or each of four PIF1 mutants with the Pr or Pfr form of phyB (B) or with single, double (2M), or triple (3M) mutants of PIF1 with the Pr or Pfr form of phyA (C). The left lane in each panel shows the input, and the other lanes show the pellet fractions from coimmunoprecipitation assays performed with in vitro synthesized bait and prey proteins. The phyA and phyB holoproteins were reconstituted by adding the chromophore. The baits were immunoprecipitated using anti-GAD antibody.
(D) LUC-PIF1-3M shows much less affinity for the Pfr form of phyA and phyB compared with LUC-PIF1 in in vivo coimmunoprecipitation assays. The input and pellet fractions from in vivo coimmunoprecipitation assays are indicated. Total protein was extracted from 4-d-old dark-grown seedlings either exposed to Rp light (R; 3000 μmol·m−2) or kept in the dark (D). Coimmunoprecipitations were performed using the anti-PIF1 antibody or with an unrelated IgG as a control. The immunoprecipitated samples were then probed with anti-phyA, anti-phyB, or anti-LUC antibody.
Furthermore, the APB motif of PIF1 was not necessary for interaction with phyA, as a truncated PIF1 (51 to 478 amino acids) without the first 50 amino acids interacted with the Pfr form of phyA in a similar manner to full-length PIF1 (see Supplemental Figure 2 online). Al-Sady et al. (2006) showed that the two Phe residues (Phe-203 and Phe-209) in PIF3 are necessary for its interaction with phyA. Interestingly, mutations in the corresponding amino acids in PIF1 (Phe-148 and Phe-155) did not disrupt the Pfr-specific binding of PIF1 to phyA (see Supplemental Figures 1B, 3A, and 3B online). However, deletion of 11 (positions 85 to 95) or 34 (positions 84 to 117) amino acid residues markedly reduced the Pfr-specific interaction of PIF1 with phyA (see Supplemental Figures 3A and 3C online). Deletion of 43 amino acid residues (positions 118 to 160) severely reduced the Pfr-specific interaction of PIF1 with phyA (see Supplemental Figures 3A and 3C online). This region of PIF1 (from residue 84 to residue 160) was scrutinized to identify specific amino acids critical for the PIF1–phyA interaction. Site-directed mutagenesis of Leu-95 to Ala showed a similar binding capacity as that of the 11– or 34–amino acid deletion mutants (Figure 5C; see Supplemental Figure 3C online). Site-directed mutagenesis of Ser-123, Gly-153, and Gly-160 to Ala in the Leu-95 mutant background did not show significant differences in binding compared with the Leu-95 single mutant (data not shown). However, site-directed mutagenesis of Asn-144 to Ala in the Leu-95 mutant background showed that these two amino acid residues were necessary for the interaction with the Pfr form of phyA in vitro (Figure 5C; see Supplemental Figure 1C online). These results suggested that the phyA binding sites were different between PIF1 and PIF3.
Although PIFs have been shown to interact with phys in experiments using in vitro transcribed and translated PIFs and phys, neither in vivo interactions nor interactions between plant-expressed proteins have been demonstrated. To investigate whether PIF1 interacts with phyA or phyB in vivo and to examine the involvement of specific amino acids in PIF1–phy interactions in vivo, we generated homozygous transgenic plants expressing LUC-PIF1-3M (a luciferase-PIF1 fusion protein with three mutations in PIF1: G47A, L95A, and N144A) in the pif1 mutant background. We performed coimmunoprecipitation assays using the anti-PIF1 antibody on samples prepared from dark- and light-exposed plants. Results showed that LUC-PIF1 could efficiently interact with both phyA and phyB from plant extracts (Figure 5D). However, coimmunoprecipitations of LUC-PIF1-3M recovered much less phyA and phyB under R light compared with LUC-PIF1 coimmunoprecipitations (Figure 5D). These results were consistent with the in vitro interactions shown in Figures 5B and 5C. Taken together, these data suggested that the three amino acids (Gly-47, Leu-95, and Asn-144) in PIF1 were critical for physical interactions with the Pfr forms of phyA and phyB both in vitro and in vivo.
Direct Interactions with the Pfr Forms of Either phyA or phyB Are Necessary for the Light-Induced Phosphorylation and Degradation of PIF1
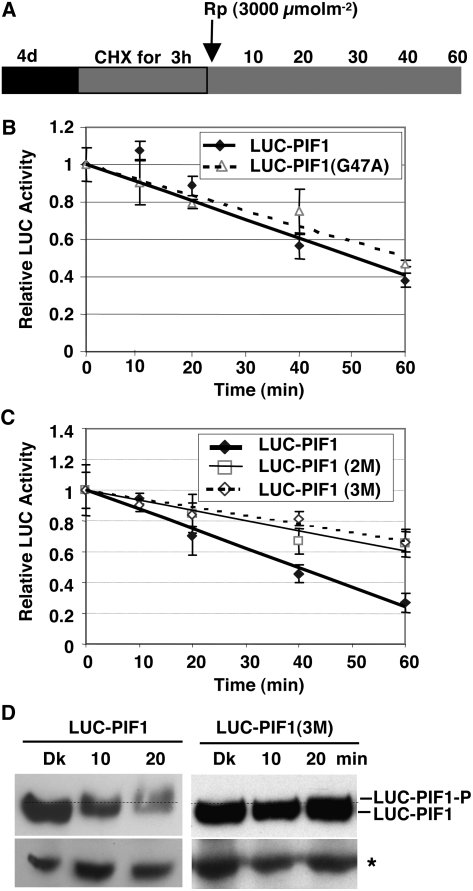
To investigate whether direct physical interactions with phys are necessary for the degradation of PIF1 in light, we generated homozygous transgenic plants expressing LUC-PIF1G47A or LUC-PIF1-2M (containing two mutations in PIF1: L95A and N144A) in the pif1 background. LUC-PIF1-3M (containing three mutations in PIF1: G47A, L95A, and N144A) is described above. Luciferase assays showed that degradation of the LUC-PIF1G47A (deficient in interaction with phyB) was slightly reduced under prolonged R light (see Supplemental Figure 4 online). The triple mutant LUC-PIF1-3M (deficient in interaction with phyA and phyB) was completely stable under FR light and only partially degraded under prolonged R light (see Supplemental Figure 4 online). To investigate the early kinetics of degradation, we performed cycloheximide chase assays for the wild type and the mutant forms of PIF1 fused to LUC after a pulse of R light followed by dark incubation (Figure 6A). The degradation rate of LUC-PIF1G47A was similar to that of wild-type LUC-PIF1 under these conditions (Figure 6B), suggesting that phyB plays a minor role in early PIF1 degradation under limited R light. However, the degradation rates of both LUC-PIF1-2M and LUC-PIF1-3M were greatly reduced after a pulse of R light compared with those of LUC-PIF1 (Figure 6C). Moreover, LUC-PIF1-3M was neither phosphorylated nor degraded up to 20 min after a pulse of R light, whereas wild-type LUC-PIF1 was both phosphorylated and degraded under these conditions (Figure 6D). These results, and those depicted in Figure 5D, suggested that direct interactions of PIF1 with phys were necessary for the light-induced phosphorylation and degradation of PIF1.
Figure 6.
Interactions with the Pfr Form of phyA and phyB Are Necessary for the Light-Induced Phosphorylation and Degradation of PIF1.
(A) Design of the cycloheximide chase assays. Relative lucerifase activity for phy interaction–deficient mutants was measured in 4-d-old dark-grown seedlings pretreated with cycloheximide (CHX) in the dark for 3 h, exposed to R (3000 μmol·m−2) light, and then incubated in the dark for the indicated times (min).
(B) and (C) Assays show the kinetics of degradation of LUC-PIF1-G47A (B) and LUC-PIF1-2M and LUC-PIF1-3M (C) compared with wild-type LUC-PIF1. LUC-PIF1G47A is deficient in phyB interaction, LUC-PIF1-2M is deficient in phyA interaction, and LUC-PIF1-3M is deficient in both phyA and phyB interaction, as shown in Figure 5. Means ± se of five biological replicates are shown.
(D) The abundance and phosphorylation status of LUC-PIF1 and LUC-PIF1-3M fusion proteins prior to and after exposure to Rp determined on protein gel blots using anti-LUC antibody. The dotted line separates the two forms of PIF1. The asterisk denotes a cross-reacting band.
The Transcriptional Activation Domain and phy Interaction Domain Overlap in the N-Terminal 150–Amino Acid Region of PIF1
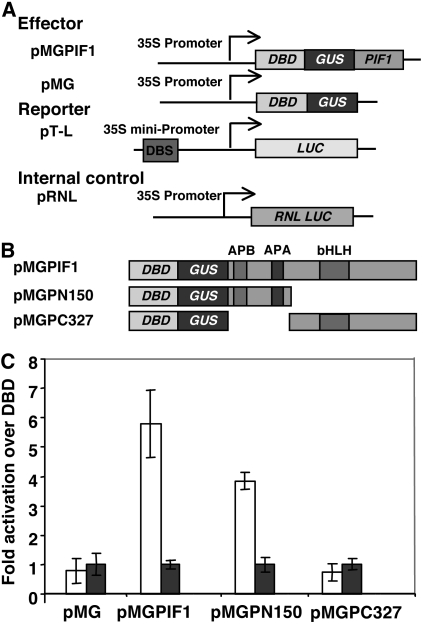
Protein degradation domains have been shown to overlap with transcription activation domains in other systems (Salghetti et al., 2000; Muratani and Tansey, 2003). To investigate whether the degradation domain and transcription activation domain of PIF1 overlap, we mapped the transcription activation domain of PIF1 using the transient assay system we developed (Huq et al., 2004). The N-terminal 150–amino acid region had the transcriptional activation activity of PIF1 (Figure 7). Strikingly, the transcription activation domain overlapped with the APB and APA motifs of PIF1 (Figure 5), which were necessary for PIF1 interaction with photoactivated phys and subsequent degradation in light.
Figure 7.
Transcriptional Activation Domains Are Located at the N Terminus of PIF1.
(A) Constructs used for the experiment. The effector constructs were designed to express a GAL4 DNA binding domain (DBD)–PIF1 fusion (pMGPIF1) or the GAL4 DNA binding domain alone (pMG). The reporter construct (pT-L) expresses a firefly luciferase (LUC) from the 35S minimal promoter fused to the gal4 DNA binding site (DBS). The internal control (pRNL) expresses a renilla luciferase (RNL LUC) from the 35S promoter.
(B) PIF1 deletion constructs used to map the transcriptional activation domains.
Each effector construct in (A) and (B) is fused to β-glucuronidase (GUS) to permit the determination of the expression level of the fusion proteins.
(C) Three-day-old etiolated Arabidopsis seedlings were cobombarded with the reporter and effector constructs. Seedlings were treated for 15 min with FR light and then incubated in darkness for 16 h. Means ± se from four biological replicates are shown. Transcriptional activity was measured in seedling extracts by a dual-luciferase assay system (Promega). Fold activation is expressed as transcriptional activation activity of DBD-GUS-PIF1 over transcriptional activity of DBD-GUS (white bars) and normalized with GUS activity for the amount of protein expressed by each construct (blue bars).
Both N- and C-Terminal Domains of PIF1 Are Necessary for the Light-Induced Degradation of PIF1
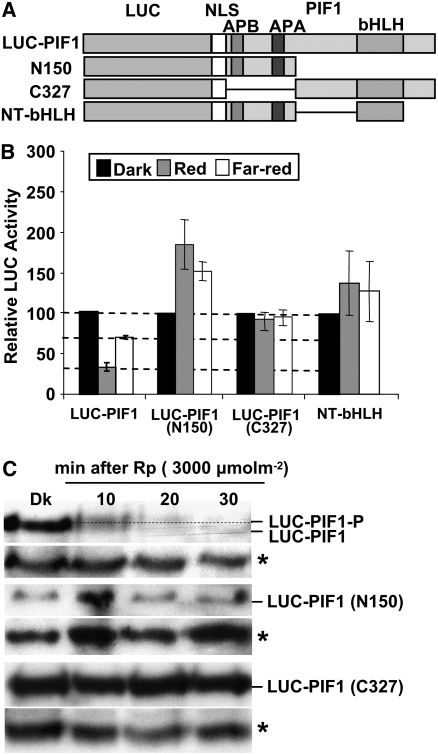
Since PIF1 was degraded under R and FR light, we initiated mapping of the degradation domain of PIF1. To this end, we generated translational fusions of LUC with one of two regions of PIF1 (amino acid residues 1 to 150, responsible for phy interaction and including the transcriptional activation domain of PIF1, and residues 151 to 478, responsible for dimerization and DNA binding) as described (Shen et al., 2005), and produced transgenic plants. To examine whether dimerization was necessary for PIF1 degradation, we also produced transgenic plants expressing LUC fused to the 150–amino acid region of PIF1 along with the bHLH domain (Figure 8A). We measured LUC activity as an indicator of fusion protein stability under dark and light conditions as described (Shen et al., 2005). All three truncated fusion proteins were stable under both R and FR light, while the full-length LUC-PIF1 fusion protein was degraded under those conditions as expected (Figure 8B). Protein gel blot analyses of two of the truncated proteins (LUC-PIF1-N150 and LUC-PIF1-C327) showed that these fusion proteins were neither phosphorylated nor degraded under R light (Figure 8C). These results strongly suggested that both the N- and C-terminal regions of PIF1 were necessary, but not sufficient, for the light-induced degradation of PIF1. In addition, since the phy interaction motifs were present in the 150–amino acid region of PIF1, these results together with the above point mutations (Figures 5 and 6) suggest that phy binding was necessary, but not sufficient, for PIF1's light-induced degradation. Moreover, the transcriptional activation domain of PIF1 was necessary, but not sufficient, to orchestrate light-induced PIF1 degradation.
Figure 8.
Both the N and C Termini of PIF1 Are Necessary for the Light-Induced Degradation of PIF1.
(A) Design of the PIF1 deletion constructs fused to LUC. The white boxes represents a nuclear localization signal (NLS).
(B) LUC activity was measured from 4-d-old dark-grown seedlings transferred to R (10 μmol·m−2·s−1) or FR (10 μmol·m−2·s−1) light for 1 h as described (Shen et al., 2005). Means ± se of five biological replicates are shown. Some constructs showed greater stability of the fusion protein in light relative to darkness for unknown reasons.
(C) Protein gel blots showing truncated PIF1 fusion proteins are neither phosphorylated nor degraded under light, but the wild-type LUC-PIF1 is both phosphorylated and degraded under light. The dotted line separates the two forms of PIF1. Asterisks denote a cross-reacting band.
DNA Binding Is Not Necessary for the Light-Induced Degradation of PIF1
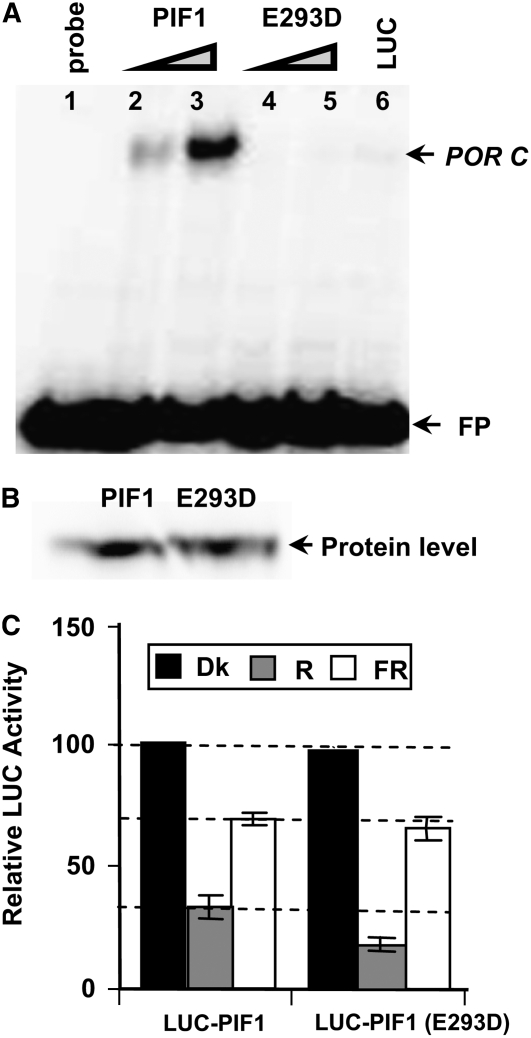
In other systems, transcription factors are often tagged for subsequent degradation by the ubi/26S proteasomal pathway while they are assembled in the transcription initiation complex bound to their DNA target (Mayr and Montminy, 2001; Muratani and Tansey, 2003). Davis et al. (1990) showed that a single amino acid substitution (E118D) in MYOD, a bHLH protein, abolished its DNA binding activity. To investigate whether DNA binding was necessary for the light-induced degradation of PIF1, we introduced the above missense mutation in the corresponding amino acid of PIF1 (PIF1E293D) and compared the DNA binding activity of the wild-type and mutant PIF1. The mutant PIF1 did not bind to the target DNA, while the wild-type PIF1 showed robust binding (Figures 9A and 9B). We made a LUC-PIF1E293D fusion construct and generated homozygous transgenic plants expressing the fusion protein in the pif1 mutant background. LUC assays showed that this mutant PIF1 (PIF1E293D) was degraded significantly more than the wild-type PIF1 under R light (Figure 9C). These data suggested that DNA binding was not necessary for, and might have retarded, light-induced PIF1 degradation.
Figure 9.
DNA Binding Is Not Necessary for the Light-Induced Degradation of PIF1.
(A) The PIF1E293D mutant does not bind to a G-box DNA sequence element (POR C; Su et al., 2001; Moon et al., 2008). In vitro translated PIF1 or PIF1E293D was incubated with a radiolabeled fragment of POR C in a DNA gel shift assay. Lane 1, free probe; lanes 2 and 3, increasing amounts of wild-type PIF1; lanes 4 and 5, increasing amounts of PIF1E293D mutant protein; lane 6, unrelated luciferase protein as a negative control. FP, free probes.
(B) Comparison of the levels of wild-type and mutant PIF1 proteins produced by in vitro transcription and translation.
(C) Relative LUC assays were performed under the conditions described for Figure 8. Means ± se of five biological replicates are shown.
Both the N and C Termini of Either phyA or phyB Are Necessary for the Pfr-Specific Interactions with PIF1
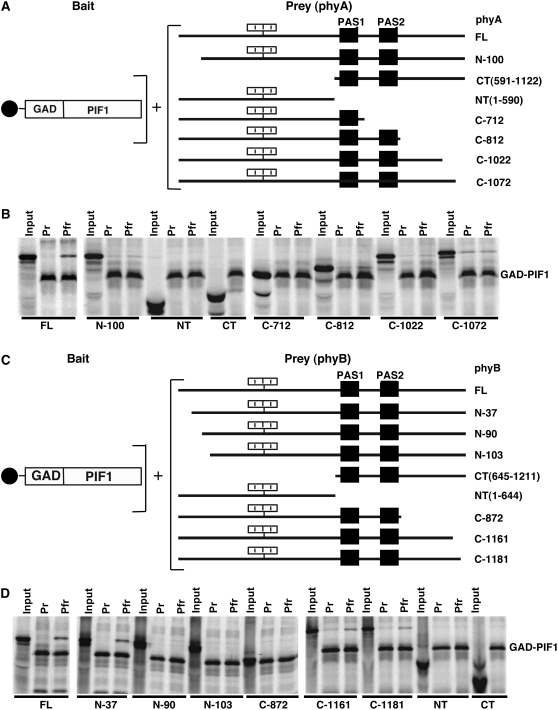
To understand the biological significance of PIF1–phy interactions, we also mapped the interaction domains in phyA and phyB, using in vitro coimmunoprecipitation assays as described (Zhu et al., 2000; Huq et al., 2004). To identify interaction domains in phys, we made seven phyA deletion mutants from either the N- or C-terminal end (Figure 10A). The deletion mutants of phyB have been described previously (Figure 10C) (Zhu et al., 2000). We used gal4 activation domain (GAD)–PIF1 as bait to immunoprecipitate full-length or deletion mutants of both phyA and phyB. PIF1 did not interact with either half of phyA (N or C terminus) separately (Figure 10B). Deletion of 100 amino acids from the N terminus of phyA abolished the Pfr-specific interaction of PIF1 with phyA. Moreover, deletion of 50 amino acids from the C terminus of phyA (Figure 10B, C-1072) abolished the conformer-specific interaction with PIF1, as this truncated protein interacted with PIF1 equally well in both the Pr and Pfr forms in a light-independent manner. These data suggested that both the N- and C-terminal domains of phyA were necessary for the Pfr-specific interaction with PIF1 (Figure 10B).
Figure 10.
Both the N and C Termini of phyA and phyB Are Necessary for Interaction with PIF1.
(A) and (C) Schematic representations of the GAD-PIF1 bait (left) and various phy preys (right) used for coimmunoprecipitation assays.
(B) and (D) Autoradiographs showing interactions of full-length PIF1 with the Pr and Pfr forms of full-length and truncated versions of phyA (B) and phyB (D). Input and pellet fractions are shown from the in vitro coimmunoprecipitation assays performed as described (see Figure 5) (Huq et al., 2004). The CT samples are from the C-terminal halves of either phyA or phyB that do not bind to chromophore and are not labeled as Pr or Pfr.
Similar to the case with phyA, PIF1 did not interact with either half of phyB (Figure 10D). Deletion of the N-terminal 37 amino acids in the full-length context slightly reduced the interaction with PIF1. However, deletion of the 90 amino acids from the N terminus completely abolished the interaction with PIF1. Deletion of the C-terminal 30 and 50 amino acids equally reduced the Pfr-specific interactions with PIF1. However, deletion of 339 amino acids from the C terminus completely abolished the interaction with PIF1. These results were similar to those obtained using PIF3 with truncated phyB proteins, in which PIF3 showed a reduced interaction with phyB proteins containing small deletions at either the N- or C-terminal domain (Zhu et al., 2000). However, unlike PIF3, PIF1 did not show any affinity for the N- or C-terminal half of phyB separately.
Overexpression of the Light-Stable Truncated Form of PIF1 Induces Constitutive Photomorphogenic Phenotypes in the Dark
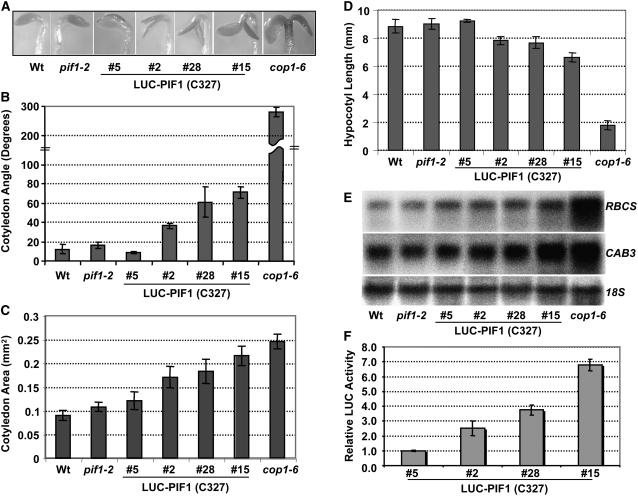
To investigate the biological functions of these various missense and truncated PIF1 mutants, we selected homozygous lines for plants expressing LUC-PIF1-3M, LUC-PIF1-E293D, LUC-PIF1-N150, and LUC-PIF1-C327 and compared the seedling phenotypes with those of LUC-PIF1 transgenic lines. As previously shown, none of these transgenic plants complemented the seed germination phenotypes of the pif1 mutant (see Supplemental Figures 5G and 6G online), possibly due to the use of the 35S promoter (Shen et al., 2005). However, both LUC-PIF1 and LUC-PIF1-3M complemented the seedling phenotypes of the pif1 mutant, including hypocotyl lengths, chlorophyll content, bleaching phenotypes, and hypocotyl negative gravitropism, to a similar extent (see Supplemental Figure 5 online). LUC-PIF1-E293D showed increased levels of chlorophyll content and shorter hypocotyls compared with the pif1 mutant (see Supplemental Figures 5A to 5D online). These data suggested that the LUC-PIF1-E293D fusion protein not only failed to complement the pif1 phenotypes but also displayed enhanced hypersensitive phenotypes compared with the pif1 mutant, possibly due to dominant negative effects. LUC-PIF1-N150 did not complement any of the above phenotypes (see Supplemental Figure 6 online). Strikingly, LUC-PIF1-C327 induced a constitutively photomorphogenic phenotype in the dark in a dose-dependent manner (Figure 11; see Supplemental Figure 6 online). These seedlings had open and expanded cotyledons and showed shorter hypocotyls and increased levels of photosynthetic gene expression compared with the wild-type seedlings in the dark. Moreover, these seedlings also showed greener and larger (more expanded) cotyledons and shorter hypocotyls compared with wild-type seedlings when grown under light (see Supplemental Figure 6 online). It is possible that the truncated form of PIF1 functions in a dominant negative manner to induce constitutive photomorphogenic phenotypes in the dark.
Figure 11.
Overexpression of the Light-Stable, Truncated Form of PIF1 (C327) Induces a Constitutive Photomorphogenic Phenotype in the Dark.
(A) Visible cotyledon-opening phenotypes of various lines grown in the dark for 4 d.
(B) to (D) Measurement of cotyledon angles (B), cotyledon areas (C), and hypocotyl lengths (D) of various lines grown in the dark for 4 d (means ± se; n ≥ 30).
(E) Photosynthetic gene expression is higher in the C327 lines compared with the wild type in the dark. RNA was extracted from 4-d-old dark-grown seedlings and probed for the indicated photosynthetic (RBCS and CAB3) or nonphotosynthetic control (18S) transcripts.
(F) Luciferase activity of various LUC fusion proteins as an indicator of C327 protein amounts in the independent transgenic lines. Relative LUC assays were performed from 4-d-old dark-grown seedlings as described (Shen et al., 2005).
DISCUSSION
Because PIFs physically interact with the photoactivated phy molecules, PIFs were thought to receive light signals from phys and induce photomorphogenesis (Ni et al., 1998, 1999; Quail, 2002). However, contrary to our expectations, the majority of the biological functions of the PIF family members are to negatively regulate phy signaling (Castillon et al., 2007; Monte et al., 2007). To remove this negative regulation, phys induce the degradation of PIFs in order to promote photomorphogenesis. Here, we present evidence that, using diverse sequences, phys interact with PIF1 to induce its phosphorylation, polyubiquitination, and subsequent degradation under both R and FR light conditions. Moreover, overexpression of a light-stable truncated form of PIF1 induced constitutively photomorphogenic phenotypes in the dark (Figure 11), suggesting that an inactivation of PIFs by higher order mutation might be sufficient to induce photomorphogenesis constitutively in the dark.
The comprehensive data presented here advances our understanding of how PIFs function to regulate photomorphogenesis in multiple ways. Our results show that PIF1, the member with the highest affinity for phys, is highly sensitive to the quality and quantity of light. The half-life of native PIF1 was ∼1 to 2 min under 1 μmol·m−2 R light (Figure 1). Other PIFs, including PIF3, PIF4, and PIF5, are degraded with varying but lower sensitivity under R and/or FR light conditions (Bauer et al., 2004; Monte et al., 2004; Lorrain et al., 2007; Nozue et al., 2007; Shen et al., 2007; Leivar et al., 2008). phyA is also degraded under R light through the ubi/26S proteasomal pathway (Shanklin et al., 1987). However, native PIF1 is much more sensitive to R light compared with all other known light-labile proteins. Because of phyA's high sensitivity and early role in responding to light signals, it played the dominant role in regulating the stability of PIF1 under low R light intensity (2 μmol·m−2) (Figure 2A). However, under high R light intensity (3000 μmol·m−2), phyB and phyD and possibly other phys also influenced PIF1 stability. The native PIF1 was also significantly more sensitive to light than the PIF1 fusion proteins originally used to demonstrate light-induced degradation of PIF1 (Shen et al., 2005; Oh et al., 2006; Castillon et al., 2007; Quail, 2007). The small difference in degradation rate might be due to the overexpression of the fusion proteins using the constitutively active 35S promoter and/or to the differential affinities of the wild-type and PIF1 fusion proteins toward phys. Taken together, these results now demonstrate that PIF1 is one of the most light-sensitive proteins known in plants, which is consistent with PIF1 having the strongest affinity for both phyA and phyB among all of the PIFs (Huq et al., 2004). The strong light sensitivity of PIF1 is also consistent with its role in regulating seed germination. In natural conditions, seeds buried under soil are exposed to a small amount of light penetrating through the soil surface, and that might be sufficient to degrade PIF1 to allow the completion of germination (Oh et al., 2004, 2007).
Recently, the early events in the light-induced degradation of PIFs have begun to be revealed. The data presented here demonstrate that PIF1 was phosphorylated and polyubiquitinated specifically under both R and FR light conditions before being degraded by the proteasomal pathway (Figures 2 and 3). Light-induced phosphorylation and polyubiquitination of PIF3 and PIF5 have recently been shown (Park et al., 2004; Al-Sady et al., 2006; Lorrain et al., 2007; Shen et al., 2007). However, these alterations were seen only under R light. Both PIF3 and PIF5 are also degraded under FR light, but the early steps in FR light–induced degradation are not yet known. Our results suggest that the early events in both R and FR light–induced degradation of PIFs might be their phosphorylation and polyubiquitination followed by their degradation by the ubi/26S proteasomal pathway.
PIFs have been shown to interact selectively with the Pfr form of phys in vitro (Ni et al., 1999; Huq et al., 2004). Sequence alignment and site-directed mutagenesis revealed that an N-terminal motif, the APB motif (see Supplemental Figure 1A online), is necessary for the physical interactions between PIF3 to PIF7 and phyB in vitro (Khanna et al., 2004; Shen et al., 2007; Leivar et al., 2008). A second motif immediately downstream of the APB motif, the APA motif (see Supplemental Figure 1B online), has been shown to mediate interactions between PIF3 and phyA (Al-Sady et al., 2006). Here, we show that while PIF1 had a functionally conserved APB motif (Figure 5), it used a novel APA motif for interaction with the Pfr form of phyA (Figure 5C; see Supplemental Figure 1C online). The APA and APB motifs were necessary for the robust interaction with phyA and phyB, respectively, both in vitro and in vivo (Figures 5C and 5D). Moreover, because the triple mutant still interacted with phyA/phyB in vivo, perhaps additional amino acid residues in PIF1 participate in physical interactions between PIF1 and phys in vivo. Combined, these data suggest that although phyB uses a highly conserved sequence motif for physical interactions with PIFs, phyA uses a more diverse sequence for physical interactions with PIFs. Identification and functional characterization of additional phyA-interacting factors might reveal whether phyA uses any conserved sequence motif for physical interaction.
The functional significance of PIF–phy physical interactions appears antagonistic. Direct interactions with phys are necessary for the light-induced phosphorylation and degradation of PIF1, because a PIF1 triple mutant deficient in phy interaction displayed reduced levels of phosphorylation and degradation under light (Figure 6). These results are consistent with recent reports that physical interactions with phys are necessary for the light-induced phosphorylation and degradation of PIF3/PIF5 (Al-Sady et al., 2006; Lorrain et al., 2007; Shen et al., 2007). However, expression of two separate regions of PIF1 (amino acids 1 to 150, containing the transcriptional activation domain as well as the APA and APB motifs, and amino acids 151 to 478, containing the dimerization domain) in transgenic plants showed that these isolated regions were neither phosphorylated nor degraded under either R or FR light conditions (Figure 8). Because the phy interaction motifs are present at the N-terminal 150–amino acid region of PIF1 (Figure 5), these results demonstrate that although the physical interactions between PIF1 and phys are necessary, they are not sufficient for the light-induced phosphorylation and degradation of PIF1. Therefore, PIFs might have additional molecular determinants for light-induced phosphorylation and degradation. Further characterization of amino acid residues using site-directed mutagenesis is necessary to identify these regions.
Our data demonstrating that the putative transcription activation domain of PIF1 was necessary, but not sufficient, for its light-induced degradation (Figure 7) suggest that not all transcription activation domains function as degrons, as hypothesized previously (Salghetti et al., 2000; Muratani and Tansey, 2003). Moreover, enhanced degradation of the PIF1 mutant that failed to bind to DNA also suggests that DNA binding may inhibit PIF1 degradation (Figure 9). These results are consistent with previous reports that a small fraction of PIF1 (20 to 30%) was not degraded even under continuous light exposure (Shen et al., 2005). Taken together, these results suggest that the light-induced degradation of PIF1 might be nucleoplasmic and is uncoupled from the transcription complex.
Although the interaction motifs in PIFs have been the focus of recent investigations, the interaction motifs in phys have not been investigated in detail. PIF3 has been shown to interact with the N- and C-terminal halves of phyB separately (Ni et al., 1999; Zhu et al., 2000). Moreover, PIF3 showed higher affinity for the Pfr form of the N-terminal half compared with the nonphotoactive C-terminal half of phyB. The Pfr form of full-length phyB showed a greater and synergistic affinity for PIF3 relative to the two isolated halves. Sequence regions of phyA for PIF3 interaction are not yet known. Here, we show that, unlike PIF3, PIF1 does not interact with the N- or C-terminal half of either phyA or phyB (Figure 10). Moreover, neither phyA nor phyB interacts with the DNA-bound PIF1 in vitro (Huq et al., 2004). Interactions between PIF3 and the N- or C-terminal half of phyB have been interpreted to explain the biological functions of the N-terminal half of phyB in transgenic plants (Ni et al., 1999; Zhu et al., 2000; Matsushita et al., 2003; Oka et al., 2004). Our data suggest that phy signaling through the direct interaction of the N-terminal half of phyB with PIF3 may not represent a general mechanism for all of the PIFs, as proposed previously.
Studies with pif monogenic mutants did not reveal any significant role of PIFs in regulating the morphological phenotypes of dark-grown seedlings. However, the hypersensitive phenotypes of pif3, pif4, and pif5 single and higher order mutants under prolonged R light have been shown to be due to an increased level of phyB (Monte et al., 2004; Leivar et al., 2008). By contrast, the chlorophyll biosynthetic and seed germination defects of the pif1 mutant are due to a misregulation of these pathways in the dark (Huq et al., 2004; Oh et al., 2006, 2007). PIF1 directly and indirectly regulates the key genes in the chlorophyll biosynthetic pathway in the dark to optimize the greening process in Arabidopsis (Moon et al., 2008). Moreover, both PIF1 and PIF3 constitutively activate transcription in the dark, which is reduced under light, presumably due to their light-induced degradation (Bauer et al., 2004; Huq et al., 2004; Shen et al., 2005; Al-Sady et al., 2008). Consistent with these results, it is striking that the overexpression of a light-stable truncated form of PIF1 induced constitutively photomorphogenic phenotypes in the dark (Figure 11). These transgenic plants showed both morphological and molecular phenotypes qualitatively similar to the cop1 mutant in a dose-dependent manner. This region of PIF1 contains the bHLH dimerization and DNA binding domains without the transcriptional activation domain (Figure 7). It is possible that this region functions in a dominant negative manner by heterodimerizing with other PIFs and titrating out their activity in the dark. These data suggest that simultaneous removal of all PIFs by light-mediated degradation might be sufficient to induce photomorphogenesis. Alternatively, photomorphogenesis might be induced in the dark by overexpression of a dominant-negative form of PIF or possibly by creating a higher order mutant of PIFs. This hypothesis is consistent with a recent report that overexpression of constitutively photoactive phyA and phyB induces photomorphogenesis in the dark (Su and Lagarias, 2007), presumably due to light-independent degradation of PIFs in the dark. Taken together, these results suggest that PIFs negatively regulate photomorphogenesis not only in the light but also in the dark.
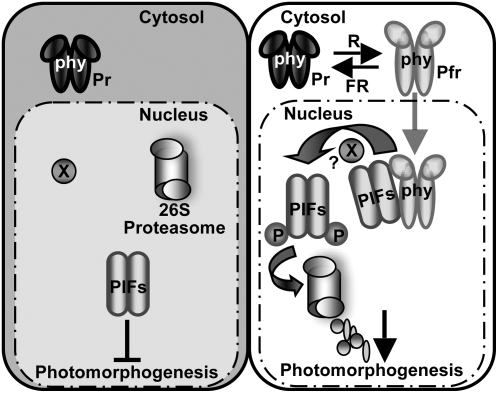
In conclusion, PIF1 and possibly other PIFs appear to play major roles in the dark to inhibit photomorphogenesis (Figure 12, left). Light-activated photoreceptors directly interact with PIFs to induce their phosphorylation, polyubiquitination, and subsequent degradation via the ubi/26S proteasomal pathway in order to promote photomorphogenesis (Figure 12, right). Because direct physical interactions of PIFs with phys are necessary for the light-induced phosphorylation of PIFs, and because phyA has been shown to function as a nonconventional Ser/Thr kinase (Yeh and Lagarias, 1998), it is possible that phys can directly phosphorylate PIFs. However, convincing in vivo evidence of phyA kinase activity is still lacking. Therefore, it remains to be determined whether the light-induced phosphorylation of PIFs represents the primary biochemical mechanism of phy signal transfer or whether phys simply function as scaffold proteins to bring the PIFs and another unknown kinase together for the phosphorylation event.
Figure 12.
Simplified Model of PIF Function in phy Signaling Pathways.
Left, in the dark, phys are localized to the cytosol, while PIFs are constitutively localized to the nucleus and negatively regulate photomorphogenesis. Right, light signals promote nuclear migration of phys by inducing the photoconversion of the Pr form to the active Pfr form. In the nucleus, the photoactivated phys interact with PIFs, resulting in the phosphorylation of PIF1 and other PIFs either directly or indirectly. The phosphorylated forms of PIFs are then polyubiquitinated by a ubi ligase and subsequently degraded by the 26S proteasome. The light-induced proteolytic removal of PIFs relieves the negative regulation, thus promoting photomorphogenesis. X indicates an unknown factor that might be involved in the light-induced phosphorylation of PIFs. P, phosphorylated form. This figure is adapted and modified from Castillon et al. (2007).
METHODS
Plant Growth Conditions and Phenotypic Analyses
Arabidopsis thaliana plants were grown in Metro-Mix 200 soil (Sun Gro Horticulture) under 24 h of light at 24 ± 0.5°C. Monochromatic R and FR light sources were as described (Shen et al., 2005). Light fluence rates were measured using a spectroradiometer (model EPP2000; StellarNet) as described (Shen et al., 2005). Seeds were surface-sterilized and plated on Murashige and Skoog (MS) growth medium containing 0.9% agar without sucrose as described (Shen et al., 2005). After 3 to 4 d of moist chilling at 4°C in the dark, seeds were exposed to 3 h of white light at room temperature in order to satisfy this requirement for the completion of germination before placing them in the dark for another 4 d. For transgenic plants, the 35S:LUC-PIF1 and 35S:LUC-GFP lines were generated as described (Shen et al., 2005). The 35S:TAP-PIF1 and 35S:TAP-GFP transgenic lines were as described (Moon et al., 2008). Mutant lines used were as follows: cop1-6 (McNellis et al., 1994), phyA-211 (Reed et al., 1994), phyB-9 (Reed et al., 1993), phyAB and phyABD in the Landsberg erecta background (Devlin et al., 1999), and pif1-2 (Huq et al., 2004). For quantitation of hypocotyl lengths, cotyledon areas, and cotyledon angles, digital photographs were taken and at least 30 seedlings were measured using the publicly available software ImageJ (http://rsb.info.nih.gov/ij/), and the experiments were repeated at least three times. The photobleaching assays for seedlings, seed germination, hypocotyl negative gravitropism, and chlorophyll measurements were performed as described (Huq et al., 2004; Shen et al., 2005, 2007).
Antibody Preparation, Protein Extraction, and Protein Gel Blotting
The amino acid sequence of PIF1 was examined for unique regions predicted to be of high antigenicity. Peptides (21-mers including a noncoded C-terminal Cys) were synthesized (United Biochemical Research) corresponding to the region N terminal to the bHLH section (NH2-EKTNVDDRKRKEREATTTDEC-COOH), and ∼5 mg of the PIF1 peptide was linked to keyhole limpet hemocyanin. An additional ∼10 mg was linked to an agarose gel (Strategic Diagnostics). Keyhole limpet hemocyanin–linked peptide was used to immunize two New Zealand White rabbits. Serum was prepared following terminal cardiac puncture and affinity-purified over the agarose-linked peptide (Strategic Diagnostics). Affinity-purified antibody was stored frozen in aliquots until use.
Four-day-old seedlings were either kept in darkness or exposed to R or FR light (the amount of light is indicated in individual figures) and incubated in the dark for various times before protein extraction. For detecting TAP-PIF1 and LUC-PIF1 proteins in transgenic plants, boiling denaturing buffer (100 mM MOPS, pH 7.6, 5% SDS, 10% glycerol, 4 mM EDTA, and 40 mM β-mercaptoethanol) was added at a 1:3 (w/v) ratio before grinding. Protease inhibitor cocktail (1×; F. Hoffmann-La Roche) and 2 mM phenylmethylsulfonyl fluoride (PMSF) were also added during extraction. For detecting native PIF1 in wild-type plants, ∼0.2 g of tissue was collected and ground in 1 mL of extraction buffer (100 mM Tris-HCl, pH 6.8, 20% glycerol, 5% SDS, 80 μM MG132, 20 mM DTT, 1 mM bromphenol blue, 2 mM PMSF, and 1× protease inhibitor cocktail [F. Hoffmann-La Roche]) and boiled for 2 min. Total protein supernatants were separated on 8% SDS-PAGE gels, blotted onto polyvinylidene difluoride (PVDF) membranes, and probed with anti-PIF1 antibody. Another membrane prepared in parallel was challenged with anti-tubulin (T6074; Sigma-Aldrich) as a loading control. The protein gel blot procedure was performed according to KPL protein detector kit (KPL) instructions, utilizing a 1:5000 dilution of anti-PIF1 and a 1:2500 dilution of the anti-tubulin antibody. Peroxidase-labeled goat anti-rabbit (anti-mouse for tubulin) antibody (KPL) in a 1:50,000 dilution was used as a secondary antibody. For other immunoblot analyses, the membranes were blocked with 1× Tris-buffered saline Tween 20 plus 0.5% nonfat milk buffer at 4°C overnight with different primary antibodies as follows: mouse monoclonal anti-PHYA (073D) (1:500), anti-PHYB (B6-B3) (1:500), and anti-Ubi (1:700; Santa Cruz Biotechnology) or rabbit anti-c-MYC (1:800; Sigma-Aldrich) and anti-luciferase (1:750; Promega). For secondary antibody, peroxidase-labeled goat anti-rabbit antibody (1:4000; Pierce Biotechnology) or anti-mouse IgG horseradish peroxidase conjugate (1:3300; Promega) was used. Membranes were developed with the SuperSignal West Pico Chemiluminescent substrate kit (Pierce Biotechnology) and visualized on x-ray film.
Immunoprecipitation and Alkaline Phosphatase Treatment
For immunoprecipitation and calf intestine alkaline phosphatase (CIAP) assays, 4-d-old dark-grown 35S:TAP-PIF1 and 35S:TAP-GFP seedlings were pretreated with MG132 to reduce ubi-mediated protein degradation. Seedling were transferred into MS-Suc liquid medium containing 30 μM MG132 or an equal volume of solvent control DMSO and incubated in the dark for 4.5 h. Total proteins were extracted from ∼0.4 g of seedlings (either kept in darkness or treated with 3000 μmol·m−2 Rp or FRp followed by dark incubation) with 1 mL of denaturing buffer (100 mM NaH2PO4, 10 mM Tris, pH 8.0, 100 mM NaCl, 8 M urea, 0.05% Tween 20, 1× protease inhibitor cocktail [Roche], 2 mM PMSF, 10 μM MG132, 25 mM β-glycerophosphate, 10 mM NaF, 2 mM Na-orthovanadate, and 100 nM calyculin A) and cleared by centrifugation at 16,000g for 15 min at 4°C. TAP-PIF1 was immunoprecipitated from supernatants with nickel-nitrilotriacetic acid agarose magnetic agarose beads (Qiagen) as described (Al-Sady et al., 2006). The pellet was resuspended in 100 μL of CIAP reaction buffer and then treated with 100 units of CIAP (F. Hoffmann-La Roche), with the same amount of boiled CIAP, or without enzyme for 60 min at 37°C. Pellets were washed with PBS, heated at 65°C in 1× SDS-Laemmli buffer for 5 min, and subjected to protein gel blot analysis with anti-c-MYC or anti-Ubi antibody as described above.
Construction of Plasmids and in Vitro/in Vivo Coimmunoprecipitation Assays
The DNA constructs for expressing full-length phyA, phyB, GAD, and GAD-PIF1 have been described previously (Huq et al., 2004). The phyB deletion constructs were as described (Zhu et al., 2000). Various fragments of PIF1 or phyA were amplified by PCR using PfuTurbo enzyme and then cloned into the pET17b vector (EMD Biosciences) for in vitro expression. The specific amino acid mutations in full-length PIF1 were introduced using a site-directed mutagensis kit (Stratagene). Restriction enzyme sites (EcoRI-SalI or EcoRI-XhoI for PIF1 and NdeI-XhoI for phyA) were introduced into the PCR primers (see Supplemental Table 1 online), and all of the constructs were sequenced completely. For in vitro coimmunoprecipitation assays, all proteins were expressed in the TnT in vitro transcription/translation system (Promega) in the presence of [35S]Met using the T7 promoter. In vitro coimmunoprecipitation experiments and sample preparation were performed as described (Ni et al., 1999; Huq et al. 2004).
For in vivo coimmunoprecipitation assays, seedlings were pretreated with MG132 as described above. Total proteins were extracted from ∼0.4 g of seedlings (either kept in darkness or treated with 3000 μmol·m−2 Rp followed by dark) with 1 mL of native extraction buffer (100 mM NaH2PO4, pH 7.8, 100 mM NaCl, 0.05% Nonidet P-40, 1× protease inhibitor cocktail [F. Hoffmann-La Roche], 2 mM PMSF, 10 μM MG132, 25 mM β-glycerophosphate, 10 mM NaF, 2 mM Na orthovanadate, and 100 nM calyculin A) and cleared by centrifugation at 16,000g for 15 min at 4°C. Anti-PIF1 antibody was incubated with Dynabeads (20 μL/μg antibody; Invitrogen) for 30 min at 4°C, and the beads were washed twice with the extraction buffer to remove the unbound antibody. The bound antibody beads were added to 500 μg of total protein extracts and rotated for another 3 h at 4°C in the dark. The beads were collected using a magnet, washed three times with wash buffer, dissolved in 1× SDS-loading buffer, and heated at 65°C for 5 min. The immunoprecipitated proteins were separated on an 8% SDS-PAGE gel, blotted onto PVDF membranes, and probed with anti-phyA, anti-phyB, or anti-LUC antibody as described above.
Cycloheximide Chase and Luciferase Assays
For cycloheximide chase assays, 4-d-old dark-grown seedlings were pretreated with 50 μM cycloheximide or solvent control DMSO in MS-Suc liquid medium for 3 h in darkness as described (Shen et al., 2005). After pretreatment, the seedlings were exposed to 3000 μmol·m−2 Rp for 1 min and then kept in darkness before harvesting at the different time points indicated in the figures. For luciferase assays, samples were collected in liquid nitrogen, and total protein was extracted using 1× luciferase cell culture lysis reagent (Promega) with 2 mM PMSF and 1× complete protease inhibitor cocktail (F. Hoffmann-La Roche). Luciferase activity was measured as described (Shen et al., 2005).
Construction of Plasmids and Transient Transcription Activation Assays
For transient transcription activation assays, the full-length PIF1 open reading frame or different fragments were cloned as SmaI-KpnI fragments into pMN6 in-frame with the Gal4 DNA binding domain (Huq et al., 2004). Full-length β-glucuronidase cDNA without the stop codon was amplified with PfuTurbo polymerase (Stratagene) using the SmaI restriction sites at both ends. This fragment was inserted into the SmaI site for in-frame fusion with either the DNA binding domain alone (pMG) or the DNA binding domain and PIF1 (pMGPIF1) reading frames. pMG alone was used as a negative control. pT-L and pRNL plasmids have been described (Huq et al., 2004). The transient experiments and dual-luciferase assays were carried out as described (Huq et al., 2004).
In Vitro Gel-Shift Assays
DNA gel-shift assays were performed as described (Huq and Quail, 2002). PIF1, PIF1E293D, and LUC were synthesized using the rabbit reticulocyte TNT system (Promega). A 70-bp POR C promoter fragment containing a G-box motif known to be a PIF1 binding site was labeled with [32P]dCTP (Su et al., 2001; Moon et al., 2008). The binding conditions and gel compositions were as described (Huq and Quail, 2002).
RNA Isolation and RNA Gel Blotting
Total RNA was isolated from 6-d-old seedlings using the Qiagen RNeasy mini kit (Qiagen). RBCS, CAB3, and 18S cDNA probes (Deng et al., 1992) were labeled ([32P]dCTP) using the random primer labeling kit (TaKaRa). RNA gel blotting was performed on 10 μg of total RNA according to the manufacturer's instructions using the NorthernMax-Gly kit (Ambion). After two low-stringency and high-stringency washes at 42°C, the membrane was dried and exposed to a phosphor screen (Kodak) at room temperature overnight. The phosphor screen was developed using the Molecular Imager FX system (Bio-Rad Laboratories).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CAB3 (At1g29910), PIF1 (At2g20180), PHYA (At1g09570), PHYB (At2g18790), POR C (At1g03630), RBCS1A (At1g67090), and TUBULIN (At1g04820).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignments of the APB and APA Motifs in PIFs.
Supplemental Figure 2. The APB Motif Is Not Responsible for the Interaction of PIF1 with phyA.
Supplemental Figure 3. The Putative APA Motif Present in PIF3 Is Not Responsible for the Pfr-Specific Interaction of PIF1 with phyA.
Supplemental Figure 4. Direct Interactions of PIF1 with phyA and/or phyB Are Necessary for the Light-Induced Degradation of PIF1.
Supplemental Figure 5. Rescue of the pif1-2 Chlorophyll Biosynthetic Phenotypes in Transgenic Seedlings Expressing Wild-Type and Point Mutant Versions of PIF1.
Supplemental Figure 6. Rescue of the pif1-2 Chlorophyll Biosynthetic Phenotypes in Transgenic Seedlings Expressing Wild-Type and Truncated Versions of PIF1.
Supplemental Table 1. Primer Sequences Used in the Experiments Described in This Article.
Supplementary Material
Acknowledgments
We thank Guy Thompson and Jennifer Moon for critical reading of the manuscript, Bassem Al-Sady for technical advice, Gary Whitelam for sharing phy mutants, Xing Wang Deng for the cop1 mutant, Peter H. Quail for initial support and for sharing antibodies against phyA and phyB, and Phi Luong, Chelsea Rasmussen, Jullie Sotillo, and Judy Liang for technical assistance. This work was supported by National Science Foundation Grant MCB-0449646 to B.D. and by National Science Foundation Grant IBN-0418653 and a set-up fund from the University of Texas at Austin to E.H.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Enamul Huq (huq@mail.utexas.edu).
Online version contains Web-only data.
References
- Al-Sady, B., Kikis, E.A., Monte, E., and Quail, P.H. (2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. [DOI] [PubMed] [Google Scholar]
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. [DOI] [PubMed] [Google Scholar]
- Chen, M., Chory, J., and Fankhauser, C. (2004). Light signal transduction in higher plants. Annu. Rev. Genet. 38 87–117. [DOI] [PubMed] [Google Scholar]
- Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr. Biol. 15 637–642. [DOI] [PubMed] [Google Scholar]
- Davis, R.L., Cheng, P.-F., Lassar, A.B., and Weintraub, H. (1990). The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell 60 733–746. [DOI] [PubMed] [Google Scholar]
- de Lucas, M., Davière, J.-M., Rodríguez-Falcón, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blázquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a novel protein with both a Zn-binding motif and a Gβ homologous domain. Cell 71 791–801. [DOI] [PubMed] [Google Scholar]
- Devlin, P.F., Robson, P.R.H., Patel, S.R., Goosey, L., Sharrock, R.A., and Whitelam, G.C. (1999). Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation and flowering time. Plant Physiol. 119 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek, P.D., and Fankhauser, C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10 51–54. [DOI] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori, T., Yamashino, T., Kato, T., and Mizuno, T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45 1078–1086. [DOI] [PubMed] [Google Scholar]
- Hiltbrunner, A., Tscheuschler, A., Viczian, A., Kunkel, T., Kircher, S., and Schafer, E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47 1023–1034. [DOI] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941. [DOI] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., and Quail, P.H. (2003). Nuclear translocation of the photoreceptor phytochrome B is necessary for its biological function in seedling photomorphogenesis. Plant J. 35 660–664. [DOI] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao, Y., Lau, O.S., and Deng, X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 217–230. [DOI] [PubMed] [Google Scholar]
- Khanna, R., Huq, E., Kikis, E.A., Al-Sady, B., Lanzatella, C., and Quail, P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, R., Shen, Y., Marion, C.M., Tsuchisaka, A., Theologis, A., Schaefer, E., and Quail, P.H. (2008). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19 3915–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Yi, H., Choi, G., Shin, B., Song, P.S., and Choi, G. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher, S., Gil, P., Kozma-Bognár, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Ádám, E., Schäfer, E., and Nagy, F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J.M., Ecker, J.R., and Quail, P.H. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, C., and Shalitin, D. (2003). Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 54 469–496. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., Allen, T., Duek, P.D., Whitelam, G.C., and Fankhauser, C. (2007). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. [DOI] [PubMed] [Google Scholar]
- Mathews, S., and Sharrock, R.A. (1997). Phytochrome gene diversity. Plant Cell Environ. 20 666–671. [Google Scholar]
- Matsushita, T., Mochizuki, N., and Nagatani, A. (2003). Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424 571–574. [DOI] [PubMed] [Google Scholar]
- Mayr, B., and Montminy, M. (2001). Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2 599–609. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.-W. (1994). Genetic and molecular analysis of anallelic series of cop1 mutants suggest functional roles for the multiple protein domains. Plant Cell 6 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, E., Al-Sady, B., Leivar, P., and Quail, P.H. (2007). Out of the dark: How the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J. Exp. Bot. 58 3125–3133. [DOI] [PubMed] [Google Scholar]
- Monte, E., Tepperman, J.M., Al-Sady, B., Kaczorowski, K.A., Alonso, J.M., Ecker, J.R., Li, X., Zhang, Y., and Quail, P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J., Zhu, L., Shen, H., and Huq, E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- Muratani, M., and Tansey, W.P. (2003). How ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4 1–10. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400 781–784. [DOI] [PubMed] [Google Scholar]
- Nozue, K., Covington, M.F., Duek, P.D., Lorrain, A.A., Fankhauser, C., Harmer, S.L., and Maloof, J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448 358–361. [DOI] [PubMed] [Google Scholar]
- Oh, E., Kim, J., Park, E., Kim, J.I., Kang, C., and Choi, G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Huc, J., Yusukeb, J., Jung, B., Paik, I., Leed, H.-S., Sun, T.-P., Kamiya, Y., and Choi, G. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by directly binding to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W.-I., and Choi, G. (2006). Light activates the degradation of PIL5 to promote seed germination through gibberellin in Arabidopsis. Plant J. 47 124–139. [DOI] [PubMed] [Google Scholar]
- Oka, Y., Matsushita, T., Mochizuki, N., Suzuki, T., Tokutomi, S., and Nagatani, A. (2004). Functional analysis of a 450–amino acid N-terminal fragment of phytochrome B in Arabidopsis. Plant Cell 16 2104–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, E., Kim, J., Lee, Y., Shin, J., Oh, E., Chung, W.I., Liu, J.R., and Choi, G. (2004). Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 45 968–975. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3 85–93. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2007). Phytochrome interacting factors. In Light and Plant Development, K.H.G. Whitelam, ed (Oxford, UK: Blackwell Publishing), pp. 81–105.
- Reed, J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. (1994). Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W., Nagpal, P., Poole, D.S., Furuya, M., and Chory, J. (1993). Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell, N.C., Su, Y.-S., and Lagarias, J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösler, J., Klein, I., and Zeidler, M. (2007). Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc. Natl. Acad. Sci. USA 104 10737–10742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salghetti, S.E., Muratani, M., Wijnen, H., Futcher, B., and Tansey, W.P. (2000). Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl. Acad. Sci. USA 97 3118–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer, E., and Nagy, F. (2006). Photomorphogenesis in Plants and Bacteria. (Dordrecht, The Netherlands: Springer).
- Shanklin, J., Jebben, M., and Vierstra, R.D. (1987). Red light-induced formation of ubiquitin-phytochrome conjugates: Identification of possible intermediates of phytochrome degradation. Proc. Natl. Acad. Sci. USA 84 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock, R.A., and Clack, T. (2004). Heterodimerization of type II phytochromes in Arabidopsis. Proc. Natl. Acad. Sci. USA 101 11500–11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Luong, P., and Huq, E. (2007). The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiol. 145 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Moon, J., and Huq, E. (2005). PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize seedling photomorphogenesis in Arabidopsis. Plant J. 44 1023–1035. [DOI] [PubMed] [Google Scholar]
- Shen, Y., Khanna, R., Carle, C.M., and Quail, P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J., Park, E., and Choi, G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49 981–994. [DOI] [PubMed] [Google Scholar]
- Su, Q., Frick, G., Armstrong, G., and Apel, K. (2001). POR C of Arabidopsis thaliana: a third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol. Biol. 47 805–813. [DOI] [PubMed] [Google Scholar]
- Su, Y.-S., and Lagarias, J.C. (2007). Light-independent phytochrome signaling mediated by dominant GAF domain tyrosine mutants of Arabidopsis phytochromes in transgenic plants. Plant Cell 19 2124–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G., and Halliday, K. (2007). Light and Plant Development. (Oxford, UK: Blackwell Publishing).
- Yeh, K.C., and Lagarias, J.C. (1998). Eukaryotic phytochromes: Light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc. Natl. Acad. Sci. USA 95 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Q., Hare, P.D., Yang, S.W., Zeidler, M., Huang, L.F., and Chua, N.H. (2005). FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J. 43 356–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Y., Tepperman, J.M., Fairchild, C.D., and Quail, P.H. (2000). Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc. Natl. Acad. Sci. USA 97 13419–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.