Abstract
The human DDB1-CUL4 ASSOCIATED FACTOR (DCAF) proteins have been reported to interact directly with UV-DAMAGED DNA BINDING PROTEIN1 (DDB1) through the WDxR motif in their WD40 domain and function as substrate-recognition receptors for CULLIN4-based E3 ubiquitin ligases. Here, we identified and characterized a homolog of human DCAF1/VprBP in Arabidopsis thaliana. Yeast two-hybrid analysis demonstrated the physical interaction between DCAF1 and DDB1 from Arabidopsis, which is likely mediated via the WD40 domain of DCAF1 that contains two WDxR motifs. Moreover, coimmunoprecipitation assays showed that DCAF1 associates with DDB1, RELATED TO UBIQUITIN–modified CUL4, and the COP9 signalosome in vivo but not with CULLIN-ASSOCIATED and NEDDYLATION-DISSOCIATED1, CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1), or the COP10-DET1-DDB1 complex, supporting the existence of a distinct Arabidopsis CUL4 E3 ubiquitin ligase, the CUL4-DDB1-DCAF1 complex. Transient expression of fluorescently tagged DCAF1, DDB1, and CUL4 in onion epidermal cells showed their colocalization in the nucleus, consistent with the notion that the CUL4-DDB1-DCAF1 complex functions as a nuclear E3 ubiquitin ligase. Genetic and phenotypic analysis of two T-DNA insertion mutants of DCAF1 showed that embryonic development of the dcaf1 homozygote is arrested at the globular stage, indicating that DCAF1 is essential for plant embryogenesis. Reducing the levels of DCAF1 leads to diverse developmental defects, implying that DCAF1 might be involved in multiple developmental pathways.
INTRODUCTION
Protein ubiquitination is an important and universal posttranslational modification in eukaryotes. In this process, a cascade of reactions is performed by three distinct enzymes, ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2), and ubiquitin protein ligase (E3). Among them, substrate specificity is largely dependent on E3 ubiquitin ligases, which mediate the recruitment of target protein and the optimal transfer of the ubiquitin moiety from E2 enzyme to the target. Therefore, eukaryotic cells contain hundreds or thousands of distinct E3 ubiquitin ligases for specific ubiquitination of diverse substrates in different biological processes (Glickman and Ciechanover, 2002; Smalle and Vierstra, 2004).
CUL4-based E3 ubiquitin ligases constitute a large subfamily of CULLIN-RING E3 ubiquitin ligases (CRLs) and consist of three core subunits: CULLIN4 (CUL4), a RING finger protein REGULATOR OF CULLINS1 (ROC1)/RING-BOX1 (RBX1), and UV-DAMAGED DNA BINDING PROTEIN1 (DDB1) (Lee and Zhou, 2007). Structurally, the arc-shaped helical N-terminal domain of CUL4 interacts extensively with the β-propeller B domain of the adapter protein DDB1 to assemble a substrate receptor complex, in which the BPA and BPC double propellers of DDB1 fold tightly into a clam-shaped pocket for substrate receptor binding. The RING finger protein ROC1/RBX1 binds to the globular C-terminal portion of CUL4 and recruits E2 enzyme to form a catalytic core. Therefore, CUL4 forms a rigid packing architecture for the precise positioning of substrate toward E2 enzyme, therefore facilitating the ubiquitin transfer (Zheng et al., 2002; Angers et al., 2006; Li et al., 2006).
The core components of CUL4-based E3 ubiquitin ligases, including CUL4 and DDB1, are highly conserved during evolution and have also been studied in Arabidopsis thaliana (Schroeder et al., 2002; Bernhardt et al., 2006; Chen et al., 2006). Arabidopsis CUL4 interacts with DDB1 and ROC1/RBX1 in a conserved manner similar to that of the human DDB1-CUL4-ROC1/RBX1 complex (Angers et al., 2006; Bernhardt et al., 2006; Li et al., 2006). Furthermore, several lines of evidence, including in vivo interactions of CUL4 with CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10) and ROC1/RBX1, and in vitro reconstruction of the COP10-DET1-DDB1 (CDD) complex with CUL4-ROC1/RBX1 to form an active E3 ubiquitin ligase, suggest the existence of a CDD-CUL4-ROC1/RBX1 E3 ubiquitin ligase complex in Arabidopsis (Chen et al., 2006). It is also shown that CUL4 binds to CULLIN-ASSOCIATED and NEDDYLATION-DISSOCIATED1 (CAND1) and the COP9 signalosome (CSN) in a mutually exclusive manner and is modified by RELATED TO UBIQUITIN (RUB), consistent with the known characteristics of the CRL E3 enzyme activity regulation mechanism (Petroski and Deshaies, 2005; Chen et al., 2006). Reducing the amount of CUL4 results in various defects in Arabidopsis development (Bernhardt et al., 2006; Chen et al., 2006). DDB1 has two homologs in Arabidopsis that are encoded by DDB1A and DDB1B, respectively. While the ddb1a mutant shows no obvious phenotype but can enhance the de-etiolated1 (det1) phenotype in the det1 ddb1a double mutant, the ddb1b null mutant is lethal (Schroeder et al., 2002). The genetic and phenotypic analysis of both core components so far has provided evidence for the essential function of CUL4 E3 ubiquitin ligases in Arabidopsis.
The substrate recruitment module for CUL4 E3 ubiquitin ligases has recently been revealed (Angers et al., 2006; He et al., 2006; Higa et al., 2006; Jin et al., 2006). Through proteomic, structural, and bioinformatics analysis, it was found that CUL4, via its linker protein, DDB1, potentially interacts with a large number of proteins containing WD40 repeats, which form a subgroup of the WD40 proteins and are variably referred to as DDB1-CUL4 ASSOCIATED FACTOR (DCAF) proteins, or DDB1 binding WD40 (DWD) proteins, or CUL4- and DDB1-associated WD40-repeat proteins (Angers et al., 2006; He et al., 2006; Higa et al., 2006; Jin et al., 2006). Structure-based sequence analysis and point mutagenesis experiments revealed the presence of one or two copies of the WDxR motif located in the WD40 domain of most DCAF proteins, and within the WDxR motif, the conserved Asp and Arg residues are major determinant features for DCAF proteins to bind DDB1 (Angers et al., 2006; Higa et al., 2006; Jin et al., 2006). Thus, the WDxR motif–containing WD40 domain is defined as the signature of potential substrate receptors of CUL4 E3 ubiquitin ligases, and as many as 90 distinct DWD proteins have been predicted in the human genome (He et al., 2006). Most recently, 85 WD40 proteins in Arabidopsis and 78 WD40 proteins in rice (Oryza sativa) have been found to contain one or two copies of a conserved 16–amino acid DDB1-interacting motif, referred to as the DWD motif, that can potentially act as substrate receptors in different CUL4 E3 ubiquitin ligases (Lee et al., 2008). These findings suggest that CUL4 may potentially constitute a large number of distinct CUL4-DDB1-DCAF/DWD E3 ubiquitin ligase complexes in vivo.
In Arabidopsis, the 85 DWD proteins can be divided into five subgroups based on the phylogenetic relationship of their DWD motifs, and representative proteins from each subgroup have been tested and confirmed for their ability to interact with DDB1 in vitro and associate with DDB1 and CUL4 in vivo (Lee et al., 2008). Among those Arabidopsis DWD proteins, so far only PLEIOTROPIC REGULATORY LOCUS1 has been shown to act as a substrate receptor to target its specific substrate ARABIDOPSIS SNF1 KINASE HOMOLOG 10 for ubiquitination and proteasome-mediated degradation (Lee et al., 2008).
In this study, we report the identification and characterization of a human DCAF1/VprBP homolog in Arabidopsis. Human DCAF1 was originally identified as a Vpr binding protein (VprBP) that interacts with a viral accessory protein (Vpr) of human immunodeficiency virus type 1 (HIV-1). Human DCAF1 is one of the well-studied DCAF proteins, which contains two WDxR motifs within its WD40 domain (Angers et al., 2006). The human CUL4-DDB1-DCAF1 E3 ubiquitin ligase is recruited by HIV-1 protein Vpr to trigger G2 arrest of HIV-1 infected cells (Belzile et al., 2007; Hrecka et al., 2007; Le Rouzic et al., 2007; Schröfelbauer et al., 2007; Tan et al., 2007; Wen et al., 2007), while the cellular function of DCAF1 and the physiological significance underlying the Vpr-DCAF1 interaction for HIV viral propagation both remains unclear. Here, we show that Arabidopsis DCAF1 also contains two WDxR motifs within its WD40 domain, which are essential for the interaction with DDB1. We also provide evidence for the existence of a CUL4-DDB1-DCAF1 E3 ubiquitin ligase in Arabidopsis, which is possibly regulated by RUB, CSN, and CAND1. Interestingly, DCAF1 was not predicted to be a DWD protein based on the criterion of a 16–amino acid DWD motif that was used in a previous report (Lee et al., 2008), suggesting that the number of DWD proteins may be underestimated and that there are potentially other WD40 proteins that can function as substrate receptors of CRL4 E3 ubiquitin ligases. The broad expression of DCAF1, the nuclear colocalization of DCAF1 with CUL4 and DDB1, the embryo lethal phenotype of dcaf1 homozygous T-DNA insertion mutants, and the multifaceted developmental defects shown by dcaf1cs cosuppression lines suggest that DCAF1 and CUL4-DDB1-DCAF1 E3 ubiquitin ligase participate in many biological processes in Arabidopsis.
RESULTS
Identification of the DCAF1 Gene in Arabidopsis
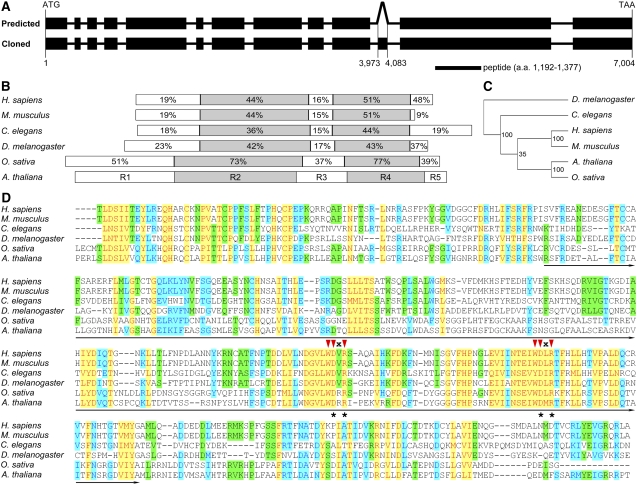
Human DCAF1 is encoded by the KIAA0800 gene, whose preferential expression in testis is transactivated specifically by a sex-determination factor SRY (sex determining region Y)-box 9 (Zhang et al., 2001; Zhao et al., 1994, 2002). Homology search in the Arabidopsis genome using the human DCAF1 amino acid sequence (Zhang et al., 2001; Angers et al., 2006) identified only one homolog, which was subsequently named Arabidopsis DCAF1. The full-length coding sequence (CDS) of the encoding gene was cloned by RT-PCR. By DNA sequencing analysis, we found that full-length DCAF1 CDS is composed of 13 exons corresponding to a protein of 1883 amino acids. However, this result differs from the prediction in the Arabidopsis Genome Initiative (AGI) database, which suggests that this gene has 12 exons and encodes a protein of 1846 amino acids. Upon detailed comparison, we found that the discrepancy is due to a missing exon of 111 bp located within the predicted 10th intron (Figure 1A). To allow further biochemical analysis, polyclonal anti-DCAF1 antibodies were raised against a peptide corresponding to amino acid 1192-1377 of DCAF1, which is encoded by nucleotide 3576-4131 located within the 12th exon (Figure 1A).
Figure 1.
Structure of the Arabidopsis DCAF1 Gene and Sequence Alignment of DCAF1 Homologs.
(A) Structure of the Arabidopsis DCAF1 gene and the location of the antigen used for antibody preparation. Exons are presented as filled black rectangles, and introns are presented as solid lines. The top diagram (predicted) depicts the predicted structure of the DCAF1 gene in the AGI database with 11 introns and 12 exons. The bottom one (cloned) presents the structure of the DCAF1 gene cloned in this study, which has an extra exon inside the predicted 10th intron, corresponding to nucleotide 3973 to 4083 of the genomic fragment. The position of the peptide used as antigen for antibody preparation is indicated underneath the gene structure. a.a., amino acids.
(B) Schematic comparison of DCAF1 homologs from representative eukaryotic organisms as labeled on the left. Based on the difference in amino acid sequence homology, the DCAF1 proteins are divided into five regions, which, in Arabidopsis, are shown by five differently colored rectangles labeled R1 through R5. The percentages of similarity in the corresponding regions from each homolog to the Arabidopsis DCAF1 are indicated in the respective region.
(C) A phylogenetic tree of DCAF1 homologs from the model organisms indicated on the right (see Methods for details on tree generation procedure). The numbers indicate the statistic values of the reliability for each node.
(D) Alignment of the R4 region of DCAF1 from the model organisms labeled on the left. The WD40 domain is underlined. The red triangles indicate the two WDxR motifs in the WD40 domain, and “x” stands for an indefinite amino acid. The asterisks indicate the Asn and Arg (on the top) within the WDxR motif, which are mutated into the Ala residue in the point mutation analysis as shown in Figure 3A. The shading mode indicates the level of conservation, with red letters in yellow shading corresponding to a high level of conservation (100%), blue letters in azure shading corresponding to a moderate level of conservation (80%), and black letters in green shading corresponding to a low level of conservation (60%).
Comparative Analysis of DCAF1 Homologs
From the GenBank reference protein database, we identified homologs of human DCAF1 from several other representative model organisms, including mouse (Mus musculus), fruit fly (Drosophila melanogaster), worm (Caenorhabditis elegans), and rice. By contrast, no DCAF1 homolog could be identified from unicellular organisms, such as budding yeast (Schizosaccharomyces cerevisiae), fission yeast (Schizosaccharomyces pombe), and bacteria (Escherichia coli). This result suggests that DCAF1 is likely to be evolutionarily conserved in multicellular eukaryotic organisms (Zhang et al., 2001; Zhao et al., 2002). Arabidopsis DCAF1 is 20% identical to its human homolog, 19% to mouse, 17% to fruit fly, 14% to worm, and 44% to rice; while the sequence similarity of Arabidopsis DCAF1 with other eukaryotic DCAF1 homologs is 36% for human, 34% for mouse, 31% for fruit fly, 28% for worm, and 60% for rice. A phylogenetic tree based on DCAF1 protein sequence homologies among different organisms is presented (Figure 1C).
As indicated from the sequence alignment (see Supplemental Figure 1 and Supplemental Data Set 1 online), two relatively conserved regions of DCAF1 divide DCAF1 proteins into five fragments, named in order as Region 1 (R1) to Region 5 (R5) (Figure 1B). R2 and R4 are the two conserved regions, and the protein sequence similarity of these regions between Arabidopsis and other eukaryotic organisms is usually ∼40 to 50% and >70% for rice (Figure 1B). These results suggest that the R2 and R4 regions might be important for the function of DCAF1 proteins.
Further sequence analysis by querying the conserved domain database at the National Center for Biotechnology Information with BLAST identified a WD40 domain located in the R4 region of DCAF1 (Marchler-Bauer et al., 2007; http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), which contains two WDxR motifs (Figure 1D). As described previously, it has been demonstrated that DCAF proteins interact physically with DDB1 through the WDxR motifs in the WD40 domain (Angers et al., 2006; Higa et al., 2006; Jin et al., 2006). Therefore, the R4 region of Arabidopsis DCAF1 is expected to mediate direct binding of DCAF1 to DDB1. However, DCAF1 does not belong to the 85 predicted Arabidopsis DWD proteins since it lacks the other sequence features of the conserved 16–amino acid DWD motif, except for the internal four amino acids constituting the WDxR motif (Figure 1D; He et al., 2006; Lee et al., 2008). Therefore, it is of great interest to test if Arabidopsis DCAF1 indeed forms a complex with DDB1 and CUL4, in the same way that its human homolog does (Angers et al., 2006; He et al., 2006; Jin et al., 2006).
Physical Interaction between DCAF1 and DDB1
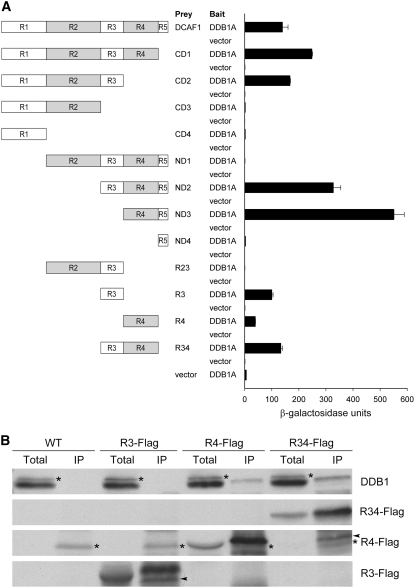
To investigate the possibility of Arabidopsis DCAF1 acting as a substrate receptor in a CUL4-DDB1–based E3 ubiquitin ligase, full-length DCAF1 and DDB1A were cotransformed into yeast for two-hybrid (Y2H) assay. As expected, increased β-galactosidase activity was observed, indicating that DCAF1 interacts directly with DDB1A in yeast (Figure 2A). To further dissect the regions of DCAF1 that mediate the interaction, we constructed a series of DCAF1 deletion mutants (Figure 2A) and tested each of them for their binding capability to DDB1A in yeast. As shown in Figure 2A, interaction tests of C-terminal deletion mutants (CD1 to CD4) with DDB1A imply that the R3 region is possibly essential for DCAF1 binding to DDB1, whereas the results of N-terminal deletion mutants (ND1 to ND4) indicate that the R4 region could bind DDB1 directly. Furthermore, the results of N-terminal deletion mutants also suggest that the R1 and R2 regions might antagonistically regulate DCAF1's interaction with DDB1, in which R1 might act as a positive regulator and R2 as a negative regulator. Then, we selected R2, R3, and R4 regions to further test their respective interactions with DDB1A. Evidently, R3, R4, and R34 (R3 and R4) all are able to bind DDB1A independently, whereas the presence of R2 blocks R3's ability to bind to DDB1A (Figure 2A), supporting the idea that the R2 region plays a negative role in the DCAF1–DDB1 interaction.
Figure 2.
Direct Interaction between DCAF1 and DDB1.
(A) Interaction analysis of DCAF1 and its deletion mutants with DDB1A by Y2H analysis. The left diagrams schematically present a series of deletion mutants corresponding to different regions in DCAF1. Empty bait vector was used as a negative control. The β-galactosidase activities resulting from the interactions are shown in the histogram. Error bars present sd (n = 4).
(B) Association analysis of Flag-tagged R3, R4, and R34 (R3 and R4) regions of DCAF1 with DDB1 by in vivo co-IP with the anti-Flag antibody. Seedling total protein extracts prepared from wild-type and 35S:R3-Flag (R3-Flag), 35S:R4-Flag (R4-Flag), and 35S:R34-Flag (R34-Flag) transgenic Arabidopsis plants were incubated with anti-Flag antibody-coupled agarose. The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against Flag (for Flag-tagged R3, R4, and R34) and DDB1. The asterisks mark the cross-reacting bands (to the left), and the arrowheads mark the partial degradation product bands (to the left).
To characterize the interaction between DCAF1 and DDB1 in Arabidopsis, we constructed 35S:R3-Flag, 35S:R4-Flag, and 35S:R34-Flag transgenic plants and studied the interactions of the Flag-tagged fusion proteins with DDB1 in vivo by a coimmunoprecipitation (co-IP) assay. No obvious growth phenotype was observed in these overexpression lines. As shown in Figure 2B, when Flag-tagged fusion proteins were immunoprecipitated from plant extracts using anti-Flag antibody, endogenous DDB1 was detected together with R4-Flag and R34-Flag but not with R3-Flag. This result demonstrates that the WD40-containing R4 region of DCAF1 is likely to interact with DDB1 in Arabidopsis, consistent with the above-mentioned interaction assays in yeast. However, a role for the R3 region could not be verified by this assay.
WDxR Motifs in the WD40 Domain Are Essential for the Interaction between DCAF1 and DDB1
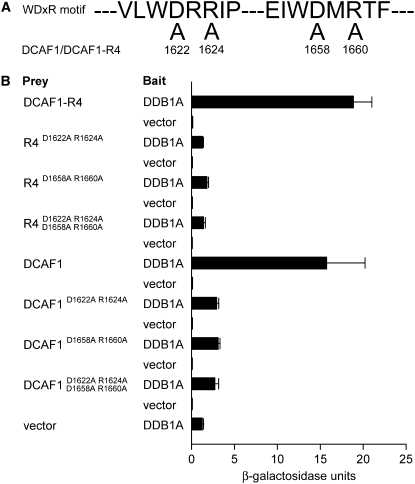
WDxR motifs in the WD40 domain have been suggested to be responsible for the physical binding of DCAF proteins to DDB1 (Angers et al., 2006). As shown in the sequence analysis, Arabidopsis DCAF1 contains two WDxR motifs in the WD40 domain of the R4 region (Figure 1D); consistently, we have demonstrated that it is the R4 region that mediates DCAF1's interaction with DDB1 in Arabidopsis (Figure 2). To further investigate the critical role of WDxR motifs in the physical interaction between DCAF1 and DDB1, we introduced point mutations at the Asp and Arg residues in the WDxR motifs of the R4 region (Figures 1D and 3A) and tested the interactions of various combinations of these DCAF1 point mutants with DDB1A in Y2H assays. As shown in Figure 3B, point mutations at either or both WDxR motifs result in a significant decrease in the interaction between either the full-length DCAF1 or its R4 region with DDB1A. These results confirm that the WDxR motifs in the WD40 domain are essential for the physical interaction between DCAF1 and DDB1, and both WDxR motifs are required for optimal interaction.
Figure 3.
Point Mutation Analysis of WDxR Motifs in DCAF1.
(A) Point mutations introduced in the two WDxR motifs of DCAF1 and its R4 region (DCAF1-R4). The numbers indicate the mutation sites (amino acid residue; D-to-A or R-to-A point mutations) in DCAF1. D1622A and R1624A are situated within the first WDxR motif, whereas D1658A and R1660A occur within the second WDxR motif.
(B) Interaction analysis of point mutants of DCAF1 or DCAF1-R4 with DDB1A by Y2H analysis. The previously shown interactions between DCAF1 or DCAF1-R4 and DDB1A (Figure 2A) were used as positive controls. The β-galactosidase activities resulting from the interactions are shown in the histogram. Empty prey vector with DDB1A as bait was used as the negative control. Error bars present sd (n = 4).
Evidence for a CUL4-DDB1-DCAF1 Complex in Vivo
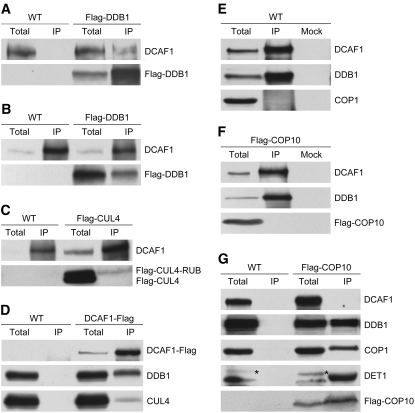
To determine whether Arabidopsis DCAF1 can form a CUL4-DDB1–based E3 ubiquitin ligase complex in vivo, we first tested the interaction between DCAF1 and DDB1 in 35S:Flag-DDB1 transgenic plants using co-IP assay. As expected, DCAF1 could be detected in the Flag-DDB1 immunocomplex pulled down with anti-Flag antibody (Figure 4A); reciprocally, Flag-DDB1 coimmunoprecipitated with DCAF1 that had been precipitated with anti-DCAF1 antibody (Figure 4B). These results confirm the interaction of Arabidopsis DCAF1 with DDB1 in vivo.
Figure 4.
Evidence for a CUL4-DDB1-DCAF1 Complex in Arabidopsis.
(A) and (B) Association of DCAF1 with Flag-DDB1 in vivo. Total seedling protein extracts prepared from wild-type and 35S:Flag-DDB1 (Flag-DDB1) transgenic Arabidopsis were incubated with anti-Flag antibody-conjugated agarose (A) or with anti-DCAF1 antibody and Protein A sepharose (B). The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against Flag (for Flag-DDB1) and DCAF1.
(C) Association of DCAF1 with RUB-modified Flag-CUL4 in vivo. Total seedling protein extracts prepared from wild-type and 35S:Flag-CUL4 (Falg-CUL4) transgenic Arabidopsis plants were incubated with anti-DCAF1 antibody and Protein A sepharose. The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against Flag (for Flag-tagged CUL4 and CUL4-RUB) and DCAF1.
(D) Association of DCAF1-Flag with both DDB1 and CUL4 in vivo. Total seedling protein extracts prepared from wild-type and 35S:DCAF1-Flag (DCAF1-Flag) transgenic Arabidopsis were incubated with anti-Flag antibody-conjugated agarose. The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against Flag (for DCAF1-Flag), DDB1, and CUL4.
(E) DCAF1 does not associate with COP1 in vivo. Total seedling protein extracts prepared from wild-type Arabidopsis were incubated with anti-DCAF1 antibody and Protein A sepharose. Incubation with Protein A sepharose alone was used as mock. The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against DCAF1, DDB1, and COP1.
(F) DCAF1 does not associate with Flag-COP10 in vivo. Total seedling protein extracts prepared from 35S:Flag-COP10 (Flag-COP10) transgenic Arabidopsis were incubated with anti-DCAF1 antibody and Protein A sepharose. Incubation with Protein A sepharose alone was used as a mock. The immunoprecipitates (IP) and the total extract (total) were subjected to immunoblot analysis with antibodies against DCAF1, DDB1, and Flag (for Flag-COP10).
(G) Flag-COP10 does not associate with DCAF1 in vivo. Total seedling protein extracts prepared from wild-type and 35S:Flag-COP10 (Flag-COP10) transgenic Arabidopsis were incubated with anti-Flag antibody-conjugated agarose. The immunoprecipitates (IP) and total extracts (total) were subjected to immunoblot analysis with antibodies against DCAF1, DDB1, COP1, DET1, and Flag (for Flag-COP10). The asterisks mark the cross-reacting bands (to the left).
Then, we tested the association between DCAF1 and CUL4 in Arabidopsis. Using the anti-DCAF1 antibody, Flag-CUL4 from 35S:Flag-CUL4 transgenic Arabidopsis coimmunoprecipitated with DCAF1. Moreover, based on the slower mobility of RUB-modified Flag-CUL4 on the protein gel (Chen et al., 2006), we concluded that most of the DCAF1-associated Flag-CUL4 had RUB modification (Figure 4C). This is consistent with the notion that the RUB/NEDD8 modification of CULLIN proteins is critical for the enzyme activity of CRL E3 ubiquitin ligases (Goldenberg et al., 2004; Petroski and Deshaies, 2005) and thus implies that Arabidopsis DCAF1 exists in a CUL4-containing E3 ubiquitin ligase complex. To further analyze the potential CUL4-DDB1-DCAF1 complex in Arabidopsis, we tested the association of CUL4, DDB1, and DCAF1 in vivo and found that CUL4 and DDB1 coimmunoprecipitated together with DCAF1-Flag from 35S:DCAF1-Flag transgenic Arabidopsis (Figure 4D). Since we detected no direct interaction between DCAF1 and CUL4 in Y2H analysis, it is most likely that DDB1 links DCAF1 and CUL4 together to form an active E3 ubiquitin ligase complex in Arabidopsis.
As introduced previously, reconstructed CDD complex can form a stable complex with CUL4-ROC1/RBX1, and in vivo, the CUL4-ROC1/RBX1-CDD complex is able to associate with the COP1 E3 complex and plays an important role in regulating COP1-mediated degradation of HY5 in darkness (Chen et al., 2006). In addition, DET1 has also been reported to associate with CUL4 in planta (Bernhardt et al., 2006). These findings imply that CUL4 E3 complexes (not just the CUL4-ROC1/RBX1 core) may associate with the CDD complex or the COP1 E3 complex in Arabidopsis. To test this, we examined DCAF1's interactions with COP1 and COP10, representative subunits of the COP1 E3 complex and CDD complex, respectively. As shown in Figures 4E and 4F, neither COP1 nor Flag-COP10 could be detected in the immunoprecipitate fractions from wild-type or 35S:Flag-COP10 transgenic Arabidopsis plants that had been immunoprecipitated with anti-DCAF1 antibody. Moreover, unlike DDB1, COP1, or DET1, DCAF1 did not coimmunoprecipitate with Flag-COP10 in fractions from 35S:Flag-COP10 transgenic Arabidopsis plants immunoprecipitated with anti-Flag antibody (Figure 4G). These results suggest that DCAF1 forms an independent complex with DDB1 and CUL4 in Arabidopsis, and there is no apparent association between the CUL4-DDB1-DCAF1 complex and the CDD or COP1 complex.
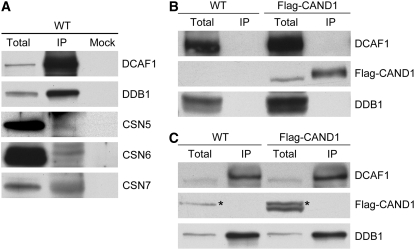
Association of the CUL4-DDB1-DCAF1 Complex with CSN and CAND1
The CRL E3 ubiquitin ligase activity is regulated by RUB, CSN, and CAND1 (Petroski and Deshaies, 2005). We have shown above that DCAF1 mainly interacts with RUB-modified CUL4 in Arabidopsis (Figure 4C). Here, we further investigate DCAF1's association with CAND1 and CSN. As shown in Figure 5A, CSN5, CSN6, and CSN7 subunits were detected in the DCAF1 immunocomplex from wild-type Arabidopsis, indicating that the CUL4-DDB1-DCAF1 complex associates with CSN; therefore, it is possible that its E3 ubiquitin ligase activity is regulated by CSN.
Figure 5.
Association Analysis of the CUL4-DDB1-DCAF1 Complex with CRL E3 Activity Regulators.
(A) Association of DCAF1 with CSN in vivo. Total seedling protein extracts prepared from wild-type Arabidopsis were incubated with anti-DCAF1 antibody and Protein A sepharose. Incubation with Protein A sepharose alone was used as a mock. The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against DCAF1, DDB1, CSN5, CSN6, and CSN7.
(B) and (C) DCAF1 does not associate with Flag-CAND1 in vivo. Total seedling protein extracts prepared from wild-type and 35S:Flag-CAND1 (Flag-CAND1) transgenic Arabidopsis were incubated with anti-Flag antibody-conjugated agarose (B) or with anti-DCAF1 antibody and Protein A sepharose (C). The immunoprecipitates (IP) and the total extracts (total) were subjected to immunoblot analysis with antibodies against DCAF1, Flag (for Flag-CAND1), and DDB1. The asterisks mark the cross-reacting bands (to the left).
Next, we tested the association between DCAF1 and Flag-CAND1 in 35S:Flag-CAND1 transgenic plants. It is clear that DCAF1 was not detected in the Flag-CAND1 immunocomplex (Figure 5B); neither was Flag-CAND1 present in the DCAF1 immunocomplex (Figure 5C). Thus, these results suggest that DCAF1 and CAND1 do not coexist in the same complex, which is consistent with previous observations that CAND1 preferentially interacts with unmodified CULLIN, inhibiting the interaction between CULLIN and its adaptor, such as DDB1, and negatively regulates CRL E3 enzyme activity by disrupting the construction of the CRL E3 complex (Feng et al., 2004; Goldenberg et al., 2004; Petroski and Deshaies, 2005). The association with CSN but not with CAND1 indicates that the CUL4-DDB1-DCAF1 complex in Arabidopsis might function as an E3 ubiquitin ligase, whose activity is potentially regulated by CSN and CAND1 in a mutually antagonistic manner.
Spatial and Temporal Expression Pattern of DCAF1 in Arabidopsis
To gain insight into the role of DCAF1 in plant development, we first analyzed its expression pattern in an organ-specific genome expression profile database from a previously published microarray study in Arabidopsis (Ma et al., 2005). In this data set, DCAF1 was found to be expressed in all organs and tissue types tested, and the results suggest that it is expressed at the highest levels in hypocotyl and root tissue of seedlings grown in white light (see Supplemental Figure 2 online).
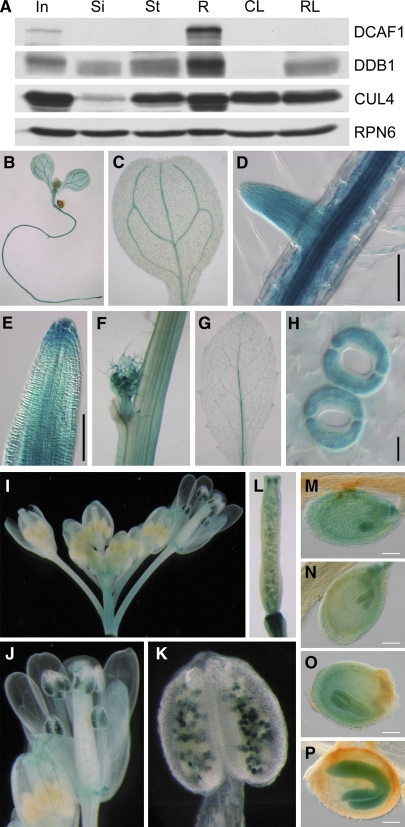
Next, we assayed the DCAF1 protein level in different organs by immunoblot analysis. We found that DCAF1 protein abundance accumulated to its highest level in root tissue and to a modest level in the inflorescence. In other organs examined, such as the stem, silique, cauline leaf, and rosette leaf, DCAF1 protein is hardly detectable (Figure 6A). Moreover, CUL4 and DDB1 proteins also accumulate to high levels in the root and inflorescence, which implies that the CUL4-DDB1-DCAF1 complex may play important roles in these organs.
Figure 6.
Expression Pattern of the DCAF1 Gene in Arabidopsis.
(A) Protein accumulation of DCAF1 in different Arabidopsis organs. Total soluble protein extracts from different organs were examined by immunoblot analysis using antibodies against DCAF1, DDB1, and CUL4. The anti-RPN6 antibody was used as a sample equal loading control.
(B) to (P) Temporal and spatial expression patterns of GUS reporter gene in DCAF1:GUS transgenic Arabidopsis. GUS activity was examined in a 7-d-old seedling (B), cotyledons (C), the primary root and lateral root (D), root tip (E), stem and axillary bud (F), rosette leaf (G), guard cell (H), inflorescence (I), mature flower (J), anther and pollen (K), silique (L), embryo at globular stage (M), embryo at heart stage (N), embryo at torpedo stage (O), and embryo at cotyledon stage (P). In, inflorescence; Si, silique; St, stem; R, root; CL, cauline leaf; RL, rosette leaf. Bars = 100 μm in (D), (E), and (M) to (P) and 10 μm in (H).
To study tissue-specific expression pattern of the DCAF1 gene in more detail, we fused a 1.93-kb promoter region upstream of ATG from the DCAF1 gene with a β-glucuronidase (GUS) reporter gene and transformed this construct into Arabidopsis plants. As shown in Figures 6B to 6P, the GUS activity was detected throughout most of the developmental stages, from seedlings to mature plants, and was also detected in most organs, including the cotyledon, root, stem, leaf, flower, and silique. Upon closer examination, we found that GUS activity was localized mainly in the vascular systems of most organs (Figures 6C, 6D, 6G, and 6J). In addition, GUS activity was observed in the stem, axillary bud, and guard cells (Figures 6F and 6H). In flowers, the GUS activity was mainly enriched in pollen of mature flower anthers, while the immature flowers in the same inflorescence did not have such GUS activity (Figures 6I to 6K). Finally, GUS activity was detectable in all stages of embryo development in siliques, from the globular stage to the cotyledon stage (Figures 6L to 6P), suggesting that DCAF1 may play important roles in the regulation of Arabidopsis embryogenesis.
Subcellular Localization of the CUL4-DDB1-DCAF1 Complex
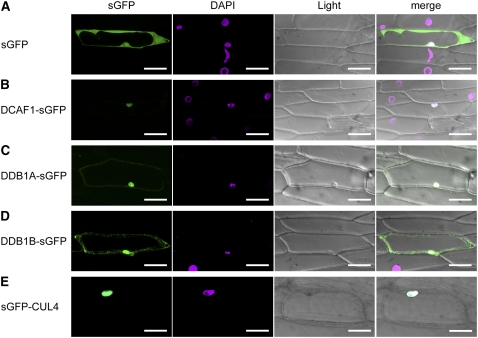
DDB1 was originally identified in the nucleus as a UV-Damaged DNA Binding protein that is absent in group E of xeroderma pigmentosa patients (Chu and Chang, 1988). Arabidopsis DDB1 was first identified in a nuclear-localized complex with DET1 (Schroeder et al., 2002) and was subsequently shown to form the CDD complex with DET1 and COP10 (Yanagawa et al., 2004). Additionally, Arabidopsis CUL4 has also been reported to be a nuclear protein (Chen et al., 2006). Thus, it seems likely that DCAF1 might be localized in the nucleus as well and might form a nuclear complex with CUL4 and DDB1 in Arabidopsis.
To study the intracellular localization of the CUL4-DDB1-DCAF1 complex, synthetic green fluorescent protein (sGFP) was fused in frame to the C termini of DCAF1, DDB1A, and DDB1B. Then, these constructs were transformed into the epidermal cells of onion (Allium cepa) with sGFP-CUL4 and sGFP as positive controls (Figures 7A and 7E). Free sGFP could be found throughout the whole cell, as predicted (Figure 7A). As previously described (Chen et al., 2006), sGFP-CUL4 was observed only in the nucleus (Figure 7E). Transiently overexpressed DCAF1-sGFP fusion protein exclusively localized to the nucleus (Figure 7B), while DDB1A-sGFP and DDB1B-sGFP were distributed in both the cytoplasm and nucleus, although the signal of DDB1A-sGFP in the cytoplasm was just barely detectable above the background (Figures 7C and 7D). These data indicate that the Arabidopsis DCAF1 protein is mainly localized in the nucleus together with DDB1 and CUL4, suggesting that the CUL4-DDB1-DCAF1 complex may function as a nuclear E3 ubiquitin ligase.
Figure 7.
Subcellular Localization of DCAF1, DDB1, and CUL4.
The following fluorescence proteins were transformed into and transiently expressed in onion epidermal cells: sGFP (A), DCAF1-sGFP (B), DDB1A-sGFP (C), DDB1B-sGFP (D), and sGFP-CUL4 (E). Signals from sGFP, 4′,6-diamidino-2-phenylindole (DAPI), bright-field (light), and the merge of the three signals (merge) are shown. Bars = 100 μm.
Identification and Phenotype Analysis of dcaf1 T-DNA Insertion Mutants
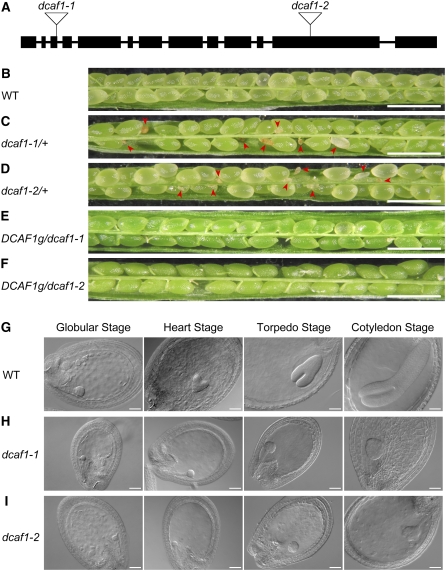
To characterize the physiological function of DCAF1 and the CUL4-DDB1-DCAF1 complex in Arabidopsis, we searched the available Arabidopsis T-DNA insertion mutagenesis collection at the SALK Institute Genomic Analysis Laboratory (Alonso et al., 2003) and obtained two independent lines that were subsequently named dcaf1-1 and dcaf1-2, with the left borders of T-DNA located in the 3rd (615 nucleotides downstream of ATG in genomic DNA) and 12th (4817 nucleotides downstream of ATG) exons, respectively (Figure 8A). The T-DNA insertion sites in DCAF1 suggest that dcaf1-1 and dcaf1-2 may be loss-of-function or severe reduction-of-function mutants with truncated expression products. PCR-based genotyping was used in attempt to identify homozygous T-DNA insertion mutants; however, no dcaf1 homozygotes were isolated despite examining >200 segregating progeny from the self-pollination of parental plants heterozygous for the T-DNA insertion. Nevertheless, the heterozygous plants do not exhibit any developmental defects when compared with wild-type plants under normal growth conditions. The ratio of heterozygotes to wild-type plants from an individual self-pollinated dcaf1/+ parental plant is close to 2:1 (Table 1), suggesting that homozygous T-DNA insertion mutations in DCAF1 may be lethal.
Figure 8.
Characterization of dcaf1 Mutants.
(A) Schematic representation of T-DNA insertions in the Arabidopsis DCAF1 gene. Exons are represented by filled black rectangles, and introns are represented by solid lines. The T-DNA insertion sites of the two mutant alleles are indicated by open inverted triangles, with the assigned allele name of each insertional mutation labeled above.
(B) to (F) Stereomicroscopy images of siliques obtained from self-pollinated wild-type (B), dcaf1-1/+ (C), dcaf1-2/+ (D), DCAF1g/dcaf1-1 (E), and DCAF1g/dcaf1-2 (F) parental plants. Red arrowheads indicate abnormal ovules. Bars = 1 mm.
(G) to (I) DIC images of cleared ovules obtained from self-pollinated wild-type (G), dcaf1-1/+ (H), and dcaf1-2/+ (I) parental plants. The four embryonic developmental stages (globular, heart, torpedo, and cotyledon) are shown from left to right. Bars = 50 μm.
Table 1.
Segregation of T-DNA in dcaf1 Heterozygous Mutants
| Genotype of Plant Used for Self-Pollinationa | Progeny with Heterozygous T-DNA Insertionb | Progeny with No T-DNA Insertionc | Total Number of Plants Analyzed |
|---|---|---|---|
| dcaf1-1/+ | 84 (59.57%) | 57 (40.43%) | 141 |
| dcaf1-2/+ | 94 (66.20%) | 48 (33.80%) | 142 |
PCR-based genotyping was performed in the progeny from self-pollinated parental plants.
Numbers of progeny with a heterozygous T-DNA insertion at the DCAF1 locus; segregation frequencies are indicated in parentheses.
Numbers of progeny without a T-DNA insertion at the DCAF1 locus; segregation frequencies are indicated in parentheses.
Mature siliques from different self-pollinated dcaf1/+ parental plants were examined. We noticed that siliques from dcaf1-1/+ and dcaf1-2/+ plants segregated both wild-type-looking developing ovules and yellowish, shrunk, and smaller developing ovules (Figures 8C and 8D, indicated by red arrowheads). By contrast, wild-type siliques uniformly produced plump, green ovules (Figure 8B). At later developmental stages, the wild-type-looking green mature ovules from dcaf1-1/+ and dcaf1-2/+ plants developed into normal matured brown seeds, whereas the yellowish ovules from the same siliques turned into drastically shrunken and red seeds (see Supplemental Figure 3 online). Stereoscopic analyses of numerous developing ovules from self-pollinated wild-type and dcaf1/+ parental plants indicate that those abnormal ovules from self-pollinated dcaf1/+ parental plants correspond roughly to 21 to ∼25% of the total developing ovules (Table 2). All of these genetic segregation data from dcaf1 heterozygous mutants agree with classic Mendelian rules, suggesting that homozygous T-DNA insertion of DCAF1 possibly leads to lethality during embryo development.
Table 2.
Seed Abortion Rates in Plants Carrying Different Genotypes of DCAF1 (P < 0.05)
| Genotype of Parental Planta | Aborted Seedsb | Normal Seedsc | Seeds Scored |
|---|---|---|---|
| Wild type | 8 (0.78%) | 1021 (99.22%) | 1029 |
| dcaf1-1/+ | 212 (21.01%) | 797 (78.99%) | 1009 |
| dcaf1-2/+ | 250 (25.23%) | 741 (74.77%) | 991 |
| dcaf1-1/DCAF1g | 34 (3.46%) | 948 (96.54%) | 982 |
| dcaf1-2/DCAF1g | 47 (3.35%) | 1354 (96.65%) | 1401 |
Developing seeds from siliques obtained from self-pollinated parental plants were analyzed under stereoscope.
Numbers of defective yellowish/pale green developing ovules that turned into aborted, shrunken, red seeds; segregation frequencies are indicated in parentheses.
Numbers of wild-type-looking green developing ovules that turned into normal mature brown seeds; segregation frequencies are indicated in parentheses.
Further examination of embryo development was performed using a microscope equipped with differential interference contrast (DIC) optics. Whole-mount cleared developing ovules at different developmental stages were analyzed (Figures 8G to 8I). Until the globular stage, fertilized ovules from self-pollinated wild-type and dcaf1/+ parental plants were virtually indistinguishable from each other, and their embryos developed normally. However, after the transition from the globular stage to the heart stage, while all ovules from wild-type plants uniformly developed into heart-stage embryos (Figure 8G), ovules from dcaf1/+ plants segregated into wild-type-looking ovules with heart-stage embryos and smaller ovules with embryos arrested at the globular stage (Figures 8H and 8I). At later developmental stages, all ovules from wild-type plants matured from the torpedo stage to the cotyledon stage (Figure 8G), whereas dcaf1/+ plants segregated two types of ovules, one of which contained developing embryos at the same stage as wild-type embryos, and the other of which contained arrested embryos at the globular stage or a slightly later stage (Figures 8H and 8I). These early-aborted ovules from self-pollinated dcaf1/+ plants correspond to about one-quarter of the total ovules from an individual silique. These observations confirm that the homozygous T-DNA insertion mutants of the DCAF1 gene are embryonic lethal.
To provide further evidence that the arrested embryos observed in dcaf1 heterozygous siliques are indeed attributable to the dcaf1 mutation, we generated a construct with a genomic fragment containing the full-length Arabidopsis DCAF1 gene (DCAF1g), including an ∼1.9-kb promoter region upstream of ATG and an ∼1.5-kb terminator region downstream of the stop codon. This construct was introduced into dcaf1-1 and dcaf1-2 heterozygous backgrounds for functional complementation tests. If loss of DCAF1 in dcaf1 mutants indeed results in embryo lethality and the DCAF1g transgene can complement the DCAF1 function in dcaf1 mutants, segregation frequencies of aborted ovules from self-pollinated DCAF1g/dcaf1-1 and DCAF1g/dcaf1-2 transgenic plants should be lower than that of self-pollinated dcaf1/+ plants. Indeed, in mature siliques from the self-pollination of DCAF1g/dcaf1-1 and DCAF1g/dcaf1-2 plants, the ratio of aborted ovules was decreased to <4%, much lower than that of the self-pollinated dcaf1 heterozygous mutants (Figures 8E and 8F, Table 2). This result corroborates the linkage between the phenotypic ovule developmental defects and the genotypic dcaf1 mutations.
Taken together, through characterization of dcaf1 T-DNA insertion mutants, we provide evidence that dcaf1 homozygous mutation leads to arrested embryo development beyond the globular stage, suggesting an important role for DCAF1 in the regulation of Arabidopsis embryo development.
Reducing the Amount of DCAF1 Results in Multifaceted Developmental Defects in Arabidopsis
Since embryonic lethality of dcaf1 homozygous mutants prevents them from being analyzed beyond embryonic development, we sought to investigate the physiological function of DCAF1 by altering its expression level in Arabidopsis. To this end, we constructed two 35S promoter–driven transgenes expressing full-length DCAF1 fused with three copies of HA tag at the N terminus (HA-DCAF1) or three copies of Flag tag at the C terminus (DCAF1-Flag), respectively. Both constructs were introduced into wild-type backgrounds. The accumulation of HA-DCAF1 and DCAF1-Flag fusion proteins in transgenic plants and the endogenous DCAF1 protein was examined using antibodies against DCAF1 or the corresponding tag. Several DCAF1 overexpression lines were isolated from 35S:DCAF1-Flag transgenic plants; however, no obvious developmental defects were observed under normal growth conditions, as was the case with DCAF1g/dcaf1 transgenic plants.
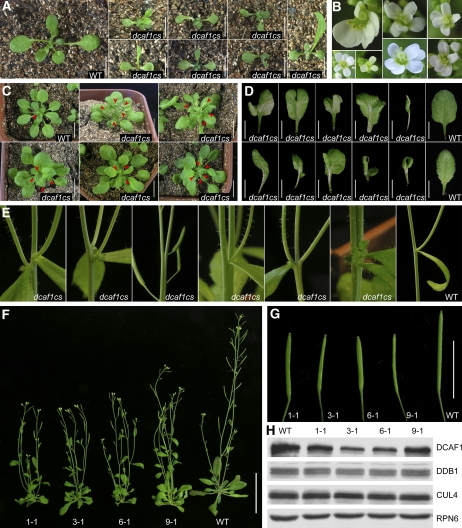
Interestingly, we also isolated several cosuppression lines from the 35S:HA-DCAF1 transgenic plants (named dcaf1cs), in which endogenous DCAF1 protein abundance was downregulated by the presence of the exogenous transgene (Figure 9H), and no HA-DCAF1 fusion protein could be detected by the anti-HA antibody. These dcaf1cs cosuppression transgenic lines exhibited multifaceted developmental defects, as shown in Figures 9A to 9G. For two-week-old seedlings, dcaf1cs plants already looked distinct from their wild-type counterparts in that they were smaller and frequently exhibited an asymmetrical development with abnormal phyllotaxy and irregularly shaped true leaves (Figure 9A). During later vegetative phases, dcaf1cs plants tended to produce smaller but more rosette leaves than wild-type plants, with various leaf developmental patterns, including lobed, curled, or asymmetrical rosette leaves (Figures 9C and 9D). In the reproductive phase, the phenotypic differences between dcaf1cs and wild-type plants became more drastic. During the transition from vegetative to reproductive development, dcaf1cs plants generally tended to produce more than two primary shoots, while only one primary shoot was produced by most wild-type plants (Figure 9C). Following stem elongation and emergence of lateral branches, most dcaf1cs plants exhibited obvious dwarfism with multiple slimmer primary shoots (Figure 9F) and irregular development of nodes and internodes, including more than two axillary buds and cauline leaves at the same node, decreased distance between two neighboring nodes, and abnormal division or combination of internodes (Figure 9E). Moreover, some dcaf1cs plants produced abnormal flowers with three, five, or six petals (Figure 9B) as opposed to the cruciate four petals in the wild-type flowers, and siliques of dcaf1cs were smaller than their wild-type counterparts (Figure 9G). In summary, different dcaf1cs lines tended to share common developmental defects, although often to different degrees of severity. The defects throughout vegetative and reproductive development displayed by dcaf1cs plants suggest that the DCAF1 protein and CUL4-DDB1-DCAF1 E3 ubiquitin ligase may participate in many developmental processes in Arabidopsis. In the future, the identification of specific E3 ubiquitin ligase substrates recruited by DCAF1 will further reveal its biological functions.
Figure 9.
Multifaceted Developmental Defects of dcaf1cs Mutants.
(A) Two-week-old wild-type and dcaf1cs plants. Bars = 0.5 cm.
(B) Abnormal flowers from dcaf1cs plants.
(C) Four-week-old wild-type and dcaf1cs plants. Red arrowheads indicate the primary shoots that were starting to bolt at the transition from vegetative to reproductive growth. Bars = 1 cm.
(D) Rosette leaves from 3-week-old wild-type and dcaf1cs plants. Bars = 0.5 cm.
(E) Abnormal development of stem, node, internode, lateral shoot, axillary bud, and cauline leaves from dcaf1cs plants, with a wild-type plant as the control.
(F) Comparison of wild-type and dcaf1cs adult plants. The numbers indicate independent dcaf1cs lines. Bar = 5 cm.
(G) Comparison of wild-type and dcaf1cs siliques. The numbers indicate siliques from independent dcaf1cs lines. Bar = 1 cm.
(H) Decrease of DCAF1 protein level in dcaf1cs mutants. Total seedling protein extracts from wild-type Arabidopsis and four independent dcaf1cs transgenic lines were examined by immunoblot analysis using antibodies against DCAF1, DDB1, and CUL4. The anti-RPN6 antibody was used as a sample equal loading control. The numbers above the blot indicate protein samples from independent dcaf1cs lines.
DISCUSSION
Implication of the Physical Interaction between DCAF1 and DDB1
Human DCAF proteins have been shown to interact directly with DDB1, which is mediated by the WDxR motifs within their WD40 domains (Angers et al., 2006; Higa et al., 2006; Jin et al., 2006). Here, we provide three lines of direct evidence supporting that Arabidopsis DCAF1 is a DDB1 binding protein with a similar manner of interaction. First, both full-length DCAF1 and the R4 region containing the WD40 domain interact with DDB1 in yeast (Figure 2A). Second, point mutations in the WDxR motifs disrupt DCAF1's interaction with DDB1 (Figure 3). Third, both DCAF1 and its WD40-containing R4 region interact with endogenous DDB1 in transgenic plants (Figures 2B and 4).
Since DCAF1 was not characterized as one of the DWD proteins in a previous study on Arabidopsis CUL4-DDB1–based E3 ubiquitin ligase complexes (Lee et al., 2008), it seems likely that DCAF1 defines a distinct group of DCAF proteins in plants. The motif that is responsible for DCAF1 interaction with DDB1 is a four–amino acid WDxR motif (Figure 1D), which is generalized as the conserved [WY][DE]x[RK] peptide at the end of the WD40 repeat in DCAF proteins (He et al., 2006; Lee et al., 2008; Jin et al., 2006). As the WDxR motif is a major determinant of DDB1-interacting WD40 proteins in human (Angers et al., 2006; Higa et al., 2006; Jin et al., 2006), we therefore propose that putative Arabidopsis DCAF proteins can be identified by the presence of a WDxR motif in their WD40 repeat domain. To this end, we manually searched for the WDxR motif in 297 Arabidopsis WD40 proteins and 223 rice WD40 proteins (Lee et al., 2008) and found 119 putative DCAF proteins with 165 WDxR motifs in Arabidopsis and 110 putative DCAF proteins with 151 WDxR motifs in rice (see Supplemental Table 1 online). All of the predicted DWD proteins in the Lee et al. (2008) study are included in these putative DCAF proteins that we identified, indicating that the previous method, which uses a 16–amino acid motif to define DWD proteins, may be an overly stringent criterion for recognizing the CUL4 E3 ligase substrate module. With the less stringent criterion of a four–amino acid WDxR motif, there could be 34 additional (see Supplemental Table 2 online) and 32 additional (see Supplemental Table 3 online) DCAF WD40 proteins in Arabidopsis and rice, respectively. It is also worth mentioning that there are other DDB1-interacting proteins, besides the DWD or WDxR motif–containing WD40 proteins, that can potentially act as substrate receptors of CUL4 E3 ubiquitin ligase complexes such as human EED and SV5-V protein (Higa et al., 2006; Li et al., 2006).
Interestingly, deleting the R1 region from the N terminus of DCAF1 disrupts the binding of DCAF1 to DDB1, whereas subsequent deletion of the R2 region rescues the DDB1 binding capability (Figure 2A). Based on these results, we hypothesize that the R1 and R2 regions modulate the interaction of DCAF1 with DDB1 in a spatially antagonistic manner. In the full-length DCAF1 molecule, under the natural configuration, the R1 region obstructs R2's inhibition of DDB1 binding. In the R1 deletion mutant, the inhibition effect of the R2 region is released by the change of molecular configuration; therefore, DCAF1–DDB1 interaction is abolished. Following deletion of the R2 region, the interacting surface is exposed again with a new conformation, and the DDB1 binding capability of DCAF1 is recovered.
CUL4-DDB1-DCAF1 E3 Ubiquitin Ligase in Arabidopsis
Human DCAF1 forms a complex with CUL4 and DDB1, which is recruited by HIV-1 Vpr protein to trigger G2 arrest of host cells (Schröfelbauer et al., 2005, 2007; Belzile et al., 2007; Hrecka et al., 2007; Le Rouzic et al., 2007; Tan et al., 2007; Wen et al., 2007). This suggests the DCAF1 homolog in Arabidopsis may also form an analogous E3 ubiquitin ligase with CUL4 and DDB1. Meanwhile, since some Arabidopsis CUL4-based complexes, whose substrate receptors have not been identified yet, have been shown to associate with the CDD complex and the COP1 complex (COP1 belongs to the predicted DWD proteins that contain the conserved DDB1 binding motif) and participate in repressing photomorphogenesis (Chen et al. 2006; Lee et al., 2008), it is interesting to determine if Arabidopsis DCAF1 is part of such a CUL4-based complex. In our studies, we successfully detected Arabidopsis DCAF1's interactions with DDB1 and CUL4 in vivo, but not with COP10 or COP1, representative subunits of the CDD complex and COP1 complex, respectively (Figure 4). In addition, no direct interaction of DCAF1 with DET1, COP10, or COP1 could be detected in yeast. These results indicate that Arabidopsis DCAF1 is able to constitute an independent CUL4-DDB1-DCAF1 complex, which is not associated with the CDD complex or COP1 complex. Nonetheless, we cannot rule out the possibility that the DET1 or CDD complex might act as a regulator of CUL4 E3 ubiquitin ligases in Arabidopsis because a recent report shows that human DET1 inhibits CUL4 E3 ubiquitin ligase activity (Pick et al., 2007).
The potential E3 ubiquitin ligase activity of the CUL4-DDB1-DCAF1 complex is supported by its association with known CRL E3 enzyme activity regulators. It is thought that CRL E3 enzyme activity is regulated through a dynamic cycle by three regulators, RUB, CSN, and CAND1 (Petroski and Deshaies, 2005). Taking the SKP1-CUL1-F-box (SCF) E3 complex for example, the CUL1-ROC1/RBX1 enzymatic core is unable to interact with the adapter protein SKP1 when it is bound by CAND1 and is held in an inactive state. The RUB modification of a C-terminal Lys residue of CUL1 is able to block the CAND1 association with CUL1, which results in the activation of the SCF E3 complex. RUB modification of CUL1, on the other hand, can be reversed by the isopeptidase activity of the CSN5 subunit of CSN, which leads to the inactivation of the SCF complex and the subsequent displacement of SKP1-F-box by CAND1 (Goldenberg et al., 2004; Petroski and Deshaies, 2005). Such an assembly and disassembly cycle is considered to be an important characteristic of CRL E3 ubiquitin ligase activity. In our studies, Arabidopsis DCAF1 is found to mainly associate with RUB-modified CUL4 and also with CSN, but not with the negative regulator CAND1 (Figures 4 and 5). This manner of interaction is consistent with the regulation mechanism of CRL E3 ubiquitin ligase activity and provides evidence for the potential E3 ubiquitin ligase activity of the CUL4-DDB1-DCAF1 complex.
Nuclear Localization of the CUL4-DDB1-DCAF1 Complex
Transient expression of sGFP fusion proteins in onion epidermal cells shows that Arabidopsis DCAF1 is predominantly localized in the nucleus, DDB1A and DDB1B are localized in both the cytoplasm and the nucleus, and CUL4 exhibits the same nuclear localization pattern as described previously (Figure 7; Chen et al., 2006). These observations imply that Arabidopsis DCAF1 mainly functions in the nucleus, consistent with the idea that the CUL4-DDB1-DCAF1 complex also works primarily as a nuclear E3 ubiquitin ligase.
In an early report, human DCAF1 was found to distribute predominantly in the cytoplasmic fractions, and its function is to interact with HIV-1 Vpr protein to block its nuclear transportation (Zhang et al., 2001). However, a recent report suggests that DCAF1 is a nuclear protein that forms a ternary complex with DDB1 and DDA1 (DET1 and DDB1 Associated 1), which is bound and modulated by the Vpr protein (Hrecka et al., 2007). Bioinformatics prediction of nuclear localization signals (NLS) (Cokol et al., 2000; http://cubic.bioc.columbia.edu/services/predictNLS/) suggests that Arabidopsis DCAF1 contains a NLS of PRKRKL in the R3 region (amino acid 1303-1308). This NLS is generalized as the [PL]RKRK[PL] peptide derived from the Apterous protein in fruit fly, the SKI oncoprotein in human, and the activator protein CHA4 (Cha4p) in budding yeast. However, using the same method, we could not find any NLS in human DCAF1 from the NLS database, which is a possible explanation for the observed cytoplasmic localization of human DCAF1.
Transiently expressed human DDB1 in fibroblasts is localized primarily in the cytoplasm, but after UV irradiation and in the presence of certain transporters, such as DDB2, it can translocate into the nucleus (Liu et al., 2000). The DDB1 homologs in chicken and fruit fly can also be transported into the nucleus from the cytoplasm (Takata et al., 2002; Fu et al., 2003), while the homologs in rice and fission yeast are predominantly localized in the nucleus (Zolezzi et al., 2002; Ishibashi et al., 2003). Taken together with DDB1's known functions in the nucleus, such as nuclear excision repair and stabilization of the genome, these findings further support a role for DDB1 as a nuclear protein. In Arabidopsis, DDB1 is a subunit of the nuclear CDD complex (Schroeder et al., 2002; Yanagawa et al., 2004), and it interacts with CUL4 (a nuclear protein as well) to form the architecture of CUL4-based E3 ubiquitin ligase complexes (Bernhardt et al., 2006; Chen et al., 2006; Lee et al., 2008). Here, our study provides direct evidence for the subcellular localization of Arabidopsis DDB1, which suggests that DDB1 might engage in diverse biological processes in both the cytoplasm and nucleus, possibly with different modes of function besides acting as the adaptor in CUL4-containing E3 ubiquitin ligases.
Human CUL4A is predominantly localized in the cytoplasm and only a small fraction (∼2 to 3%) resides in the nucleus; furthermore, unlike DDB1, CUL4A's intracellular distribution does not change following UV irradiation (Chen et al., 2001), which suggests that the small amount of CUL4A in the nucleus is enough for its nuclear functions with DDB1. By contrast, Arabidopsis CUL4 is mainly localized in the nucleus (Chen et al., 2006), which is reconfirmed by the observations from our studies (Figure 7). Therefore, it appears that most Arabidopsis CUL4-DDB1–based E3 ubiquitin ligases work as nuclear complexes, including the CUL4-DDB1-DCAF1 complex.
Biological Functions of the CUL4-DDB1-DCAF1 E3 Complex
Identification of substrate receptors and studying how they recognize specific target proteins in diverse biological processes are the two keys to understanding the functions of CUL4-containing E3 ubiquitin ligases. Indeed, with the identification of different DCAF proteins as substrate receptors and the discovery of various target substrates, the function of human CUL4-DDB1-DCAF E3 ubiquitin ligases has been implicated in several important cellular processes (Higa and Zhang, 2007). For example, human CUL4-DDB1-DCAF1 E3 ubiquitin ligase is recruited by the HIV-1 Vpr protein to mediate the ubiquitination and proteasomal degradation of human UNG2 and SMUG1, for the regulation of virus replication (Schröfelbauer et al., 2005, 2007; Angers et al., 2006; He et al., 2006; Jin et al., 2006).
Here, we show that Arabidopsis DCAF1 can potentially be the substrate receptor in a nuclear CUL4-DDB1-DCAF1 E3 ubiquitin ligase, which shares the same architecture with its human counterpart. To act as a substrate receptor, DCAF1 needs to have a protein–protein interaction domain to recognize specific target proteins. Besides the DDB1 binding WD40 domain, the LIS1 Homology (LisH) motif is the only known protein interacting motif in the DCAF1 molecule (located between 1089 and 1115 amino acids). Crystal structural analysis shows that the LisH motif is a thermodynamically stable dimerization domain, which mediates microtubule association, protein interaction, and intracellular localization (Emes and Ponting, 2001; Kim et al., 2004; Gerlitz et al., 2005; Mateja et al., 2006). From the AGI protein database, we identified 30 unique genes encoding LisH motif–containing proteins, including TOPLESS (TPL)/WUS-Interacting Protein 1, TONNEAU1, and LEUNIG (LEU) (Emes and Ponting, 2001; Kim et al., 2004). As previously reported, the N-terminal LisH motif of TPL, a transcription corepressor-like protein, interacts with the conserved C-terminal domain of WUSCHEL (WUS) to maintain the shoot apical meristem (Kieffer et al., 2006), and the LisH motif–containing LUFS domain of LEU mediates the interaction with an adaptor protein SEUSS for transcriptional repression in flower development (Sridhar et al., 2004). These findings raise the possibility that the LisH motif may enable DCAF1 to interact with other LisH motif–containing proteins or with the known LisH motif–interacting proteins and subsequently target them for ubiquitination and degradation. We used a Y2H assay to test DCAF1's interaction with the above-mentioned LisH-associating proteins as well as with several other LisH-containing proteins. Our preliminary results show that DCAF1 potentially interacts with WUS and a Ran binding protein M (RanBPM) related protein (see Supplemental Figure 4 online). Therefore, it is of great interest to further characterize the interaction between DCAF1 and its possible target proteins, which should help elucidate the biological function of DCAF1.
The embryonic lethality at the globular embryo stage in Arabidopsis dcaf1 homozygous mutants suggests that DCAF1, and possibly also the CUL4-DDB1-DCAF1 complex, plays essential roles in plant embryogenesis. In addition, the ubiquitous expression pattern of DCAF1:GUS implies that DCAF1 might participate in many biological processes, which is further supported by the multiple developmental defects shown by dcaf1cs mutants. Some of the phenotypes of dcaf1cs are also observed in cul4 knockdown mutants, including aberrant leaf development, increased number of secondary organs, and adult dwarfism and emaciation (Bernhardt et al., 2006; Chen et al., 2006), indicating that DCAF1 and CUL4 work together, presumably in the form of a CUL4-DDB1-DCAF1 complex, in many aspects of Arabidopsis development. The multiple primary shoots and asymmetrical leaf pattern exhibited by dcaf1cs suggest that DCAF1 is involved in the maintenance and differentiation of stem cells at the shoot apical meristem. In the future, identifying specific DCAF1-interacting proteins, which might be targets of CUL4-DDB1-DCAF1 E3 ubiquitin ligase, will shed light on the molecular mechanism of the role of DCAF1 in plant development.
METHODS
Phylogenetic Analysis
DCAF1 homologs were identified from GenBank using the protein basic local alignment search tool (BLASTp) (http://www.ncbi.nlm.nih.gov/BLAST/). Sequence alignment was performed by the ClustalW method of the MegAlign program in the Lasergene 5.06 software package (DNASTAR). The protein weight matrix was Gonnet Series with 10.00 of the gap penalty, 0.20 of the gap length penalty, 30% of the delay divergent sequences, and 0.50 of the DNA transition weight. Alignment shading was performed using GeneDoc 3.2.0 software (http://www.nrbsc.org/gfx/genedoc/index.html). The aligned sequences were analyzed using the PHYLIP 3.64 software package (http://evolution.genetics.washington.edu/phylip.html). An unrooted tree was constructed using the Neighbor program based on the neighbor-joining algorithm; human DCAF1 was taken as the defined outgroup. The bootstrap confidence values were calculated with 1000 replicates using the Seqboot program, and a consensus tree was created with the Consense program. The phylogenetic tree was graphically visualized as the rectangular cladogram by the TreeView 1.6.6 software (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
Plant Materials and Growth Conditions
All of the wild-type Arabidopsis thaliana plants used in this study were of the Columbia-0 ecotype. Transgenic Arabidopsis obtained from others are 35S:Flag-CUL4 (Chen et al., 2006), 35S:Flag-CAND1 (Feng et al., 2004), 35S:Flag-COP10 (Yanagawa et al., 2004), and 35S:Flag-DDB1 (Lee et al., 2008).
Arabidopsis seeds were surface sterilized by washing in 15% sodium hypochlorite solution for 10 min, rinsed five times with sterile water, spread on Murashige and Skoog (MS) plates (Sigma-Aldrich) containing 1% sucrose, and cold-treated at 4°C for 2 to 4 d. Then, seeds were placed in a standard continuous white light growth chamber and grown for 7 to 10 d at 22°C. To obtain adult plants, seedlings were transferred to soil and grown in a standard long-day (16 h light, 22°C; 8 h dark, 19°C) green house.
Cloning of Arabidopsis DCAF1 Gene and Isolation of dcaf1 T-DNA Insertion Mutants
The CDS of DCAF1 was amplified by RT-PCR from total mRNA extracted from wild-type Arabidopsis seedlings using fragmental cloning strategy with the following primers: YAF2, 5′-CGGCATATGAGGCCTATGGACGGGCAAGAGCAT-3′; YAR, 5′-GATCCCGGGAGAACCAGTTCCTGCTCCTAAG-3′; YDF, 5′-CGGCATATGGACCGATCAGCTCCTGAAGTC-3′; and YBR1, 5′-CATCCCGGGCCCTTAGGAAGAACGAATGTTGTCTC-3′.
The NdeI/SmaI fragments of the YAF2/YAR PCR product (YA2) and the YDF/YBR1 PCR product (YD1) were individually cloned into pGBKT7 (Clontech). Then, the NdeI/AatII fragment from pGBKT7-YA2 was subcloned into pGBKT7-YD1 to produce pGBKT7-DCAF1 that contains the full-length DCAF1 CDS.
To obtain a genomic fragment containing the full-length DCAF1 gene, the following primers were used to amplify three individual fragments from the total genomic DNA of wild-type Arabidopsis: GF1, 5′-CAGTCTAGAACTAGTGTGGTCGCACTGCCAGTTCTTT-3′; GR1, 5′-TACGGATCCGCACCTTCGTGAACCCTTCCTC-3′; GF2, 5′-TACGGATCCAGGGTGCGTAAGATTGTGGATAC-3′; GR2, 5′-CAGAAGCTTTGTAGCCCAGAGTTGCTGAATT-3′; GF3, 5′-CAGAAGCTTCACTTGTTCAGCCGTATGTCTC-3′; and GR3, 5′-TACGGTACCAGGGACTTACAACGGAGAAGAT-3′.
The XbaI/BamHI fragment of the GF1/GR1 PCR product (G1), the BamHI/HindIII fragment of the GF2/GR2 PCR product (G2), and the HindIII/KpnI fragment of the GF3/GR3 PCR product (G3) were individually cloned into pBSK (Stratagene). Then, the SpeI/Aro13HI fragment from pBSK-G1 and the MluI/KpnI fragment from pBSK-G3 were subcloned into pBSK-G2 to produce pBSK-DCAF1g that contains the complete genomic fragment of DCAF1 spanning 1916 bp upstream of ATG to 1467 bp downstream of the stop codon.
Through a database search, we obtained two T-DNA insertion mutants for DCAF1 gene from the SALK collection (Alonso et al., 2003), which were named dcaf1-1 and dcaf1-2, respectively. Plants heterozygous for the T-DNA insertion were identified by PCR-based genotyping with primers as follows: 010755FP, 5′-TTGCCAATAACCCTAATCCTAAT-3′; 010755RP, 5′-TGCATAAGCTTAGCAGATAGTCCT-3′; 114078FP, 5′-CAGGCATCAGGTCTTGGTGAT-3′; and 114078RP, 5′-ACCGTTTAGGTTCAGGACAGAC-3′.
Generation of Transgenic Arabidopsis Plants
Fragmental (R3, R4, and R34) and full-length CDS of DCAF1 were fused in frame to the 5′ end of a Flag-tag (three copies) coding sequence. Then, the KpnI/SpeI fragments containing R3-Flag, R4-Flag, and R34-Flag were individually subcloned into pJim19(Bar), a plant binary expression vector containing a basta resistance gene, a cauliflower mosaic virus 35S promoter, and the 3′ untranslated region of nopaline synthase, while the KpnI/SpeI fragment containing full-length DCAF1-Flag was subcloned into a different plant binary vector, pJim19(Gen), which has a gentamycin resistance gene as a selective marker. Wild-type Arabidopsis plants were used in the transformation. Transgenic plants containing 35S:R3-Flag, 35S:R4-Flag, or 35S:R34-Flag transgenes were selected on MS plates containing 20 mg/L glufosinate-ammonium (Riedel-de Haën), and the 35S:DCAF1-Flag transgenic plants were selected with 100 mg/L of gentamycin (Ameresco). The anti-Flag antibody (Sigma-Aldrich) was used to check the level of fusion protein in total protein extracts from inflorescence for each independent transgenic line.
The KpnI/SpeI fragment containing the full-length DCAF1 genomic DNA from pBSK-DCAF1g was subcloned into pCAMBIA-1300 binary vector (CAMBIA), and the resulting construct was transformed into heterozygous dcaf1 mutants. Transgenic plants were selected on MS plates containing 25 mg/L of hygromycin (Ameresco). Subsequently, dcaf1 heterozygous mutants carrying the complementing DCAF1g transgene (DCAF1g/dcaf1-1 and DCAF1g/dcaf1-2) were identified by PCR-based genotyping.
The full-length DCAF1 CDS from pGBKT7-DCAF1 was fused in frame to the 3′ end of the HA-tag (three copies). Then, the SpeI/ApaI fragment containing HA-DCAF1 was subcloned between the cauliflower mosaic virus 35S promoter and the 3′ untranslated region of nopaline synthase in a plant binary vector derived from pCAMBIA-1200. The 35S:HA-DCAF1 transgenic plants with wild-type background were selected with 25 mg/L of hygromycin. The transgene expression levels of independent transgenic lines were monitored by anti-HA (Sigma-Aldrich) and anti-DCAF1 antibodies in total protein extracts from seedlings.
To clone the promoter region of DCAF1, we designed two primers: ProF, 5′-CAGAAGCTTAAGTTACCTGATCCGGTGGTCG-3′; ProR, 5′-CAGCCATGGATTGCCCGTCCATGATGAATACC-3′. They were used to amplify a HindIII/NcoI genomic fragment containing 1931 bp upstream of ATG, which was then inserted into pCAMBIA-1301 (CAMBIA). The construct was transformed into wild-type Arabidopsis plants, and the DCAF1:GUS transgenic plants were selected with 25 mg/L of hygromycin.
Immunoblot Analysis and Antibodies
Arabidopsis tissues were homogenized in liquid nitrogen, and total proteins were extracted in buffer containing 150 mM NaCl, 10 mM MgCl2, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1×complete protease inhibitor (Roche). Extracts were centrifuged twice at 13,000 rpm for 10 min at 4°C, and protein concentration in the supernatant was determined by Bradford assay (Bio-Rad). Protein samples were boiled in sample buffer, run on SDS-PAGE gels, and blotted onto polyvinylidene difluoride membranes (Millipore). The protein blots were probed with specific primary antibodies.
A BamHI/XhoI fragment containing nucleotide 3574-4131 of the DCAF1 CDS (encoding amino acid 1192-1377), an NheI/XhoI fragment containing nucleotide 1831-2490 of the DDB1A CDS (encoding amino acid 611-830), and an EcoRI/XhoI fragment containing nucleotide 1054-1632 of the DET1 CDS (encoding amino acid 352-543) were individually cloned into the pET-28a prokaryotic expression vector (Novagen). The 6×His-tag fusion proteins were expressed in Escherichia coli strain BL21 (DE3) and purified with Ni-NAT agarose (Qiagen). Polyclonal antibodies were raised by immunizing rabbits with the purified fusion proteins as antigens.
Other primary antibodies used in this study include anti-CUL4 (Chen et al., 2006), anti-CSN5 (Kwok et al., 1998), anti-CSN6 (Peng et al., 2001), anti-CSN7 (Karniol et al., 1999), anti-RPN6 (Kwok et al., 1999), and anti-Flag (Sigma-Aldrich). Each immunoblot assay was repeated at least twice.
Y2H Assay and in Vivo Co-IP
The Y2H procedure and β-galactosidase activity assay were performed according to the Yeast Protocols Handbook (Clontech), except that the LexA fusion constructs and the activation domain constructs were cotransformed into yeast strain L40 (Invitrogen). The StuI/ApaI fragments containing the full-length CDS, deletion mutants, and point mutants of DCAF1 were all subcloned into pB42AD (Clontech) to make prey constructs. DDB1A was cloned into pLexA (Clontech) to make a bait construct as described previously (Chen et al., 2006).
The co-IP experiments were performed as previously described (Feng et al., 2003) with minor modifications. Each immunoprecipitation was repeated at least twice. The lysis/binding buffer consisted of 150 mM NaCl, 10 mM MgCl2, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10 mM NaF, 2 mM Na3VO4, 25 mM β-glycerolphosphate, 10% glycerol, 0.1% Nonidet P-40, 1 mM PMSF, and 1×complete protease inhibitor (Roche). The washing buffer consisted of 150 mM NaCl, 10 mM MgCl2, 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% glycerol, 0.1% Nonidet P-40, and 1 mM PMSF.
Transient Expression in Onion Epidermal Cells
Full-length coding sequences of DCAF1, DDB1A, and DDB1B were individually subcloned in frame to the 5′ end of sGFP CDS in pUC18-sGFP (Chiu et al., 1996; Niwa et al., 1999) to generate pUC18-DCAF1-sGFP, pUC18-DDB1A-sGFP, and pUC18-DDB1B-sGFP. The pUC18-sGFP-CUL4 construct was described previously (Chen et al., 2006). The expression of sGFP fusion proteins was driven by the 35S constitutive promoter. Living onion (Allium cepa) epidermal cells were bombarded with 5 μg superhelical plasmid DNA using the Biolistic PDS-1000/He Gene Gun System (Bio-Rad). Bombarded epidermal cells were incubated for 16 to 24 h at 22°C under continuous white light. The cell layers were then mounted in DAPI staining buffer containing 0.1% DAPI in 5% DMSO and 1% Tween 20 and examined by laser scanning confocal microscopy (LSM 510 META microscope; Zeiss). GFP fluorescence was imaged using excitation with the 488-nm line of the argon laser and a 505- to 530-nm band-pass emission filter, while DAPI fluorescence was imaged using excitation with the 405-nm line of the argon laser and a 420- to 480-nm band-pass emission filter. Imaging of GFP and DAPI fluorescence was performed sequentially.
Whole-Mounted Clearing of Ovules
To study embryonic development, siliques from self-pollinated Arabidopsis plants were dissected and cleared in Herr's solution containing lactic acid, chloral hydrate, phenol, clove oil, and xylene (2:2:2:2:1, w/w) (Ding et al., 2006). Cleared ovules were further removed from siliques in a drop of the same clearing solution, whole-mounted, and observed under an Axio Imager.M1 microscope (Zeiss) equipped with DIC optics. Micrographs were captured using an AxioCam MRc5 digital camera (Zeiss) and processed with AxioVision 4.6.3.0 software (Zeiss). Embryos of DCAF1:GUS transgenic plants were examined after staining for GUS activity.
Histochemical Staining for GUS Activity
GUS staining was performed as previously described (Byrne et al., 2003). Different tissues of DCAF1:GUS transgenic plants were directly immersed in the reaction buffer containing 100 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 100 mg/L chloramphenicol, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl β-d-glucuronic acid. Samples were incubated in the dark for 24 h at 37°C, rinsed with sterile water, and then treated with 100% ethanol at room temperature to extract chlorophyll.
For observation of the whole mounts, sample clearing was performed as previously described (Malamy and Benfey, 1997). Samples were first transferred into 0.24 n HCl in 20% methanol and incubated for 15 min at 57°C. This solution was replaced by 7% NaOH in 60% ethanol for another 15 min of incubation at room temperature. Samples were then rehydrated in an ethanol series, 40%, 20%, and 10% ethanol for 5 min each, and infiltrated in 5% ethanol and 25% glycerol for 15 min. Finally, samples were mounted in 50% glycerol on glass microscope slides and observed with a stereomicroscope or a microscope equipped with DIC optics.
Accession Numbers
Sequence data for the DCAF1 homologs used in the phylogenetic analysis were obtained from the GenBank database with the following accession numbers: NP_611592 (fruit fly), NP_501725 (worm), NP_055518 (human), NP_001015507 (mouse), and NP_001054357 (rice). The cDNA sequences used for Arabidopsis gene cloning were deposited in the AGI and GenBank data libraries under the following accession numbers: DCAF1 (At4g31160, NM_119266), DDB1A (At4g05420, NM_116781), DDB1B (At4g21100, NM_118228), WUS (At2g17950, NM_127349), and RanBPM related (At1g06060, NM_100487). The accession numbers of the SALK T-DNA insertion mutants of DCAF1 are Salk_010755 (dcaf1-1) and Salk_114078 (dcaf1-2), respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of DCAF1 Homologs.
Supplemental Figure 2. Expression Profiles of the DCAF1 Gene from a Prior Published Microarray Analysis.
Supplemental Figure 3. Matured Siliques from the dcaf1 Heterozygotes.
Supplemental Figure 4. Interaction Tests between Potential DCAF1 Target Proteins and DCAF1.
Supplemental Table 1. Numbers of Putative DCAF Proteins and [WY][DE]x[RK] Motifs in Arabidopsis and Rice.
Supplemental Table 2. Putative DCAF Proteins in Arabidopsis and the Numbers of [WY][DE]x[RK] or DWD Motifs in Each Protein.
Supplemental Table 3. Putative DCAF Proteins in Rice and the Numbers of [WY][DE]x[RK] or DWD Motifs in Each Protein.
Supplemental Methods. Microarray Analysis.
Supplemental Data Set 1. Alignment of DCAF1 Homologs.
Supplementary Material
Acknowledgments
We thank Yan Guo for providing the reconstructed pCAMBIA-1200 binary vector and Junli Zhou for critical reading of the manuscript. We also thank other members in Deng's lab at the National Institute of Biological Sciences for their constructive discussion and kind help. This study was supported by a grant from the National 863 High-Tech Project of National Ministry of Science and Technology, People's Republic of China (2003AA 210070) and in part by a National Science Foundation 2010 grant (MCB-0519970). H.C. was a Peking-Yale Joint Center Monsanto Fellow.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Xing Wang Deng (xingwang.deng@yale.edu).
Online version contains Web-only data.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Angers, S., Li, T., Yi, X., MacCoss, M.J., Moon, R.T., and Zheng, N. (2006). Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443 590–593. [DOI] [PubMed] [Google Scholar]
- Belzile, J.P., Duisit, G., Rougeau, N., Mercier, J., Finzi, A., and Cohen, E.A. (2007). HIV-1 Vpr-mediated G2 arrest involves the DDB1-CUL4A(VPRBP) E3 ubiquitin ligase. PLoS Pathog. 3 e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt, A., Lechner, E., Hano, P., Schade, V., Dieterle, M., Anders, M., Dubin, M.J., Benvenuto, G., Bowler, C., Genschik, P., and Hellmann, H. (2006). CUL4 associates with DDB1 and DET1 and its downregulation affects diverse aspects of development in Arabidopsis thaliana. Plant J. 47 591–603. [DOI] [PubMed] [Google Scholar]
- Byrne, M.E., Groover, A.T., Fontana, J.R., and Martienssen, R.A. (2003). Phyllotactic pattern and stem cell fate are determined by the Arabidopsis homeobox gene BELLRINGER. Development 130 3941–3950. [DOI] [PubMed] [Google Scholar]
- Chen, H., Shen, Y., Tang, X., Yu, L., Wang, J., Guo, L., Zhang, Y., Zhang, H., Feng, S., Strickland, E., Zheng, N., and Deng, X.W. (2006). Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18 1991–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Zhang, Y., Douglas, L., and Zhou, P. (2001). UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 276 48175–48182. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6 325–330. [DOI] [PubMed] [Google Scholar]
- Chu, G., and Chang, E. (1988). Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242 564–567. [DOI] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y.H., Liu, N.Y., Tang, Z.S., Liu, J., and Yang, W.C. (2006). Arabidopsis GLUTAMINE-RICH PROTEIN23 is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III. Plant Cell 18 815–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emes, R.D., and Ponting, C.P. (2001). A new sequence motif linking lissencephaly, Treacher Collins and oral-facial-digital type 1 syndromes, microtubule dynamics and cell migration. Hum. Mol. Genet. 10 2813–2820. [DOI] [PubMed] [Google Scholar]
- Feng, S., Ma, L., Wang, X., Xie, D., Dinesh-Kumar, S.P., Wei, N., and Deng, X.W. (2003). The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell 15 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., Shen, Y., Sullivan, J.A., Rubio, V., Xiong, Y., Sun, T.P., and Deng, X.W. (2004). Arabidopsis CAND1, an unmodified CUL1-interacting protein, is involved in multiple developmental pathways controlled by ubiquitin/proteasome-mediated protein degradation. Plant Cell 16 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, D., Wakasugi, M., Ishigaki, Y., Nikaido, O., and Matsunaga, T. (2003). cDNA cloning of the chicken DDB1 gene encoding the p127 subunit of damaged DNA-binding protein. Genes Genet. Syst. 78 169–177. [DOI] [PubMed] [Google Scholar]
- Gerlitz, G., Darhin, E., Giorgio, G., Franco, B., and Reiner, O. (2005). Novel functional features of the Lis-H domain: Role in protein dimerization, half-life and cellular localization. Cell Cycle 4 1632–1640. [DOI] [PubMed] [Google Scholar]
- Glickman, M.H., and Ciechanover, A. (2002). The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82 373–428. [DOI] [PubMed] [Google Scholar]
- Goldenberg, S.J., Cascio, T.C., Shumway, S.D., Garbutt, K.C., Liu, J., Xiong, Y., and Zheng, N. (2004). Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119 517–528. [DOI] [PubMed] [Google Scholar]
- He, Y.J., McCall, C.M., Hu, J., Zeng, Y., and Xiong, Y. (2006). DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4–ROC1 ubiquitin ligases. Genes Dev. 20 2949–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa, L.A., Wu, M., Ye, T., Kobayashi, R., Sun, H., and Zhang, H. (2006). CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat. Cell Biol. 8 1277–1283. [DOI] [PubMed] [Google Scholar]
- Higa, L.A., and Zhang, H. (2007). Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrecka, K., Gierszewska, M., Srivastava, S., Kozaczkiewicz, L., Swanson, S.K., Florens, L., Washburn, M.P., and Skowronski, J. (2007). Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. USA 104 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi, T., Kimura, S., Yamamoto, T., Furukawa, T., Takata, K., Uchiyama, Y., Hashimoto, J., and Sakaguchi, K. (2003). Rice UV-damaged DNA binding protein homologues are most abundant in proliferating tissues. Gene 308 79–87. [DOI] [PubMed] [Google Scholar]
- Jin, J., Arias, E.E., Chen, J., Harper, J.W., and Walter, J.C. (2006). A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23 709–721. [DOI] [PubMed] [Google Scholar]
- Karniol, B., Malec, P., and Chamovitz, D.A. (1999). Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell 11 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer, M., Stern, Y., Cook, H., Clerici, E., Maulbetsch, C., Laux, T., and Davies, B. (2006). Analysis of the transcription factor WUSCHEL and its functional homologue in Antirrhinum reveals a potential mechanism for their roles in meristem maintenance. Plant Cell 18 560–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, M.H., Cooper, D.R., Oleksy, A., Devedjiev, Y., Derewenda, U., Reiner, O., Otlewski, J., and Derewenda, Z.S. (2004). The structure of the N-terminal domain of the product of the lissencephaly gene Lis1 and its functional implications. Structure 12 987–998. [DOI] [PubMed] [Google Scholar]
- Kwok, S.F., Solano, R., Tsuge, T., Chamovitz, D.A., Ecker, J.R., Matsui, M., and Deng, X.W. (1998). Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, S.F., Staub, J.M., and Deng, X.W. (1999). Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J. Mol. Biol. 285 85–95. [DOI] [PubMed] [Google Scholar]
- Lee, J., and Zhou, P. (2007). DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26 775–780. [DOI] [PubMed] [Google Scholar]
- Lee, J.H., Terzaghi, W., Gusmaroli, G., Charron, J.B., Yoon, H.J., Chen, H., He, Y.J., Xiong, Y., and Deng, X.W. (2008). Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20 152–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic, E., Belaidouni, N., Estrabaud, E., Morel, M., Rain, J.C., Transy, C., and Margottin-Goguet, F. (2007). HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6 182–188. [DOI] [PubMed] [Google Scholar]
- Li, T., Chen, X., Garbutt, K.C., Zhou, P., and Zheng, N. (2006). Structure of DDB1 in complex with a paramyxovirus V protein: Viral hijack of a propeller cluster in ubiquitin ligase. Cell 124 105–117. [DOI] [PubMed] [Google Scholar]
- Liu, W., Nichols, A.F., Graham, J.A., Dualan, R., Abbas, A., and Linn, S. (2000). Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem. 275 21429–21434. [DOI] [PubMed] [Google Scholar]
- Ma, L., Sun, N., Liu, X., Jiao, Y., Zhao, H., and Deng, X.W. (2005). Organ-specific expression of Arabidopsis genome during development. Plant Physiol. 138 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy, J.E., and Benfey, P.N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124 33–44. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., et al. (2007). CDD: A conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35 D237–D240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateja, A., Cierpicki, T., Paduch, M., Derewenda, Z.S., and Otlewski, J. (2006). The dimerization mechanism of LIS1 and its implication for proteins containing the LisH motif. J. Mol. Biol. 357 621–631. [DOI] [PubMed] [Google Scholar]
- Niwa, Y., Hirano, T., Yoshimoto, K., Shimizu, M., and Kobayashi, H. (1999). Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J. 18 455–463. [DOI] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski, M.D., and Deshaies, R.J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6 9–20. [DOI] [PubMed] [Google Scholar]
- Pick, E., Lau, O.S., Tsuge, T., Menon, S., Tong, Y., Dohmae, N., Plafker, S.M., Deng, X.W., and Wei, N. (2007). Mammalian DET1 regulates Cul4A activity and forms stable complexes with E2 ubiquitin-conjugating enzymes. Mol. Cell. Biol. 27 4708–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, D.F., Gahrtz, M., Maxwell, B.B., Cook, R.K., Kan, J.M., Alonso, J.M., Ecker, J.R., and Chory, J. (2002). De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 12 1462–1472. [DOI] [PubMed] [Google Scholar]
- Schröfelbauer, B., Hakata, Y., and Landau, N.R. (2007). HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl. Acad. Sci. USA 104 4130–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröfelbauer, B., Yu, Q., Zeitlin, S.G., and Landau, N.R. (2005). Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 79 10978–10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle, J., and Vierstra, R.D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55 555–590. [DOI] [PubMed] [Google Scholar]
- Sridhar, V.V., Surendrarao, A., Gonzalez, D., Conlan, R.S., and Liu, Z. (2004). Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 101 11494–11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata, K., Ishikawa, G., Hirose, F., and Sakaguchi, K. (2002). Drosophila damage-specific DNA-binding protein 1 (D-DDB1) is controlled by the DRE/DREF system. Nucleic Acids Res. 30 3795–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, L., Ehrlich, E., and Yu, X.F. (2007). DDB1 and Cul4A are required for human immunodeficiency virus type 1 Vpr-induced G2 arrest. J. Virol. 81 10822–10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, X., Duus, K.M., Friedrich, T.D., and de Noronha, C.M. (2007). The HIV1 protein Vpr acts to promote G2 cell cycle arrest by engaging a DDB1 and Cullin4A-containing ubiquitin ligase complex using VprBP/DCAF1 as an adaptor. J. Biol. Chem. 282 27046–27057. [DOI] [PubMed] [Google Scholar]
- Yanagawa, Y., Sullivan, J.A., Komatsu, S., Gusmaroli, G., Suzuki, G., Yin, J., Ishibashi, T., Saijo, Y., Rubio, V., Kimura, S., Wang, J., and Deng, X.W. (2004). Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 18 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., Feng, Y., Narayan, O., and Zhao, L.J. (2001). Cytoplasmic retention of HIV-1 regulatory protein Vpr by protein-protein interaction with a novel human cytoplasmic protein VprBP. Gene 263 131–140. [DOI] [PubMed] [Google Scholar]
- Zhao, L.J., Mukherjee, S., and Narayan, O. (1994). Biochemical mechanism of HIV-I Vpr function. Specific interaction with a cellular protein. J. Biol. Chem. 269 15577–15582. [PubMed] [Google Scholar]
- Zhao, L.J., Zhang, S., and Chinnadurai, G. (2002). Sox9 transactivation and testicular expression of a novel human gene, KIAA0800. J. Cell. Biochem. 86 277–289. [DOI] [PubMed] [Google Scholar]
- Zheng, N., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416 703–709. [DOI] [PubMed] [Google Scholar]
- Zolezzi, F., Fuss, J., Uzawa, S., and Linn, S. (2002). Characterization of a Schizosaccharomyces pombe strain deleted for a sequence homologue of the human damaged DNA binding 1 (DDB1) gene. J. Biol. Chem. 277 41183–41191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.