Abstract
Mitochondrial fission is achieved partially by the activity of self-assembling dynamin-related proteins (DRPs) in diverse organisms. Mitochondrial fission in Arabidopsis thaliana is mediated by DRP3A and DRP3B, but the other genes and molecular mechanisms involved have yet to be elucidated. To identify these genes, we screened and analyzed Arabidopsis mutants with longer and fewer mitochondria than those of the wild type. ELM1 was found to be responsible for the phenotype of elongated mitochondria. This phenotype was also observed in drp3a plants. EST and genomic sequences similar to ELM1 were found in seed plants but not in other eukaryotes. ELM1:green fluorescent protein (GFP) was found to surround mitochondria, and ELM1 interacts with both DPR3A and DRP3B. In the elm1 mutant, DRP3A:GFP was observed in the cytosol, whereas in wild-type Arabidopsis, DRP3A:GFP localized to the ends and constricted sites of mitochondria. These results collectively suggest that mitochondrial fission in Arabidopsis is mediated by the plant-specific factor ELM1, which is required for the relocalization of DRP3A (and possibly also DRP3B) from the cytosol to mitochondrial fission sites.
INTRODUCTION
Plant mitochondria, in addition to playing roles in respiration and metabolism, also play plant-specific roles, such as photorespiration and redox regulation, in photosynthetic cells (reviewed in Mackenzie and McIntosh, 1999; Raghavendra and Padmasree, 2003; Noguchi and Yoshida, 2008). Mitochondria are not synthesized de novo but are created by the division of existing mitochondria. Such division has been occurring throughout each lineage of eukaryotes, probably since the first endosymbiotic birth of the original mitochondrion. Mitochondrial fission is needed not only for maintenance of number of mitochondria during the host cell cycle but also for the proper mitochondrial morphology and distribution within a single cell (Bleazard et al., 1999; Sesaki and Jensen, 1999; Okamoto and Shaw, 2005). Mitochondrial fission has also been shown to play a role in apoptosis, longevity, and lethal disease in yeasts and animals (Frank et al., 2001; Li et al., 2004; Jagasia et al., 2005; Scheckhuber et al., 2007; Waterham et al., 2007; reviewed in Chan, 2006a, 2006b). Although mitochondrial fission is a fundamental cell process in higher plants, studies of plant mitochondrial fission have just begun.
The molecular mechanisms involved in mitochondrial fission have been studied extensively in the yeast Saccharomyces cerevisiae. In this organism, Dnm1p (a yeast dynamin-related protein [DRP]) exists as a dimer and/or oligomer in the cytoplasm that is recruited to mitochondrial fission sites. This relocalization of Dnm1p depends on the mitochondrial outer-membrane protein Fis1p and the cytosolic molecular adapter Mdv1p (and its paralog Caf4p), which can bind to both Dnm1p and Fis1p (Tieu and Nunnari, 2000; Tieu et al., 2002; Karren et al., 2005; Bhar et al., 2006; Naylor et al., 2006; reviewed in Okamoto and Shaw, 2005; Hoppins et al., 2007). Dnm1p and Mdv1p are thought to form higher-order multimer complexes, named fission complexes, that surround and pinch off the mitochondria. In vitro, isolated DRPs can make spiral polymers that tubulate liposomes in the presence of guanosine-5-O-(3-thio)triphosphate (a nonhydrolyzable GTP [GTPγS]) (Yoon et al., 2001; Ingerman et al., 2005).
The Arabidopsis thaliana DRPs involved in mitochondrial fission are DRP3A and DRP3B (formerly, Arabidopsis dynamin-like protein 2A [ADL2a] and ADL2b, respectively) (Arimura and Tsutsumi, 2002; Hong et al., 2003; Arimura et al., 2004a; Logan et al., 2004; Mano et al., 2004). The Arabidopsis genome has at least one possible ortholog of yeast Fis1p, named BIGYIN (Scott et al., 2006). However, Arabidopsis has no genes similar to yeast Mdv1p. Here, we use a genetic approach to show that a hypothetical protein of unknown function encoded by the Arabidopsis genome is involved in mitochondrial fission and is required for the correct localization of DRP3A to mitochondrial fission sites.
RESULTS
Isolation and Mapping of the Mitochondrial Fission Mutants
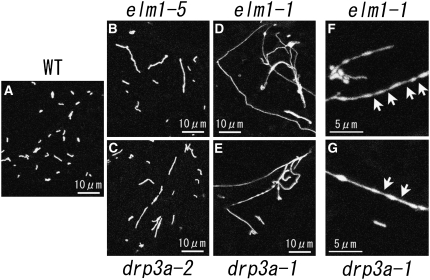
In an earlier study, seeds of transgenic Arabidopsis with mitochondrial-targeted green fluorescent protein (Mt-GFP) were treated with ethyl methanesulfonate to induce mutagenesis, and the leaves of 19,000 M2 plants were screened by fluorescence microscopy for abnormally shaped mitochondria (Feng et al., 2004). In this study, two of these mitochondrial morphology mutants were selected and mapped. Both mutants had severely elongated mitochondria that occasionally occurred as an interconnected network (Figures 1D to 1G), similar to the mitochondria of yeast mutants with defective mitochondrial fission genes (Bleazard et al., 1999; Sesaki and Jensen, 1999).
Figure 1.
Mitochondrial Morphologies in drp3a and elm1 Mutants.
Mitochondria in leaf epidermal cells of wild-type plants (A) and in weak (elm1-5 [B] and drp3a-2 [C]) and severe (elm1-1 [D] and ]F] and drp3a-1 [E] and [G]) mutants transformed with Mt-GFP. Arrows indicate the constricted sites of elongated mitochondria.
One of these genes responsible for the elongated mitochondrial phenotype was mapped to the long arm of chromosome 4. Because this region contained the DRP3A gene, the genomic sequence of DRP3A of the mutant was determined. A G-to-A substitution changed an Arg residue at position 82 in the conserved GTPase domain to a His residue, and we named this mutant drp3a-1 (see Supplemental Figure 1 online). Sequencing of other mutants found previously (Feng et al., 2004) with slightly elongated mitochondria enabled us to find an additional allele, drp3a-2 (Figure 1; see Supplemental Figure 1 online), which had a G-to-A substitution that changed Ala at position 208 to Thr.
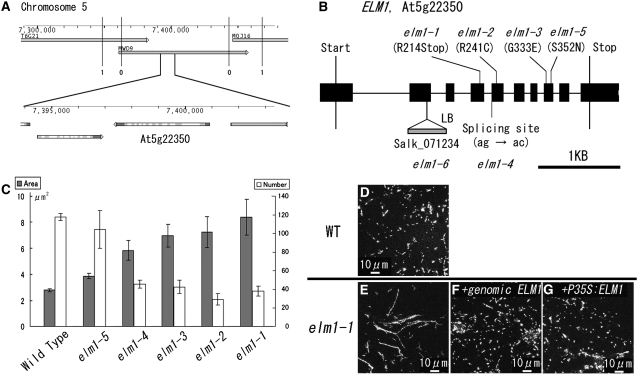
The second locus responsible for the severe mitochondrial elongation phenotype was mapped to the short arm of chromosome 5. This region does not have any sequences that are similar to the yeast and animal genes involved in mitochondrial fission and mitochondrial morphology. We named the mutant elm1 because of its phenotype, elongated mitochondria. By fine-mapping and sequencing, the elm1-1 mutant was found to have a G-to-A substitution in At5g22350, changing Trp at position 214 to a stop codon in the predicted amino acid sequence (Figures 2A and 2B; see Supplemental Figure 2 online). Four additional mutants with slightly elongated mitochondria had independent nucleotide substitutions in this same gene (Figure 2B; see Supplemental Figure 2 online). We named these five mutant alleles elm1-1 to elm1-5, according to the severity of the phenotype (Figure 2C). A T-DNA insertion line of this gene was obtained from the ABRC. MitoTracker staining of leaf epidermal cells of this line showed an interconnected network of mitochondria similar to that of the elm1-1 mutant (Figure 3A). We named this mutant elm1-6. Introduction of a fusion gene P35S:ELM1-ORF (for open reading frame) or a genomic fragment that includes At5g22350 from 950 bp upstream to 345 bp downstream (genomic ELM1) into elm1-1 restored the wild-type mitochondrial phenotype (Figures 2D to 2G). Together, these results show that At5g22350 is identical to ELM1.
Figure 2.
ELM1 Is the Gene Responsible for the Elongation of Mitochondria in elm1 Mutants.
(A) Map-based cloning of ELM1. The numbers of recombination events between the molecular markers and the ELM1 locus are shown under the vertical bars (0 and 1). T6G21, MWD9, and MQJ16 are overlapping BAC clones. Numbers on the gauges show the physical distance in base pairs from the northern end of chromosome 5. The figure was modified from the Arabidopsis Genome Initiative Map of Map Viewer in The Arabidopsis Information Resource home page.
(B) The positions of base pair substitutions in elm1-1 through elm1-5 mutants and the DNA insertion in elm1-6. ELM1 consists of 10 exons (black boxes) and nine introns (black lines).
(C) Mitochondrial sizes and numbers in epidermal cells of wild-type and elm1 mutants. Gray bars show the average planar area (in μm2) of the mitochondria (n > 90). White bars show the average numbers of mitochondria in a 120 μm × 120 μm area (n = 3). All error bars show se.
(D) to (G) Complementation test of mitochondrial elongation in elm1-1. The wild type (D), elm1-1 (E), and elm1-1 (F) transformed with a genomic fragment of the ELM1 gene with 950 bp upstream region of the start codon to 345 bp downstream of the stop codon (genomic ELM1) and elm1-1 (G) with P35S:ELM1-ORF. Both restored mitochondrial morphology to that of the wild type.
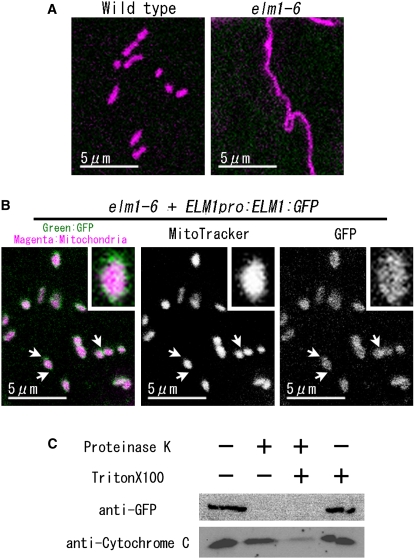
Figure 3.
ELM1:GFP Localizes to the Outer Surface of Mitochondria.
(A) and (B) ELM1:GFP rescued the elongation of mitochondria in elm1-6 and localized on mitochondria. Mitochondria in epidermal cells of the cotyledon were visualized as magenta by MitoTracker Orange CMTMRos staining.
(B) A genomic fragment of ELM1 fused to GFP (ELM1pro:ELM1:GFP) was introduced into elm1-6. Insets show 4× magnified images of mitochondria. Arrows show constricted sites and end sites of mitochondria where green signals seem more intense.
(C) Protease protection assay. Isolated mitochondria from elm1-6 transformed with ELM1pro:ELM1:GFP were incubated in the presence or absence of Proteinase K or Triton X-100, as indicated. The samples were subjected to SDS-PAGE, and protein gel blotting was done with anti-GFP and anti-rice cytochrome C antiserum. A representative result of three technical repeats and two biological repeats is shown.
Plant Growth and Other Mitochondrial Phenotypes of elm1-1
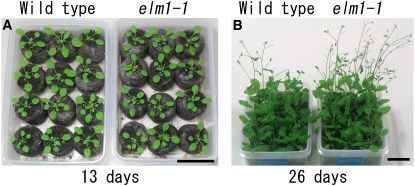
The elm1-1 plants grow more slowly and are slightly smaller than those of the wild type, but bolt earlier (Figure 4). All of the elm1 mutants can complete their life cycle and have normal fertility under standard growth conditions.
Figure 4.
Plant Growth of the elm1-1 Mutant.
Wild-type and elm1-1 plants on the 13 d (A) and 26 d (B) after germination. The wild type is a Mt-GFP transgenic line. Bars = 3 cm.
Although elm1-1 mutants have an interconnected network of mitochondria, each cell still has several small discrete mitochondria (Figure 1; see Supplemental Figure 3 online), suggesting that the cells have residual mitochondrial fission. Mitochondria in elm1-6 were stained with MitoTracker Orange CMTMRos, which accumulates in mitochondria in a membrane potential–dependent manner (Figure 3A), and this indicated that the mutants maintained an inner-membrane potential. In the elm1-1 mutant, both disconnected small mitochondria and interconnected mitochondria moved similar to mitochondria in the wild type (see Supplemental Movie 2 online) and changed their morphology dynamically (see Supplemental Figure 3 and Supplemental Movie 1 online). Mitochondria also transiently appeared in the shape of beads on a string (arrows in Figure 1; see Supplemental Figure 3 online). The constricted sites may be sites of ongoing but hampered mitochondrial fission.
ELM1-Like Genes Were Detected in Plants but Not in Other Eukaryotes
The predicted ELM1 protein has 427 amino acid residues with a mass of ∼48 kD. We identified homologs of ELM1 in the Arabidopsis genome (AT5G06180, 53.6% identity) as well as in other higher plants (see Supplemental Figure 2 online). Although homologous genes were not found in other eukaryotes, the C-terminal two-thirds of the ELM1 sequence matched a domain of unknown function (see Supplemental Figure 2 online) (DUF1022) that was present in a number of hypothetical proteobacterial proteins. The evolutionary relationship between ELM1 and these bacterial ELM1-like sequences is not clear. SOSUI and DAS transmembrane domain searches (Cserzo et al., 1997; Hirokawa et al., 1998) predicted no apparent transmembrane domain in the ELM1 sequence.
The elm1 Mitochondrial Phenotype Is Similar to That of drp3a
As described above, mitochondria in elm1 mutants are longer in shape (and sometimes interconnected in elm1-1) but fewer in number than those in the wild type. The reciprocal relationship between the number and size (planar area) of mitochondria was demonstrated in wild-type and elm1 cells (Figure 2C, from weak elm1-5 to severe elm1-1). The mitochondrial morphologies in weak (elm1-5) and strong (elm1-1) alleles of elm1 were quite similar to those of weak (drp3a -2) and severe (drp3a-1) alleles of drp3a mutants, respectively (Figure 1). These elongated mitochondria have about the same diameter (0.5 to 1 μm) as those of the wild type but vary in length from 1 to >50 μm. Interconnected networks of mitochondria could be seen in both of the severe alleles, drp3a-1 and elm1-1. At high magnification, the mitochondria of drp3a-1 mutants sometimes appeared as beads on a string, as described above for elm1-1 (arrows in Figure 1; see Supplemental Figure 3 online). The similar mitochondrial phenotypes of the drp3a and elm1 mutants imply that DRP3A and ELM1 are involved in the same or related steps of mitochondrial fission that seem to occur after the initial constriction of mitochondria.
ELM1 Localizes to Mitochondria
To determine the subcellular localization of ELM1, the DNA fragment ELM1pro:ELM1:GFP was introduced into elm1-6. We selected elm1-6 because its mitochondrial phenotype is as severe as that of elm1-1 and it does not have Mt-GFP. The elongated and interconnected mitochondrial phenotype in elm1-6 was rescued by the introduction of ELM1pro:ELM1:GFP, suggesting that ELM1:GFP could function like the native ELM1 (Figures 3A and 3B). While ELM1:GFP signals colocalized with MitoTracker signal in the cells, the signals did not completely coincide. The observation that green signal (GFP) surrounds the magenta signal (MitoTracker) suggests that ELM1 localizes to the outer membrane or the surface of the mitochondrial outer membrane. Some of the GFP fluorescence seemed to be slightly concentrated at the ends and constricted sites of mitochondria (Figure 3B, arrows). Similarly, DRP3A:GFP and DRP3B:GFP localized to the ends and constricted sites of mitochondria (Figure 5A) (Arimura and Tsutsumi, 2002; Arimura et al., 2004a). The ends and constricted sites of mitochondria seem to show the past and ongoing sites of mitochondrial fission.
Figure 5.
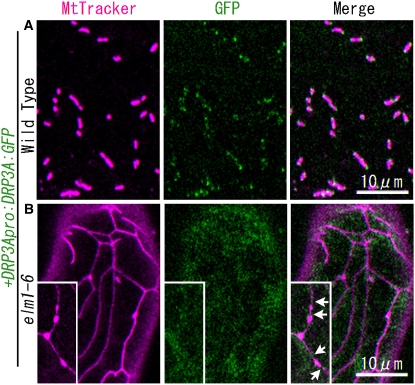
DRP3A:GFP Does Not Localize to Mitochondrial Ends and Constricted Sites in the elm1-6 Mutant.
Confocal laser scanning microscopy images of the cotyledon leaf epidermal cells of wild-type plants (A) and elm1-6 T-DNA insertion mutants (B) transformed with the construct DRP3Apro:DRP3A:GFP. Mitochondria were visualized by MitoTracker Orange staining. Insets in (B) show beads-on-a-string-shaped mitochondria that transiently appeared in elm1-6 from a separate microscope field. Arrows show the constricted sites on mitochondria in elm1-6. Bar in the bottom panel indicates scale of insets.
To determine the intramitochondrial localization of ELM1, mitochondria isolated from elm1-6 plants transformed with ELM1pro:ELM1:GFP were incubated with Proteinase K. While proteinase K degraded the ELM1:GFP, it did not affect cytochrome c that localizes between the inner and outer mitochondrial membranes (Figure 3C, lane 2). When the mitochondrial membranes were broken down with a detergent (Triton X-100), Proteinase K degraded both ELM1:GFP and cytochrome c (Figure 3C, lane 3). These results suggest that ELM1:GFP localizes to the outer surface of mitochondria.
ELM1 Interacts with DRP3A and DRP3B
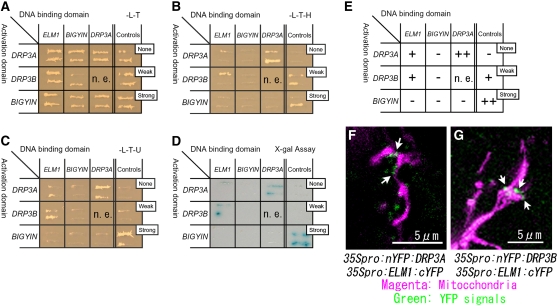
To test the possibility that ELM1, DRP3A, DRP3B, and BIGYIN interact with each other, yeast two-hybrid (Y2H) experiments were performed using full-length fragments of DRP3A, DRP3B, and ELM1. In the case of BIGYIN, an N-terminal predicted-cytosolic portion of the protein without the transmembrane domain was used. As shown in Figures 6A to 6E, ELM1 interacts with DRP3A and with DRP3B, weakly but repeatedly, when checked by three reporter genes (HIS3, URA3, and LacZ) expressed from independent Gal4 promoters. The Y2H experiment also showed that DRP3A strongly interacts with itself (Figures 6A to 6E), which was expected because DRPs form dimers and higher-ordered polymers (reviewed in Okamoto and Shaw, 2005).
Figure 6.
ELM1 Interacts with DRP3A and DRP3B.
(A) to (C) Y2H analysis. Yeast strain Mav203 was transformed with paired constructs of protein fused to the GAL4 activation domain (left) or the GAL4 DNA binding domain (top). Controls show the combinations with no, weak, and strong interactions, as indicated (see Methods). Transformants were streaked onto a SD/-Leu/-Thr plate (A), confirming the introduction of both plasmids into the cells; a SD/-Leu/-Thr/-His plate (B), determining the interaction between fusion proteins; and a SD/-Leu/-Thr/-Ura plate (C), determining the interaction between fusion proteins by Ura synthase gene expression.
(D) X-Gal assay to check the interaction of fusion proteins by β-galactosidase expression.
(E) Summary of the Y2H analysis from (A) to (D). –, no signal; +, weak signal; ++, strong signal; n.e., not examined.
(F) and (G) Fluorescence images of Arabidopsis leaf epidermal cells transiently transformed with BiFC formation constructs as indicated, together with a construct for expressing mitochondrial-targeted DsRed by particle bombardment. nYFP, N-terminal half of YFP; cYFP, C-terminal half of YFP. If the two parts of YFP (nYFP and cYFP) come together by interactions between each fused protein (DRP3A and ELM1 in [F]; DRP3B and ELM1 in [G]), they give off a yellow–green fluorescence on excitation. We used a GFP filter set, which made the fluorescence appear green. Arrows show the green spots that indicate protein interaction on mitochondria.
The interactions between ELM1 and DRP3A and between ELM1 and DRP3B were confirmed by bimolecular fluorescence complementation (BiFC) (Bracha-Drori et al., 2004). Arabidopsis leaves were bombarded with two transient expression plasmids, one expressing the N-terminal half of yellow fluorescent protein (YFP) fused to DRP3A (or DRP3B) and the other expressing the C-terminal half of YFP fused to ELM1. If the two parts of YFP come together, they give off a yellow–green fluorescence on excitation. We used a GFP filter set, which made the fluorescence appear green. The leaves were simultaneously bombarded with a plasmid expressing mitochondrial-targeted red fluorescent protein (mitochondrial presequence of Arabidopsis ATPase δ-prime subunit fused to DsRed) to identify transformed cells and also to identify mitochondria in the cells. As shown in Figures 6F and 6G, green puncta were observed on mitochondria. These results, together with the Y2H results, suggest that ELM1 interacts with DRP3A and with DRP3B on mitochondria.
ELM1 Is Required for the Localization of DRP3A to Mitochondria
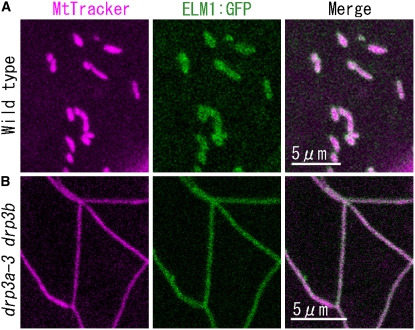
We transformed the wild type, the elm1-6 mutant, and the drp3a-3 drp3b double T-DNA insertion mutant (of SALK_066958 [drp3a-3] and SALK_017492 [drp3b] [see Supplemental Figure 4 online], in which mitochondria are elongated and interconnected [Figure 7B]), with ELM1:GFP. In all cases, ELM1:GFP signal was observed on mitochondria (Figure 7), indicating that the localization of ELM1 to the mitochondrial outer surface does not require DRP3A or DRP3B. In wild-type plants transformed with DRP3A:GFP, the fluorescent signal localized to the ends and constricted sites of mitochondria (Figure 5A), while in elm1-6 transformed with DRP3A:GFP, it remained in the cytosol (Figure 5B), suggesting that ELM1 is required for the subcellular transfer of DRP3A from the cytosol to mitochondrial fission sites.
Figure 7.
ELM1:GFP Localizes on Mitochondria in Both the Wild Type and the drp3a-3 drp3b Mutant.
Confocal laser scanning microscopy images of the cotyledon leaf epidermal cells of the wild type (A) and a drp3a-3 drp3b double T-DNA insertion homoline (B) introduced with the construct ELM1pro:ELM1:GFP and stained with MitoTracker Orange.
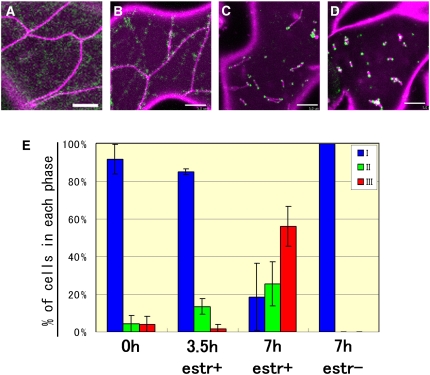
Similar results were obtained with a double transgenic elm1-6 line carrying both an estradiol-inducible ELM1 gene (pER8:ELM1) and DRP3Apro:DRP3A:GFP (Figure 8). In the absence of estradiol, these cells had elongated and network-shaped mitochondria and cytosolic DRP3A:GFP signals (Figure 8A). In the presence of estradiol, two types of cells were observed, cells with green spots localized to network-shaped or elongated mitochondria (Figures 8B and 8C) and cells with green spots on constricted sites and ends of particle-shaped or fragmented mitochondria (Figure 8D), like those of wild-type plants transformed with DRP3Apro:DRP3A:GFP (Figure 5A). We did not see cells with particulate mitochondria without localization of DRP3A:GFP to mitochondria. Green spots were observed to localize on the mitochondria before the mitochondria divided (Figure 8E). These results suggest that the relocalization of DRP3A from the cytosol to mitochondrial fission sites depends on the presence of ELM1.
Figure 8.
Induction of ELM1 Gene Expression in the elm1-6 Mutant Relocates DRP3A:GFP from the Cytosol to Mitochondria and Restores Wild-Type Mitochondrial Morphology.
(A) to (D) Representative images of mitochondria (magenta, stained by MitoTracker Orange) and DRP3A:GFP (green) in elm1-6 transformed with pER8:ELM1 and DRP3A:GFP before (A) and after ([B] to [D]) addition of 17-β estradiol. Bars = 5 μm.
(E) The percentage of transformed cells in each phase of mitochondrial fission before and after addition of 17-β estradiol (estr). Phase I (A) is characterized by cells with elongated mitochondria and cytosolic DRP3A:GFP. In phase II, DRP3A:GFP relocalizes to the elongated mitochondria (shown in [B] and [C]). In phase III, DRP3A:GFP is found on the ends of mitochondrial particles (D). Average percentages of cells in each phase for each time point are shown by bars, and lines indicate sd. More than 50 cells were counted each time in triplicate.
elm1-1 Has Normal Peroxisome Morphology
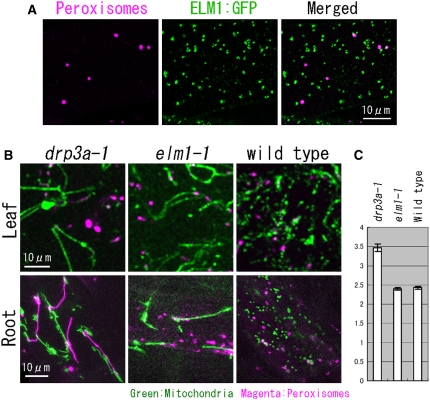
DRP3A is also found on peroxisomes and plays a role in peroxisome division (Mano et al., 2004). To determine whether ELM1 also functions in peroxisome division, we looked for peroxisomal localization of ELM1:GFP and studied peroxisome morphology in elm1-1 and drp3a-1 mutants transformed with ELM1pro:ELM1:GFP. In F1 plants derived from a cross between a transgenic plant transformed with ELM1pro:ELM1:GFP and another transgenic plant expressing DsRed1 fused to a peroxisomal targeting signal, ELM1:GFP signals were observed on peroxisomes (Figure 9A) but only as a small portion of the total ELM1:GFP signal, which was primarily associated with mitochondria (Figures 3B and 7A). Consistent with the weak peroxisomal localization of ELM1:GFP, peroxisomes of elm1-1 appeared very similar to those of the wild-type (Figures 9B and 9C). By contrast, peroxisomes were enlarged in leaves and elongated in roots of the drp3a-1 mutant. Chloroplast morphology was similar to the wild type in both elm1-1 and drp3a-1 mutants. Although the localization of ELM1 on peroxisomes should be examined in more detail in the future, these results collectively suggest that ELM1 does not have a clear function in peroxisome morphology.
Figure 9.
Loss of ELM1 Does Not Change Peroxisome Morphology.
(A) Confocal laser scanning image of the leaf epidermis of a transgenic plant with ELM1pro:ELM1:GFP and peroxisomal-localized DsRed.
(B) Mitochondria and peroxisomes in leaf epidermis and root epidermis in drp3a-1, elm1-1, and the wild type were highlighted by GFP (mitochondria) and DsRed (peroxisomes).
(C) The average planar area of peroxisomes in leaf epidermis in drp3a-1, elm1-1, and wild-type plants (n > 60). Error bars show se.
DISCUSSION
Role of ELM1 in Mitochondrial Fission
Our results suggest that ELM1 localizes on the outer surface of mitochondria and is required for the localization of DRP3A to mitochondrial fission sites. The finding that expression of ELM1 caused DRP3A:GFP to form foci on the ends and at constricted sites of mitochondria in a double transgenic elm1-6 line carrying both an estradiol-inducible ELM1 gene (pER8:ELM1) and DRP3Apro:DRP3A:GFP (Figure 8) suggests that ELM1 is required for the formation of DRP3A multimers as well as the mitochondrial localization of DRP3A. In the BiFC experiments with ELM1 and DRP3A (Figures 6F and 6G), punctate fluorescent signals were observed on mitochondria. This result suggests that ELM1 interacts with DRP3A multimers at the sites of mitochondrial fission. It is unclear whether ELM1 is also required for subsequent steps in mitochondrial fission. DRP3B, a putative paralog of DRP3A (with 76.7% identity), also interacted with ELM1 on mitochondria (Figure 6G), suggesting that DRP3B may also localize to mitochondrial fission sites in an ELM1-dependent manner.
The Role of Mitochondrial Fission in Plant Growth
Mitochondrial fission is thought to be required for mitochondrial inheritance, cell viability, and development (Chan, 2006a). Even though each of the elm1 mutants had drastically altered mitochondria caused by defects of mitochondrial fission, their growth was only slightly retarded and their appearance and fertility were similar to those of the wild-type plants. Also, following cell division, each of the daughter cells appeared to inherit mitochondria because none of the mutant cells lacked mitochondria or had uneven amounts of mitochondria. Mitochondria in elm1 mutants also appeared to retain mitochondrial membrane potential (Figure 3A) and normal mitochondrial movement (see Supplemental Figure 3 and Supplemental Movie 1 online). Therefore, mitochondrial fission defects in elm1 mutants seem not to cause severe defects of mitochondrial inheritance, cell viability, or plant growth. However, our results do not establish whether mitochondrial fission is required for mitochondrial inheritance, cellular activity, and plant growth because elm1 mutants have residual mitochondrial fission activity. The residual mitochondrial fission activity may be caused by an ELM1 homolog or by a completely independent mitochondrial fission system.
ELM1 Is Involved in Mitochondrial Fission but Not in Peroxisome Division
Some dynamin-related proteins required for mitochondrial fission are also involved in peroxisome division in Arabidopsis, human, and yeast (Koch et al., 2003; Mano et al., 2004; Kuravi et al., 2006). Although a small portion of ELM1:GFP signal was observed on peroxisomes (Figure 9), ELM1 seems not to be required for peroxisome division. It is not surprising that the mechanisms by which DRP3A localizes to peroxisomes and mitochondria are different because mitochondrial fission must occur frequently to balance mitochondrial fusion, whereas fusion of peroxisomes occurs far less frequently (Arimura et al., 2004b).
Comparison of Yeast, Human, and Arabidopsis Mitochondrial Fission Mechanisms
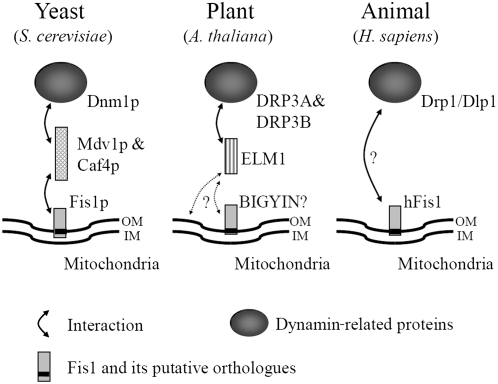
The function and localization of ELM1 (Figure 10) are similar to those of Mdv1p (and Caf4p) in yeast, proteins thought to be required for the localization of Dnm1p to mitochondria (Tieu and Nunnari, 2000; Cerveny et al., 2001; Tieu et al., 2002; Griffin et al., 2005). Mdv1p is known as a molecular adaptor protein between Dnm1p and Fis1p, a mitochondrial outer-membrane embedded protein (Mozdy et al., 2000). Mdv1p has three domains: an N-terminal domain is required for binding to the N-terminal soluble domain of Fis1p; a coiled-coil domain in the middle is needed for the self-oligomerization of Mdv1p; and a C-terminal WD40 repeat is required for binding to Dnm1p. Although ELM1 does not have known functional domains in its sequence, it should have at least two domains, one for binding to DRP3A and DRP3B, and another for mitochondrial localization. ELM1 might localize to mitochondria by interacting with BIGYIN (a possible ortholog of yeast Fis1; Scott et al., 2006), in the same way that MDV1p interacts with Fis1p in yeast. Although our Y2H experiments did not detect an interaction between ELM1 and BIGYIN, they do not rule out the possibility of an interaction because the BIGYIN expressed in yeast might be improperly folded or unstable. Although no orthologs of Mdv1p have been identified in mammalian cells, mammals might have a specific factor that is functionally similar to Mdv1p in the same way that ELM1 is specific to plants. Mitochondrial fission in yeast, animals, and plants may have a common origin, as shown by the highly conserved DRPs. However, our results show that mitochondrial fission in Arabidopsis is mediated not only by conserved factors but also by a plant-specific factor.
Figure 10.
Comparison of Mitochondrial Fission Factors in Yeast, a Plant, and an Animal.
In yeast, a dynamin-related protein, Dnm1p, which is a mitochondrial fission executor, is recruited from the cytosol to mitochondria in a manner dependent on Fis1p and an adaptor protein, Mdv1p (and a Mdv1p-homolog, Caf4p). In the plant Arabidopsis, dynamin-related proteins, DRP3A and DRP3B, interact with ELM1 for their localization to mitochondria. Although ELM1 does not have sequence similarity to Mdv1p or Caf4p, its role may be similar to the roles of Mdv1p and Caf4p. In human, localization of dynamin-related protein, Drp1/Dlp1, to mitochondria depends on hFis1. Other proteins, such as Mdv1p, Caf4p, and ELM1, may also be involved. A black bar in Fis1 shows putative transmembrane domains. OM, outer membrane; IM, inner membrane.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) and its transformant with Mt-GFP were used as wild-type plants in this article (Feng et al., 2004). All Arabidopsis plants were grown at 22°C under diurnal 14 h light (100 μmol/m2s). The T-DNA insertion lines SALK_071234 (elm1-6), SALK_066958 (drp3a-3), and SALK_045136 (drp3b) were provided by the ABRC at Ohio State University.
Map-Based Cloning of ELM1
The elm1-1 and drp3a-1 mutations were mapped with molecular markers based on simple sequence polymorphisms (Bell and Ecker, 1994). By rough mapping, elm1-1 and drp3a-1 were mapped to the short arm of chromosome 5 and long arm of chromosome 4, respectively. Because the DRP3A gene was found near the drp3a-1 locus, we sequenced and found mutations in the amplified DNA fragment of DRP3A genes from drp3a-1 and drp3a-2. Further fine-mapping for elm1-1 was performed using the In/Del polymorphisms list on The Arabidopsis Information Resource website (http://www.Arabidopsis.org/Cereon). Using >800 F2 plants generated by crossing elm1-1 with ecotype Landsberg erecta, elm1-1 was mapped in a region of 117 kb, which is on a single BAC clone (MWD9) that includes 41 genes. Of these genes, 26 genes were amplified from the DNA extracted from elm1-1 and wild-type plants and sequenced. At5g22350 (ELM1) was also amplified and sequenced from other elongated mitochondria morphology mutants, and we found other single nucleotide mutations in the same gene from four other mutants (elm1-2 to elm1-5, Figure 2).
DNA Constructs and Plant Transformation
Plant binary vectors based on Gateway cloning technology (Invitrogen) were used for most molecular manipulations. All Arabidopsis DNA fragments described in the results were first amplified by PCR with high-fidelity PCR enzyme (KOD polymerase [TOYOBO] and Phusion polymerase [Finnzymes]) and cloned into pENTR/D-TOPO or pENTR/SD/D-TOPO. All of the cloned plasmids were confirmed by sequencing. Most destination vectors used in this report were provided by the Plant System Biology at Flanders Institute for Biotechnology, Ghent University (Karimi et al., 2002). The DNA fragment ELM1pro:ELM1:GFP includes a genomic fragment from 950 bp upstream of the ATG codon to the termination codon of ELM1 and GFP fused to the terminator sequence of cauliflower mosaic virus 35S. DRP3Apro:DRP3A:GFP contains the promoter region (3025 bp upstream region) of DRP3A, fused to DRP3A-ORF, GFP, and the cauliflower mosaic virus 35S terminator. Oligonucleotide primers used in this work are presented in Supplemental Table 1 online.
Microscopy Observations
Small sections (<5 mm2) were cut out of cotyledon or leaf tissue with a laser blade and were inspected with a confocal laser scanning microscope system (Bio-Rad microradiance with Nikon TE-2000) or a confocal spectral laser scanning microscope (Nikon C1Si). The latter system was only used to generate Figure 9A. In both systems, a 488-nm Ar/Kr laser was used for excitation of GFP and 543-nm He/Ne was used for DsRed and MitoTracker Orange. In the former system, emission signals were detected using a 500- to 530-nm filter for GFP and a 540- to 600-nm filter for DsRed and MitoTracker. The latter filter can detect the signals DsRed and MitoTracker but excludes red autofluorescence from chloroplasts. All images were inspected with a ×100 1.40 numerical aperture oil objective lens (Nikon). All of the data were processed using Adobe Photoshop 7.0 (Adobe Systems).
Mitochondrial Fractionation, Protease Protection Assay, and Immunoblot Analysis
Five grams of seedlings of the three-week-old transformant with ELM1pro:ELM1:GFP was homogenized on ice in 50 mL buffer (150 mM TrisHCl, pH 7.5, 1 mM EDTA, 0.5 M Sucrose, 4 mM cysteine, 0.2% BSA, and 1% DMSO solution of Proteinase Inhibitor Cocktail for plant and tissue extracts [Sigma-Aldrich]). The homogenate was filtered by Miracloth (Calbiochem) and centrifuged at 2000g for 10 min at 4°C, and the supernatant was recentrifuged as before. The second mitochondria-containing supernatant was centrifuged at 14,000g for 15 min at 4°C. The mitochondrial pellet was suspended in buffer without Proteinase Inhibitor Cocktail. Each mitochondrial sample was examined in the presence or absence of 50 μg/mL of Proteinase K (Invitrogen) and in the presence or absence of 1% Triton X-100 (Sigma-Aldrich) for 15 min on ice. Immunoblot analysis was performed essentially as described previously (Nakazono et al., 2000), except that the dilutions of the antibodies were 1:5000 for anti-GFP (MBL) and 1:5000 for anti-rice cytochrome C (Shibasaka et al., 1994).
Y2H Assay
A yeast two-hybrid assay was performed using the ProQuest two-hybrid system (Invitrogen). The ORF of cDNAs listed in Figure 6 was first amplified by RT-PCR with Superscript III (Invitrogen) and high-fidelity PCR enzyme (KOD polymerase [TOYOBO] and Phusion polymerase [Finnzymes]) with total RNA extracted from whole plants of 2-week-old Arabidopsis Col-0 and with primers listed in Supplemental Table 1 online. They were cloned into a gateway entry clone pENTR/SD/D-TOPO by TOPO reaction (Invitrogen). All of the cloned plasmids were confirmed by sequencing. For BIGYIN, the cloned ORF excludes the C-terminal 30 amino acids that code for the transmembrane domain. Each ORF was transferred by LR reaction (Gateway system; Invitrogen) to pDEST 32 for fusing to the GAL4 DNA binding domain or transferred to pDEST 22 for fusing to the GAL4 activation domain. Paired constructs (shown in Figure 6) were introduced into the strain Mav203 of Saccharomyces cerevisiae and selected on SD/-Leu/-Thr (synthetic defined plate deficient for both Leu and Thr) plates. The interactions were examined by X-gal assay (according to the supplier's protocol) and by assessing growth on SD/-Leu/-Thr/-His and SD/-Leu/-Thr/-Ura plates. The weak, strong, and negative control plasmids were provided by the ProQuest Two-Hybrid System (Invitrogen). Interaction between Krev1 and RalGDS was examined as the control of strong interaction (Herrmann et al., 1996). The controls for weak and no interaction were done with the combinations between Krev1 and RalGDS-m1 (weak) or RalGDS-m2 (no interaction) mutants that affect the interaction with Krev1 (Serebriiskii et al., 1999).
BiFC Formation
For BiFC (split YFP) studies, full-length DRP3A, DRP3B, and ELM1 clones were recombined into four vectors that fused each half of YFP to either the N or C terminus of the test protein (Walter et al., 2004). The appropriate destination vectors were kindly provided by T. Nakagawa (Shimane University). Arabidopsis leaf epidermal cells were transformed transiently with each combination of plasmids and with mitochondrial-targeted DsRed by the particle bombardment methods described previously (Arimura et al., 2004b). YFP and DsRed expression were examined 48 h after bombardment by fluorescent microscopy (using a Nikon TE1000 microscope). The signals were captured by the confocal laser scanning unit (Micro radiance; Bio-Rad) attached to the Nikon TE1000 microscope.
ELM1 Gene Induction by Estradiol
The ORF of ELM1 was cloned into the estradiol-inducible Ti vector, pER8 (Zuo et al., 2000), between its XbaI and SpeI sites. The resulting plasmid was introduced into the elm1-6 mutant along with DRP3Apro:DRP3A:GFP. Selection was done with hygromycin (Wako). To induce the ELM1 gene and to visualize mitochondria, small laser-cut leaves of the transformants were incubated in water with 500 μM 17-β estradiol (Wako) and 50 μM MitoTracker Orange (Molecular Probes).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: ELM1 (At5g22350; accession number AB379589), DRP3A (previous name is ADL2a) (At4g33650), DRP3B (previous name is ADL2b) (At2g14120), and BIGYIN (At3g57090).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Two Mutants with Elongated Mitochondria Have Nucleotide Substitutions in the DRP3A Gene.
Supplemental Figure 2. Amino Acid Sequence Alignments of ELM1 and Its Homologs in Other Plants.
Supplemental Figure 3. Mitochondria in elm1-1 Move and Change Their Morphology.
Supplemental Figure 4. Full-Length ORF Transcripts of DRP3A and DRP3B Were Not Detected in T-DNA Insertion Mutants.
-
Supplemental Table 1. Oligonucleotide Primer Sequences.
Supplemental Movie 1. Serial Images of Mitochondria (Labeled with Mt-GFP) in an elm1-1 Mutant Taken by Confocal Laser Scanning Microscopy.
Supplemental Movie 2. Serial Images of Mitochondria (Labeled with Mt-GFP) in a Wild-Type Plant Taken by Confocal Laser Scanning Microscopy.
Supplementary Material
Acknowledgments
We thank M. Karimi (Flanders Institute for Biotechnology, Ghent University, Belgium) and T. Nakagawa (Shimane University, Japan) for their kind donation of Gateway destination vectors and M. Shibasaka (Okayama University, Japan) for his kind donation of antiserum against rice cytochrome C. We also thank the ABRC at Ohio State University for providing the seeds of Arabidopsis T-DNA insertion mutants. We appreciate K. Matsuoka and H. Takanashi for their help during the study. We also thank E.A. Amiott and J.M. Shaw (University of Utah, Salt Lake City, UT) for discussion and comments on this study. This work was supported by Grants-in-Aid for Scientific Research on Priority Area (Grant 18075005) and Scientific Research (A) (Grant 18208002) to N.T. and Young Scientists (Grant 17780002) to S.A. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Nobuhiro Tsutsumi (atsutsu@mail.ecc.u-tokyo.ac.jp).
Online version contains Web-only data.
References
- Arimura, S., Aida, G.P., Fujimoto, M., Nakazono, M., and Tsutsumi, N. (2004. a). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45 236–242. [DOI] [PubMed] [Google Scholar]
- Arimura, S., and Tsutsumi, N. (2002). A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 99 5727–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura, S., Yamamoto, J., Aida, G.P., Nakazono, M., and Tsutsumi, N. (2004. b). Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA 101 7805–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bhar, D., Karren, M.A., Babst, M., and Shaw, J.M. (2006). Dimeric Dnm1-G385D interacts with mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 281 17312–17320. [DOI] [PubMed] [Google Scholar]
- Bleazard, W., McCaffery, J.M., King, E.J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J.M. (1999). The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha-Drori, K., Shichrur, K., Katz, A., Oliva, M., Angelovici, R., Yalovsky, S., and Ohad, N. (2004). Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40 419–427. [DOI] [PubMed] [Google Scholar]
- Cerveny, K.L., McCaffery, J.M., and Jensen, R.E. (2001). Division of mitochondria requires a novel DNM1-interacting protein, net2p. Mol. Biol. Cell 12 309–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D.C. (2006. a). Mitochondria: Dynamic organelles in disease, aging, and development. Cell 125 1241–1252. [DOI] [PubMed] [Google Scholar]
- Chan, D.C. (2006. b). Mitochondrial fusion and fission in mammals. Annu. Rev. Cell Dev. Biol. 22 79–99. [DOI] [PubMed] [Google Scholar]
- Cserzo, M., Wallin, E., Simon, I., vonHeijne, G., and Elofsson, A. (1997). Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: The dense alignment surface method. Protein Eng. 10 673–676. [DOI] [PubMed] [Google Scholar]
- Feng, X.G., Arimura, S., Hirano, H.Y., Sakamoto, W., and Tsutsumi, N. (2004). Isolation of mutants with aberrant mitochondrial morphology from Arabidopsis thaliana. Genes Genet. Syst. 79 301–305. [DOI] [PubMed] [Google Scholar]
- Frank, S., Gaume, B., Bergmann-Leitner, E.S., Leitner, W.W., Robert, E.G., Catez, F., Smith, C.L., and Youle, R.J. (2001). The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell 1 515–525. [DOI] [PubMed] [Google Scholar]
- Griffin, E.E., Graumann, J., and Chan, D.C. (2005). The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann, C., Horn, G., Spaargaren, M., and Wittinghofer, A. (1996). Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J. Biol. Chem. 271 6794–6800. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics 14 378–379. [DOI] [PubMed] [Google Scholar]
- Hong, Z., Bednarek, S.Y., Blumwald, E., Hwang, I., Jurgens, G., Menzel, D., Osteryoung, K.W., Raikhel, N.V., Shinozaki, K., Tsutsumi, N., and Verma, D.P.S. (2003). A unified nomenclature for Arabidopsis dynamin-related large GTPases based on homology and possible functions. Plant Mol. Biol. 53 261–265. [DOI] [PubMed] [Google Scholar]
- Hoppins, S., Lackner, L., and Nunnari, J. (2007). The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76 751–780. [DOI] [PubMed] [Google Scholar]
- Ingerman, E., Perkins, E.M., Marino, M., Mears, J.A., McCaffery, J.M., Hinshaw, J.E., and Nunnari, J. (2005). Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagasia, R., Grote, P., Westermann, B., and Conradt, B. (2005). DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C-elegans. Nature 433 754–760. [DOI] [PubMed] [Google Scholar]
- Karimi, M., Inze, D., and Depicker, A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 193–195. [DOI] [PubMed] [Google Scholar]
- Karren, M.A., Coonrod, E.M., Anderson, T.K., and Shaw, J.M. (2005). The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J. Cell Biol. 171 291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch, A., Thiemann, M., Grabenbauer, M., Yoon, Y., McNiven, M.A., and Schrader, M. (2003). Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278 8597–8605. [DOI] [PubMed] [Google Scholar]
- Kuravi, K., Nagotu, S., Krikken, A.M., Sjollema, K., Deckers, M., Erdmann, R., Veenhuis, M., and van der Klei, I.J. (2006). Dynamin-related proteins Vps1p and Dnm1p control peroxisome abundance in Saccharomyces cerevisiae. J. Cell Sci. 119 3994–4001. [DOI] [PubMed] [Google Scholar]
- Li, Z., Okamoto, K., Hayashi, Y., and Sheng, M. (2004). The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119 873–887. [DOI] [PubMed] [Google Scholar]
- Logan, D.C., Scott, I., and Tobin, A.K. (2004). ADL2a, like ADL2b, is involved in the control of higher plant mitochondrial morphology. J. Exp. Bot. 55 783–785. [DOI] [PubMed] [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano, S., Nakamori, C., Kondo, M., Hayashi, M., and Nishimura, M. (2004). An Arabidopsis dynamin-related protein, DRP3A, controls both peroxisomal and mitochondrial division. Plant J. 38 487–498. [DOI] [PubMed] [Google Scholar]
- Mozdy, A.D., McCaffery, J.M., and Shaw, J.M. (2000). Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazono, M., Tsuji, H., Li, Y.H., Saisho, D., Arimura, S., Tsutsumi, N., and Hirai, A. (2000). Expression of a gene encoding mitochondrial aldehyde dehydrogenase in rice increases under submerged conditions. Plant Physiol. 124 587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor, K., Ingerman, E., Okreglak, V., Marino, M., Hinshaw, J.E., and Jodi, N. (2006). Mdv1 interacts with assembled Dnm1 to promote mitochondrial division. J. Biol. Chem. 281 2177–2183. [DOI] [PubMed] [Google Scholar]
- Noguchi, K., and Yoshida, K. (2008). Interaction between photosynthesis and respiration in illuminated leaves. Mitochondrion 8 87–99. [DOI] [PubMed] [Google Scholar]
- Okamoto, K., and Shaw, J.M. (2005). Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39 503–536. [DOI] [PubMed] [Google Scholar]
- Raghavendra, A.S., and Padmasree, K. (2003). Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8 546–553. [DOI] [PubMed] [Google Scholar]
- Scheckhuber, C.Q., Erjavec, N., Tinazli, A., Hamann, A., Nystrom, T., and Osiewacz, H.D. (2007). Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 9 99–105. [DOI] [PubMed] [Google Scholar]
- Scott, I., Tobin, A.K., and Logan, D.C. (2006). BIGYIN, an orthologue of human and yeast FIS1 genes functions in the control of mitochondrial size and number in Arabidopsis thaliana. J. Exp. Bot. 57 1275–1280. [DOI] [PubMed] [Google Scholar]
- Serebriiskii, I., Khazak, V., and Golemis, E.A. (1999). A two-hybrid dual bait system to discriminate specificity of protein interactions. J. Biol. Chem. 274 17080–17087. [DOI] [PubMed] [Google Scholar]
- Sesaki, H., and Jensen, R.E. (1999). Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaka, M., Ushimaru, T., Ookubo, K., Tsuchida, S., and Tsuji, H. (1994). Increase in cytochrome c and a 11.9 kDa protein in submerged rice seedlings after exposure to air. Bull. RIB Okayama Univ. 2 149–157. [Google Scholar]
- Tieu, Q., and Nunnari, J. (2000). Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002). The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J. Cell Biol. 158 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M., Chaban, C., Schutze, K., Batistic, O., Weckermann, K., Nake, C., Blazevic, D., Grefen, C., Schumacher, K., Oecking, C., Harter, K., and Kudla, J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40 428–438. [DOI] [PubMed] [Google Scholar]
- Waterham, H.R., Koster, J., van Roermund, C.W.T., Mooyer, P.A.W., Wanders, R.J.A., and Leonard, J.V. (2007). A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356 1736–1741. [DOI] [PubMed] [Google Scholar]
- Yoon, Y., Pitts, K.R., and McNiven, M.A. (2001). Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12 2894–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J.R., Niu, Q.W., and Chua, N.H. (2000). An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.