Abstract
Polyamines (PAs) exert a protective effect against stress challenges, but their molecular role in this remains speculative. In order to detect the signaling role of apoplastic PA-derived hydrogen peroxide (H2O2) under abiotic stress, we developed a series of tobacco (Nicotiana tabacum cv Xanthi) transgenic plants overexpressing or downregulating apoplastic polyamine oxidase (PAO; S-pao and A-pao plants, respectively) or downregulating S-adenosyl-l-methionine decarboxylase (samdc plants). Upon salt stress, plants secreted spermidine (Spd) into the apoplast, where it was oxidized by the apoplastic PAO, generating H2O2. A-pao plants accumulated less H2O2 and exhibited less programmed cell death (PCD) than did wild-type plants, in contrast with S-pao and samdc downregulating plants. Induction of either stress-responsive genes or PCD was dependent on the level of Spd-derived apoplastic H2O2. Thus, in wild-type and A-pao plants, stress-responsive genes were efficiently induced, although in the latter at a lower rate, while S-pao plants, with higher H2O2 levels, failed to accumulate stress-responsive mRNAs, inducing PCD instead. Furthermore, decreasing intracellular PAs, while keeping normal apoplastic Spd oxidation, as in samdc downregulating transgenic plants, caused enhanced salinity-induced PCD. These results reveal that salinity induces the exodus of Spd into the apoplast, where it is catabolized by PAO, producing H2O2. The accumulated H2O2 results in the induction of either tolerance responses or PCD, depending also on the levels of intracellular PAs.
INTRODUCTION
Plant polyamine (PA) metabolism is highly modulated by endogenous cues and environmental factors, especially those imposing stress (Bouchereau et al., 1999). Salt stress in many plant species has been linked to readjustment of PA titers, mostly by changing the levels of the diamine putrescine (Put), the triamine spermidine (Spd), and the tetramine spermine (Spm), through activation of PA biosynthetic genes. Arginine decarboxylase (ADC; EC 4.1.1.19) was vital for Arabidopsis thaliana adaptation during salt stress (Kasinathan and Wingler, 2004), and ADC activity increased in salt-tolerant Oryza sativa cultivars during prolonged salt stress (Chattopadhyay et al., 1997). S-Adenosyl-l-methionine decarboxylase (SAMDC; EC 4.1.1.50), spermidine synthase (SPDS; EC 2.5.1.16), and spermine synthase (SPMS; EC 2.5.1.22), enzymes that synthesize higher PAs, have also been implicated in salt tolerance (Kasukabe et al., 2004). Transgenic plants overexpressing genes encoding PA-synthesizing enzymes, such as adc (Capell et al., 2004), odc (Kumria and Rajam, 2002), samdc (Wi et al., 2006), and spds (Kasukabe et al., 2004), exhibited increased stress tolerance; on the contrary, decreases in ADC (Kasinathan and Wingler, 2004) or SPMS (Yamaguchi et al., 2006), through the use of antisense technology and mutants, exerted opposite effects. What is more interesting is that in plants overexpressing spds, a subset of stress-inducible genes was modulated (Kasukabe et al., 2004). Recently, overexpression of stress-related transcription factors, such as Hahb-4 and Ca PF1, led to tolerance, through their ability to retain PA homeostasis (Manavella et al., 2006; Tang et al., 2007; Agalou et al., 2008). However, most of the previous studies did not give any conclusive evidence for an underlying mechanism by which PAs exert their tolerance effects.
A nodal point in the reaction of plant cells to stress is reactive oxygen species (ROS) generation. The identification of ROS-generating enzymes, such as the plant homolog of respiratory burst NADPH oxidase, revealed that plant cells, like their mammalian counterparts, can initiate and amplify ROS production for signaling purposes. In addition, apoplastic ROS generation can be mediated by the cell wall oxidase peroxidase, amine oxidases, and oxalate oxidase. The apoplastic flavoprotein polyamine oxidase (PAO; EC 1.5.3.3), which oxidizes Spd and Spm, yields Δ1-pyrroline and 1,5-diazabicyclononane, respectively, along with 1,3-diaminopropane (1,3-Dap) and hydrogen peroxide (H2O2). Overexpression of the maize (Zea mays) pao gene in tobacco (Nicotiana tabacum) plants, and the associated decrease in PAs, resulted in H2O2 generation, which then induced the antioxidant genes necessary for cell survival/adaptation (Rea et al., 2004; Moschou et al., 2008). However, despite the enhanced antioxidant machinery, further increase in the intracellular ROS, by directly providing H2O2 or prooxidants such as methylviologen or menadione in the transgenics, resulted in an oxidative burst that transgenics were unable to cope with. The detrimental effect of the oxidative burst was due to the inability of transgenic cells to further respond, whereas induction of antioxidant enzymes in the wild type was evident soon after treatment. Thus, although the higher levels of H2O2 generated by the overexpression of PAO in the transgenic plants were successfully regulated by the concomitant activation of the antioxidant machinery, further increase of ROS was detrimental to cellular functions and induced programmed cell death (PCD) (Moschou et al., 2008).
In a recent study, Yoda et al. (2006) identified PAO as one of the key elements in the execution of the PCD that takes place during the hypersensitive reaction (HR), but the role of PAO during abiotic stress is largely unknown. In addition, levels of higher PAs (i.e., Spd and Spm) are tightly regulated, and PAO is considered to contribute to this. Moreover, increased diamine oxidase (DAO; EC 1.4.3.6) activity found in transgenic tobacco plants overexpressing samdc resulted in tolerance to salinity (Waie and Rajam, 2003). However, it is not clear whether the enhanced tolerance is due to increased PA catabolism or increased PA titers (the latter being the main characteristic caused by the overexpression of a biosynthetic enzyme).
To unravel the possible contribution of apoplastic PAs to the signaling of tolerance responses, we have developed and used transgenic tobacco plants with modified PAO levels, through sense (Moschou et al., 2008) and antisense (this work) technology. The results presented here show that upon abiotic stress, Spd is secreted into the apoplast and catabolized by PAO to produce H2O2, which acts as a second messenger either to trigger additional cell-protective stress responses or to induce PCD in a dose-responsive manner. To further establish the potential role of the ratio of PA anabolism to catabolism in plant tolerance, we developed and used transgenic tobacco plants with downregulated SAMDC via RNA interference (RNAi). Results obtained with these transgenic tobacco plants showed that lowering the PA anabolism-to-catabolism ratio is sufficient to promote PCD under abiotic stress, reinforcing the view that the regulation of PA catabolism in the apoplast is a key factor that exerts a specific role in abiotic stress responses.
RESULTS
Production and Screening of Antisense pao Tobacco Transgenic Lines: Downregulation of the pao Gene Results in Decreased PAO Protein and PAO Activity
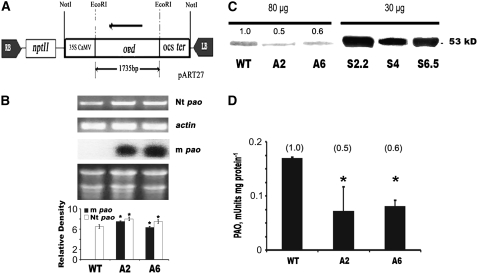
We have already developed transgenic tobacco (cv Xanthi) plants carrying the full-length maize pao (m pao) cDNA in the sense orientation (S-pao), under the control of the 35S promoter (Moschou et al., 2008). In this work, we have generated plants expressing antisense pao (A-pao) cDNA (Figure 1A). All eight regenerated transgenic lines exhibited decreased PAO levels (Figure 1; see Supplemental Figure 1 online). The pao antisense and endogenous transcript levels were assessed by RNA gel blotting and RT-PCR, respectively. The m pao and tobacco pao (Nt pao) cDNAs shared 77.5% sequence similarity over 0.3 kb in their C termini, whereas in the remaining part of the cDNAs sequence similarity was lower (60%). Interestingly, the levels of both the antisense and endogenous pao transcripts were higher than in the wild type (Figure 1B; m pao and Nt pao, respectively). This was correlated with low PAO protein levels (Figure 1C; see Supplemental Figure 1 online) and reduced enzymatic activities (Figure 1D; see Supplemental Figure 1 online). The reduction in PAO protein levels in the A2 and A6 lines (50 and 60%, respectively), correlated well with reduced PAO activity, although the Nt pao transcript levels increased, suggesting that the low endogenous PAO levels are the result of translational inhibition rather than posttranscriptional inactivation, as suggested previously by Purnell et al. (2005) (Figure 1).
Figure 1.
A-pao Generation and Analysis of PAO mRNA, Protein, and Activity Levels.
(A) Construct used for the production of stable A-pao lines. RB, right border; nptII, neomycin phosphotransferase II; 35S CaMV, 35S promoter from the Cauliflower mosaic virus; pao, m pao cDNA expressed in the antisense direction; ocs ter, octopine synthase transcription terminator; LB, left border.
(B) RT-PCR (top) showing endogenous pao (Nt pao) and actin mRNA levels and RNA gel blot analysis of the antisense m pao transcript (middle, with gel showing equal loading below). Densitometric analysis (bottom) is shown for both m pao and Nt pao. In both cases, mRNAs were equalized against actin.
(C) PAO protein levels in wild-type, A2, and A6 antisense lines (80 μg total protein/well). Numbers above the bands correspond to the relative intensities of the signals compared with wild-type plants. For comparison, a protein gel blot of PAO from the sense lines S2.2, S4, and S6.5 (Moschou et al., 2008) is shown (30 μg total protein/well).
(D) PAO activity levels in wild-type, A2, and A6 antisense lines (milliunits/mg protein). The numbers above the bars represent relative activities of the lines with respect to wild-type plants. Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
A-pao Plants Exhibit Increased Soluble Spd Levels
PA titers in all fractions, namely soluble (S), soluble hydrolyzed (SH), and pellet hydrolyzed (PH), were determined in A-pao plants (see Supplemental Figure 1 online). S-Spd was 2-fold greater in line A2 and slightly higher in A6 than in wild-type plants (Figure 2). SH and PH levels did not change significantly (Figure 2). To the contrary, S-pao transgenic tobacco plants contained significantly lower S-PA and SH-PA levels (Figure 2) (Moschou et al., 2008).
Figure 2.
S-, SH-, and PH-PA Titers in Wild-Type, A2, A6, S2.2, S4, and S6.5 Transgenic Lines.
PAs are expressed in nanomoles per gram fresh weight (FW). Closed bars, Put; open bars, Spd; striped bars, Spm. Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
PAO Affects Seed Germinability, Biomass Production, and Tolerance to Salinity
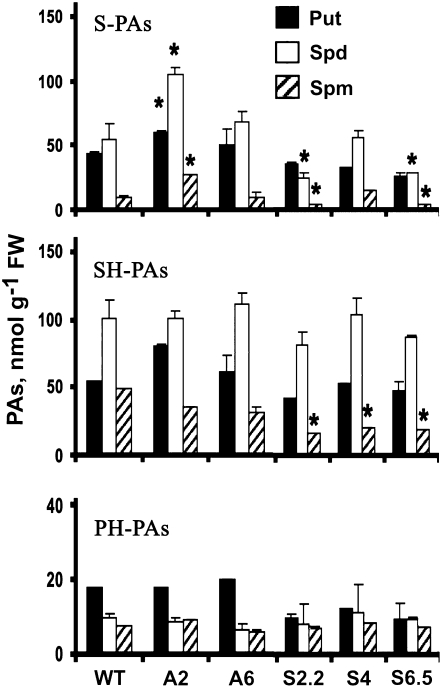
Transgenic tobacco A-pao and S-pao plants were tested to examine whether deregulation of PAO affects physiological characteristics, such as seed germinability, biomass production, and tolerance to salinity. Transgenic T2 seeds were grown in vitro on Murashige and Skoog (MS) medium supplemented with NaCl. A2 seeds germinated faster (Figure 3A; days 7 and 9) and showed increased viability, mostly at 300 mM NaCl, at which level a 2-fold increase in seed germination was observed on day 14. To the contrary, S2.2 seeds exhibited a profound decrease in germination, reaching a 2.6-fold decrease at 300 mM (Figure 3A). Although a 100% germination of wild-type, A2, and S2.2 seeds was achieved by day 14 at 200 mM, the rate of germination differed significantly (Figure 3A). Furthermore, A2 plants grown in vitro on MS medium supplemented with NaCl showed increased biomass production of ∼40% at 100 mM and 17% at 200 mM (Figure 3B). At 300 mM NaCl, biomass increased only slightly. The same trend was observed for the A6 and A8 lines (see Supplemental Figure 2A online). Conversely, S2.2 plants grown under the same conditions produced ∼20% less biomass than the wild type (Figure 3B).
Figure 3.
Wild-Type, S2.2, and A2 Transgenic Line Phenotypes under Salinity Stress.
(A) Seed viability in wild-type, A2, and S2.2 plants in MS medium supplemented with 300 mM NaCl (top), and rate of seed germination at 200 and 300 mM NaCl (bottom).
(B) Plant growth phenotype of wild-type, A2, and S2.2 transgenic lines treated with 200 mM NaCl (top), and fresh weight of wild-type, A2, and S2.2 transgenic lines treated with 0, 100, 200, and 300 mM NaCl (bottom). Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
Salinity Induces the Exodus of PAs into the Apoplast, Where They Are Catabolized by PAO
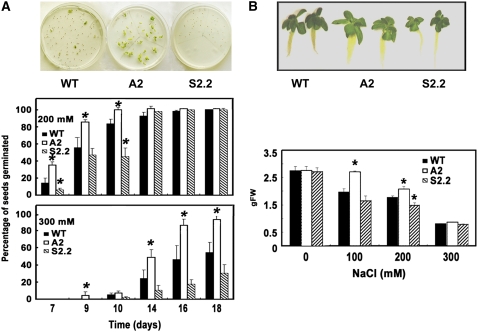
Recently, Yoda et al. (2003, 2006) showed that the HR-like response to biotic stress induces Spd secretion into the apoplast. We examined whether a similar phenomenon takes place under abiotic stress. Thus, apoplastic fluids from hydroponically grown wild-type, A2, and S2.2 plants were extracted and PAs were measured after confirming the purity of the apoplastic fluids by the malate dehydrogenase and glucose-6-phosphate dehydrogenase assay (Figure 4A). The most abundant PA in the apoplastic compartment was Spd (2.5 and 6 μM for the wild type and A2, respectively); Spd could not be detected in the apoplast of S2.2 plants (Figure 4A). Upon salinity stress, Spd gradually increased (12 ± 0.7 and 15 ± 0.9 μM in the wild type and A2, respectively), whereas it accumulated to only 3 μM in S2.2 plants (Figure 4A). To the contrary, high amounts of 1,3-Dap accumulated in the apoplastic compartment of S2.2 plants (15 ± 1.0 μM), whereas at the same time, 1,3-Dap was threefold and fivefold lower in the wild type and A2, respectively, compared with S2.2 (Figure 4A).
Figure 4.
Apoplastic S-Spd and 1,3-Dap Titers, ROS Epifluorescence, and Localization in Salt-Treated Cell Suspensions and Plants.
(A) Apoplastic S-Spd and 1,3-Dap levels in wild-type, A2, and S2.2 transgenic lines at 72 h after treatment with 0 (−) and 200 mM (+) NaCl. RLU, relative units.
(B) ROS epifluorescence in cell suspension cultures from wild-type, A2, and S2.2 transgenic lines at 24 h after treatment with 0 or 200 mM NaCl using the highly specific ROS probe DCFMA (top), and relative ROS fluorescence as measured by pixel analysis (bottom).
(C) Quantitative in situ ROS localization in the apoplast of wild-type, A2, and S2.2 transgenic lines using transmission electron microscopy and cerium chloride, which precipitates in the presence of H2O2, forming black adducts. Plants were treated with 0 or 200 mM NaCl, and bars indicate 0.4 μm. Ap, apoplast; Cl, chloroplast; CT, cytoplasm; Vc, vacuole.
Data in (A) and (B) are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05. Data in (C) represent a single representative experiment replicated three times.
PAO Affects ROS Generation and Accumulation
Since salinity induces the generation of ROS (Xiong et al., 2002; Skopelitis et al., 2006), it was of interest to test how PAO alteration would affect ROS levels in A-pao and S-pao transgenic suspension cultures in the presence of NaCl. Although no significant differences could be observed among wild-type, A2, and S2.2 cells under control conditions (Figure 4B), in the presence of 200 mM NaCl, the A2 cells accumulated less ROS than the S2.2 cells (Figure 4B). Interestingly, ROS levels (suspension cells; Figure 4B) positively correlated with 1,3-Dap titers, as they were dramatically higher in S2.2 plants (Figure 4A). This suggests that the source of ROS accumulation is Spd oxidation.
To reinforce the validity of these results in planta, in situ detection of H2O2 by transmission electron microscopy was also performed (Figure 4C). The results confirmed the presence of H2O2 detected as precipitates of electron-dense cerium perhydroxides, localized predominantly within the cell walls and in the apoplast (Figure 4C). The formation of cerium perhydroxides, in analogy to in situ H2O2 localization, was remarkably greater in S2.2 cells compared with wild-type cells, in contrast with A2 plants (Figure 4C). These results present clear evidence that apoplastic PAO activity contributes to H2O2 generation under salt stress.
H2O2 Produced by PAO in the Apoplast Participates in the Induction of the PCD Syndrome during Abiotic Stress
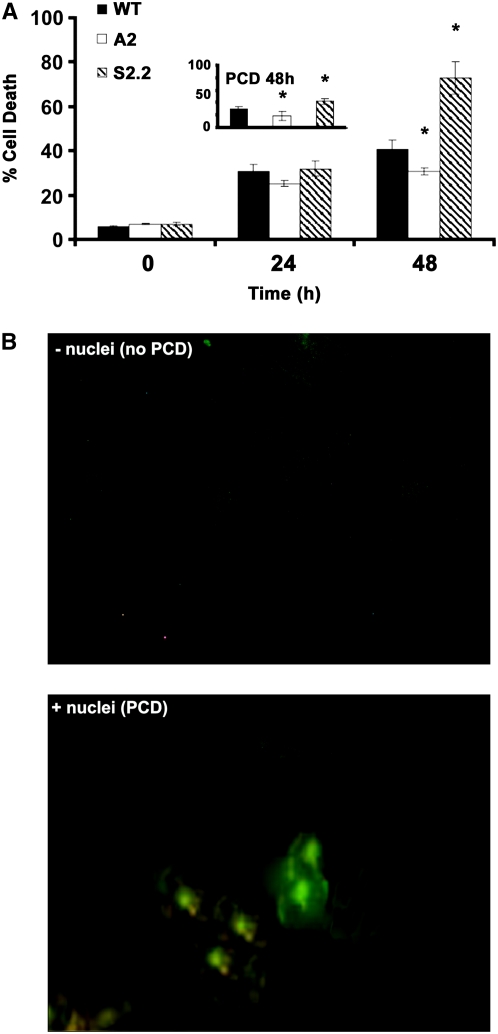
To test whether the stress-induced secretion of cellular Spd in the apoplast produces sufficient ROS to induce PCD under salt stress, cells from the wild type, A-pao, and S-pao were assessed. The S2.2 cells reached a 35% greater rate of cell death compared with wild-type cells, in contrast with A2 cells (Figure 5A). The observed cell death was indeed PCD, as indicated by the percentage of cells positive for the TdT-mediated dUTP nick-end labeling (TUNEL) reaction, with corresponding values of ∼60 and 75% for the wild type and S2.2, respectively (Figure 5). These results suggest that, under salt stress conditions, the transgenic S-pao was significantly more susceptible than the wild type, in contrast with A-pao cells.
Figure 5.
Cell Death Rate and PCD in Cell Suspension Cultures Exposed to Salinity.
(A) Rates of cell death measured using Evans blue or PCD at 48 h (inset) after treatment with 200 mM NaCl in wild-type, A2, and S2.2 transgenic plants. Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
(B) PCD observed in wild-type cells (−NaCl, top panel; +NaCl, bottom panel) using the TUNEL assay, whereby nuclei are observed only in cells undergoing PCD treated with 200 mM NaCl (bottom panel).
Apoplastic PA-Derived ROS Affect Defense Responses in a Dose-Dependent Manner
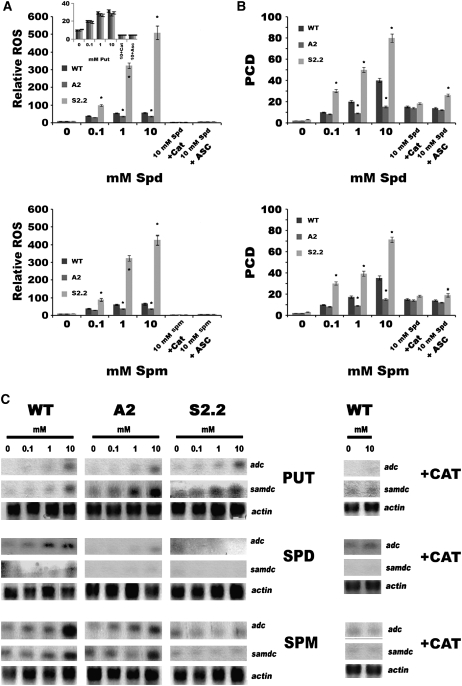
To further confirm that Spd (or Spm) catabolism is the source of the observed ROS, we supplied cell suspension cultures with exogenous Put, Spd, and Spm. The ROS levels and PCD rates were estimated at 24 h after treatment (Figure 6A). Spd oxidation showed a positive dose-dependent relationship to ROS production (Figure 6A). A2 plants accumulated ∼30% less ROS than wild-type plants, whereas S2.2 plants accumulated 3- to 10-fold higher ROS levels (Figure 6A; 0.1 and 10 mM Spd, respectively). As expected, Put-derived ROS were at the same levels in the three genotypes (Figure 6A, inset). Interestingly, ROS promoted PCD depending on their concentration (Figure 6B), while the addition of ascorbate or catalase prevented this effect (Figures 6A and 6B). Moreover, it is worth noting that ROS levels in S2.2 plants were significantly higher than in wild-type plants even at 0.1 mM Spd. The same pattern was observed when using Spm as substrate (Figures 6A and 6B).
Figure 6.
ROS Levels and PCD in Cell Suspension Cultures and Expression Patterns of adc and samdc in Wild-Type, A2, and S2.2 Transgenic Lines after Treatment with 0.1, 1, and 10 mM Put, Spd, and Spm.
(A) Epifluorescent relative ROS quantification in cell suspensions treated with 0.1, 1, and 10 mM Spd and Put (inset) at 1 h after treatment using the highly specific ROS probe DCFMA.
(B) Percentage of cells undergoing PCD in cell suspensions at 24 h after treatment as measured using the TUNEL assay, whereby positive nuclei are observed only in cells undergoing PCD.
(C) RNA gel blot analysis of adc and samdc in fully developed leaves at 6 h after treatment. Equal loading against actin is shown, and simultaneous addition of catalase with the corresponding PA represents the negative control.
Data in (A) and (B) are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05. Data in (C) represent a single representative experiment replicated twice. In all cases, treatments with catalase (CAT) and ascorbate (ASC) represent negative controls.
Furthermore, we attempted to mimic the apoplastic PAO-dependent ROS generation machinery in planta by providing the apoplast with exogenous PAs and measuring the effect of the produced ROS on the transcription of two well-known stress-responsive genes, adc and samdc. Thus, Put, Spd, and Spm were infiltrated in leaves of wild-type, A2, and S2.2 plants and the induction of the two transcripts was estimated at 6 h after treatment (Figure 6C). In wild-type plants, all three PAs showed a dose-dependent inductive effect on adc and samdc mRNA accumulation (Figure 6C), with the most striking increase exerted by Spm (Figure 6C, Spm). Put caused a moderate increase of the corresponding mRNAs and a significant induction only at 10 mM (Figure 6, Put). In A2 plants, the same trend of increase was found, with differences in the induction levels, consistent with the ROS-dependent induction of these genes (Figure 6C, A2). Interestingly, in S2.2 plants, only Put exerted induction of the corresponding mRNAs, whereas Spd and Spm exerted an opposite effect (Figure 6C, S2.2). Moreover, simultaneous infiltration of catalase with the corresponding PAs inhibited induction of the two genes, suggesting ROS dependence of the effect (Figure 6C).
Transcript Levels of PA Biosynthetic Genes Are PAO Dependent
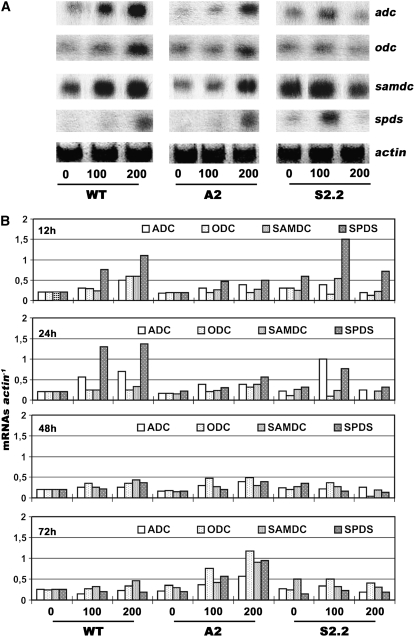
Since PAO catabolizes Spd and Spm, producing ROS, we examined whether this could trigger PA biosynthetic gene expression to reestablish PA homeostasis and to retain a stable PA-to-ROS ratio. Under control conditions, downregulation of PAO activity reduced only slightly the adc, samdc, and spds mRNA abundance (Figure 7). More specifically, in the A2 line, an ∼20% decrease in adc, samdc, and spds transcripts was found (Figures 7A and 7B), whereas odc transcript abundance remained stable and in some cases rather increased (Figure 7B; e.g., 72 h). On the other hand, a 40% increase in adc, a 50% increase in samdc, and a twofold increase in spds mRNA levels were found in S2.2 plants. These data suggest that the overall PA biosynthetic machinery is under feedback regulation by PAs, probably through a H2O2 signal.
Figure 7.
Expression Patterns of adc, odc, samdc, and spds in Wild-Type, A2, and S2.2 Transgenic Plants under Control and Salinity Conditions using 100 and 200 mM NaCl.
(A) RNA gel blot analysis of the expression of adc, odc, samdc, and spds genes at 12 h after the onset of salt stress.
(B) mRNA level quantification (mRNA/actin) at 12, 24, 48, and 72 h after treatment. Data represent a single representative experiment replicated three times.
When wild-type and transgenic tobacco plants were salt-treated, mRNAs of all PA biosynthetic genes were readjusted a few hours after treatment, depending on the pao levels and the stress magnitude. The most dramatic increase was that of the spds transcript, exhibiting a fivefold increase in wild-type plants (200 mM NaCl) in the first few hours after treatment, a continuing increase up to 24 h, and a decrease thereafter (Figure 7B). Moreover, the abundance of odc and samdc transcripts increased profoundly at 12 h, declined thereafter, but remained higher than basal levels. On the contrary, in A2, the abundance of transcripts was lower and the transcripts increased constantly (rather than following a burst-type pattern, as in the wild type) (Figure 7B). These results suggest that the perception of the stress signal in A-pao lines is slower and that the controlled ROS accumulation could stimulate the induction of the above mRNAs, as described in the previous section. In S2.2 plants, the burst of spds mRNA accumulation was higher compared with that in the wild type at 12 h after treatment, but when exposed to 200 mM NaCl, there was a failure in the overall PA biosynthetic transcript accumulation, similar to that observed when higher PAs were exogenously supplied to these plants (Figures 6C and 7B). Even at 100 mM, a higher rate of transcript decline was evident in S2.2 plants compared with wild-type plants.
Levels of PA Biosynthetic Enzymatic Proteins Do Not Necessarily Follow the Transcription Patterns
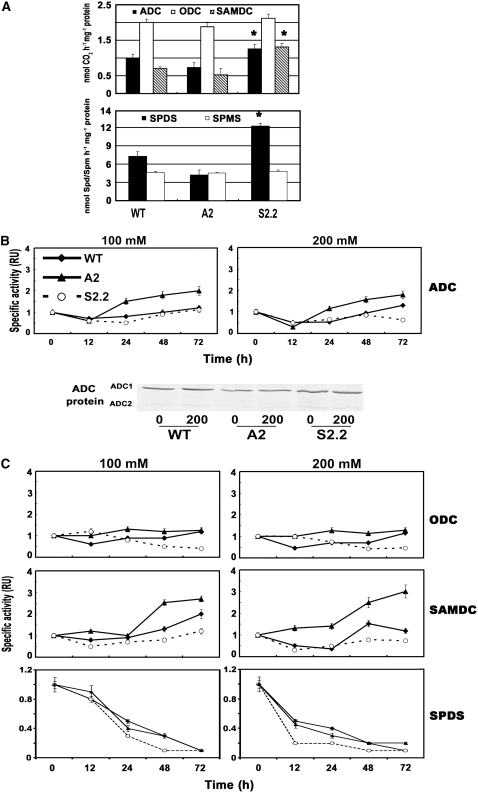
We then explored the mode of regulation of biosynthetic PA genes, since the differential accumulation of ROS could have different effects in their respective protein synthesis and/or activity. Under control conditions, the patterns of PA biosynthetic enzymatic activities mirrored the corresponding transcript levels (Figure 8A). More specifically, ADC activity was increased by 25% in S2.2 plants and decreased by 25% in A2 plants (Figure 8A, top). The same trend was observed for ADC protein (Figure 8B). ODC activity levels, on the other hand, did not exhibit any significant differences (Figure 8A). The activity of SAMDC showed a twofold increase in S2.2 and a 20% decrease in A2 compared with wild-type plants (Figure 8A). SPDS showed a 70% increase in S2.2 and a 75% decrease in A2, whereas SPMS was only slightly affected in the transgenic plants.
Figure 8.
ADC, ODC, SAMDC, SPDS, and SPMS Specific Activities and ADC Protein Levels in Control and Salinity Conditions of Wild-Type, A2, and S2.2 Transgenic Plants.
(A) Specific activities in control plants of ADC, ODC, and SAMDC (nmol CO2·h−1·mg−1 protein), SPDS (nmol Spd·h−1·mg−1 protein), and SPMS (nmol Spm·h−1·mg−1 protein).
(B) Time course of ADC specific activity, at 100 and 200 mM NaCl, and ADC (cytosolic [ADC1] and chloroplastic [ADC2]) protein levels in 0 and 200 mM NaCl at 72 h after the onset of stress.
(C) Time course of ODC, SAMDC, and SPDS specific activity at 100 and 200 mM NaCl.
All data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
Under stress conditions, the transcript levels did not always correspond to activity levels, at least early after treatment (Figures 8B and 8C). A 30% increase in ADC activity levels was found at 24 to 48 h after treatment, depending also on the magnitude of stress, following an initial decrease (Figure 8B). Although the same pattern of increase was observed in the A2 line, it was higher and more profound, whereas ADC in the S2.2 line rather declined (except for a minor increase at 100 mM after 24 h). Although ADC mRNA levels increased rapidly after treatment, the corresponding activity started to increase after an initial decline (Figure 8B). In accordance with the previous results, ADC protein (both ADC1 [cytosolic] and ADC2 [chloroplastic]) accumulated only in wild-type and A2 stressed plants (Figure 8B).
ODC specific activity showed the same initial temporal decrease in wild-type plants and returned to basal levels after ∼72 h (Figure 8C). In S2.2 plants, a profound decrease in activity levels of ODC was found, whereas no significant differences were found in A2 plants in comparison with the respective untreated plants (Figure 8C). Thus, the slight increase in transcript levels led to only a similar increase (∼10%) in activity levels (Figures 7B and 8C).
SAMDC specific activity showed a twofold increase with 100 mM NaCl at 72 h after treatment and a 50% increase with 200 mM at 48 h after treatment in wild-type plants (Figure 8C). Interestingly, SAMDC activity increased more sharply in A2 plants, whereas no decrease in activity levels, like those observed at initial time points with 200 mM for the wild type, were observed (Figure 8C). On the other hand, no increase in SAMDC activity levels was found in S2.2 plants (Figure 8C).
SPDS specific activity constantly decreased and was almost nullified at 72 h after treatment, irrespective of the genotype examined (Figure 8C). Nevertheless, SPDS activity decreased more sharply with 200 mM in S2.2 compared with the wild type, whereas the opposite was true for the A2 line (Figure 8C). SPMS, on the other hand, could not be detected after treatment in any of the genotypes examined (see Supplemental Figure 3 online).
pao Is an Abiotic Stress-Inducible Gene, Although the Total PAO Activity Remains Constant
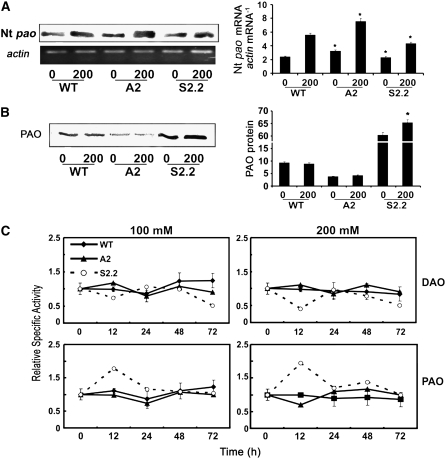
Endogenous pao transcript levels increased under salt stress conditions, exhibiting an approximately twofold increase in wild-type, A2, and S2.2 plants (Figure 9A). Although pao mRNA accumulated, PAO protein and activity in the wild type and A2 remained fairly constant during stress, irrespective of the salt concentration, whereas PAO protein and activity increased in S2.2 (Figures 9B and 9C). Although DAO activity did not significantly change in wild-type and A2 plants, it decreased in S2.2, mostly with 200 mM at 12 h after treatment (Figure 9C).
Figure 9.
Levels of PAO mRNA, Protein, and Activity and DAO Activity in Wild-Type, A2, and S2.2 Transgenic Plants under Control and Salinity Conditions.
(A) RT-PCR for the endogenous pao mRNA levels, product hybridization against an Nt pao 32P-labeled probe, and densitometric analysis (right) in wild-type, A2, and S2.2 lines treated with 0 or 200 mM NaCl, normalized to actin.
(B) PAO protein levels and densitometric analysis (right) in wild-type, A2, and S2.2 lines treated with 0 or 200 mM NaCl.
(C) DAO and PAO activity levels over time in wild-type, A2, and S2.2 lines treated with the indicated levels of NaCl.
Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
Salinity Induces Significantly Higher S-PA Accumulation in A-pao Plants
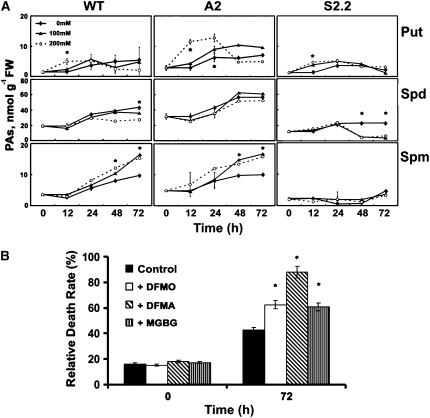
To assess possible differential accumulation of PAs in transgenic plants under stress, PA titers were estimated and compared with the corresponding biosynthetic and catabolic enzymatic activities (Figure 10). Soon after the onset of stress, Put increased in all transgenic lines and in wild-type plants (Figure 10A). This increase was more profound in A2 plants, exhibiting an up to fourfold transient increase (Figure 10A, 24 h) followed by a decrease (Figure 10A), in accordance with the higher ADC activity in these plants (Figure 8B). On the other hand, Spd rather decreased in all genotypes, mainly at 24 h after treatment, except for a slight increase in A2 plants with 100 mM at 48 h after treatment (Figure 10A). Spd was more profoundly decreased in S2.2 plants, consistent with both the lower overall biosynthetic activities and the higher catabolic activities (Figures 9 and 10A). Moreover, the lack of increase in Spd titers could be due to its redistribution to the apoplast, where it is catabolized. In contrast with the Spd pattern, Spm significantly accumulated, mostly at 48 h after treatment (Figure 10A). In A2 plants, Spm accumulation was higher compared with that in wild-type plants, whereas in S2.2, no significant increase was found and it decreased at 200 mM (Figure 10A). These results are in accordance with the overall biosynthetic and catabolic capacity of the plants examined (Figures 7 to 9 and 10A) and reinforce the view that PAO levels affect both PA biosynthesis and PA levels. Moreover, additional A-pao lines exhibited the same pattern of PA increase (see Supplemental 2B online).
Figure 10.
S-Put, S-Spd, and S-Spm Accumulation and Effect of PA Biosynthesis Inhibitors on Cell Death Rate.
(A) S-PA accumulation over time in wild-type, A2, and S2.2 hydroponically grown transgenic plants supplemented with 0, 100, and 200 mM NaCl.
(B) Effect of the ADC, ODC, and SAMDC inhibitors DFMA, DFMO, and MGBG, respectively, on cell death rate of wild-type hydroponically grown plants supplemented with 200 mM NaCl.
Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
The increased tolerance of A-pao plants could be exerted through their increased PA levels or, alternatively, through the decreased ROS levels generated via the apoplastic oxidative PA deamination. Although PAO-overexpressing plants accumulate higher ROS levels under normal growth conditions, these ROS are sufficient to induce the increase of the overall antioxidant machinery and to sustain ROS homeostasis (Moschou et al., 2008). PA depletion, on the other hand, could also affect tolerance to abiotic stress. In an attempt to exclude the ROS produced through PAO action and to concentrate on the intracellular PA content, we performed experiments with a relatively high supply of exogenous H2O2 to minimize the effect of endogenous ROS levels. The antioxidant machinery of S-pao plants, although enhanced in control conditions, failed to respond to this external stimulus, suggesting that intracellular PAs are crucial in inhibiting PCD and in permitting cell survival (Moschou et al., 2008).
In a further attempt to test the validity of our results with the transgenic tobacco plants, we used PA biosynthetic inhibitors in wild-type plants, namely, difluoromethylornithine (DFMO), difluoromethylarginine (DFMA), and methylglyoxalbisguanylhydrazone (MGBG), for ODC, ADC, and SAMDC enzymes, respectively. A decrease in ODC slightly decreased tolerance, whereas a decrease in ADC by DFMA had a strong effect on tolerance. Moreover, MGBG caused a minor loss of tolerance, equal to that of ODC (Figure 10B). One would expect that the decrease in PA levels could delay the production of ROS through PAO action in the apoplast. Interestingly, although the intracellular pool of PAs was reduced by the inhibitors, the rate of Spd exodus into the apoplast remained quite stable (10.5 ± 0.8 and 10.2 ± 2 μM for the treated and nontreated cultures, respectively). These observations suggest that the reduction of PA anabolism within cells, with a constant PA catabolism in the apoplast, is able to enhance the detrimental effects of ROS.
Downregulation of samdc Affects Biomass Production and Tolerance to Salinity
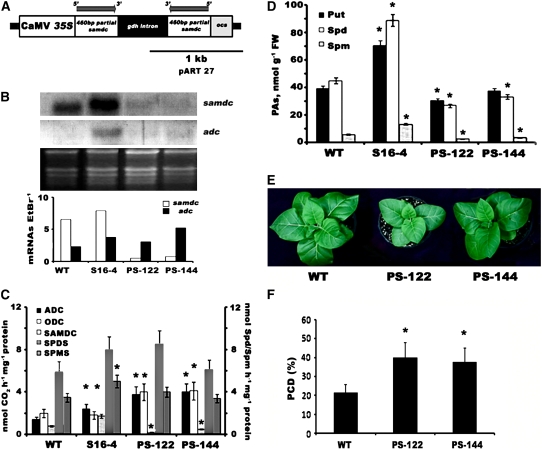
To further elucidate the need to retain high intracellular PA titers during stress, we followed an opposite strategy to that mentioned previously. We developed transgenic tobacco plants downregulating samdc via RNAi, using tobacco samdc (Figure 11A), and subjected them to salinity treatment. Moreover, we used transgenic tobacco plants with upregulated samdc (S16-4 line; Wi et al., 2006) as controls. Most of the overexpressing (Wi et al., 2006) and underexpressing samdc plants showed normal phenotypes and significantly altered samdc transcript and activity levels (Figures 11B and 11C). More specifically, in both PS-122 and PS-144 RNAi transgenic lines, samdc transcript and activity decreased >10-fold compared with the wild type; thus, they possessed only residual SAMDC activity (Figures 11B and 11C). On the contrary, adc transcript and activity were significantly increased in these samdc downregulated lines (Figures 11B and 11C). As described for the S-pao plants, ODC, SPDS, and SPMS activities also increased (Figure 11C). These results strengthen the hypothesis that there is a PA sensor in the cells that activates a feedback mechanism. Unexpectedly, S16-4 plants showed a marked increase of the corresponding activities (with the exception of ODC), involving a different pathway for this increase. Interestingly, PAO and DAO activities in both PS-122 and PS-144 lines did not change significantly compared with wild-type plants.
Figure 11.
Characterization of SAMDC Tobacco RNAi Lines: samdc and adc mRNA Levels; ADC, ODC, SAMDC, SPDS, and SPMS Specific Activities; PA Titers; Plant Phenotypes; and PCD Rates under Salinity.
(A) Construct used for the generation of stable tobacco RNAi samdc lines. CaMV 35S, 35S promoter from the Cauliflower mosaic virus; 460bp partial samdc, fragment of the samdc cDNA in either the sense or antisense orientation; gdh intron, Glu dehydrogenase intron; ocs, octopine synthase transcription terminator.
(B) RNA gel blot analysis of samdc and adc mRNA levels and quantification of mRNA levels (10 μg RNA/well) for the wild type, samdc-overexpressing line S16-4, and RNAi lines PS-122 and PS-144.
(C) ADC, ODC, and SAMDC (nmol CO2·h−1·mg−1 protein), SPDS (nmol Spd·h−1·mg−1 protein), and SPMS (nmol Spm·h−1·mg−1 protein) specific activities.
(D) Soluble PAs (nmol/g fresh weight [FW]).
(E) Wild-type, PS-122, and PS-144 plants after a 2-week exposure to 200 mM NaCl.
(F) Relative PCD rates (percentage of TUNEL-positive nuclei) in wild-type and PS-122 and PS-144 cell suspension cultures under salinity stress.
Data are means ± se of three independent experiments, and asterisks indicate statistical significance at P < 0.05.
With respect to PA levels, Spd and Spm were reduced significantly in lines PS-122 and PS-144, consistent with the role of SAMDC in regulating the higher PA content. More specifically, Put decreased by 30 and 5%, Spd decreased by 73 and 33%, and Spm decreased by 100 and 65% in PS-122 and PS-144 lines, respectively (Figure 11D).
PS-122 and PS-144 transgenic T2 plants grown in vitro and supplemented with 200 mM NaCl showed increased susceptibility and decreased biomass production upon salinity (Figure 11E). Moreover, cell suspension cultures derived from these genotypes showed significantly increased salinity-induced PCD (at 200 mM NaCl) compared with the wild type (Figure 11F). Furthermore, the PA biosynthetic machinery of the RNAi samdc lines failed to respond, in agreement with the corresponding observations for the S-pao plants (see Supplemental Figure 3 online). Finally, no significant differences in the PA catabolic activity were observed during salt stress for any of the genotypes examined, while Spd exodus to the apoplast remained constant in these plants.
DISCUSSION
That PAs play a pivotal role in plant adaptation to stress conditions has been shown by classical experimental approaches (Alcazar et al., 2006). Stress-tolerant genotypes show enhanced PA biosynthesis in response to abiotic stress compared with stress-sensitive genotypes. In spite of the fact that genetic manipulation of PA biosynthesis leads to enhanced stress tolerance against multiple stresses, the role of PA catabolism in this response is obscure (Cona et al., 2006). In this work, a series of transgenic tobacco plants overexpressing and downregulating pao and samdc (Moschou et al., 2008; this work) were used. As expected, PAO increase in S-pao plants resulted in significantly lower Spd and Spm titers, while decrease of PAO activity in A-pao plants resulted in increased Spd and Spm titers (Figure 2). As A-pao plants were more tolerant to salinity in contrast with S-pao plants (Figure 3), this led us to hypothesize that pao may exert a specific role in abiotic stress adaptation, and it was tempting to examine the effect of PA catabolism on plant adaptation to stress.
Previously, Yoda et al. (2006) showed the involvement of PAO in vitro in PCD induction during the HR-like response. Similar to these results, in which Spd was secreted from the cells upon elicitation, we show here that abiotic stress triggers Spd exodus to the apoplast in planta. Thus, under salinity, A-pao plants showed significantly higher levels of apoplastic Spd and lower titers of 1,3-Dap, in contrast with S-pao plants (Figure 4). Since 1,3-Dap and H2O2 are produced equimolarly, S-pao plants accumulated higher ROS levels, particularly in the apoplast, consistent with the localization of PAO protein in this compartment (Figure 4). The stress-induced constitutive supply of the apoplast with Spd in S-pao plants initiated a futile cycle of Spd exodus and catabolism, resulting in a remarkable ROS accumulation, similar to that induced by the HR (Bestwick et al., 1997). ROS produced during the HR response were both necessary and sufficient to induce PCD rapidly (Yoda et al., 2006). Thus, it was of interest to examine whether the Spd-derived apoplastic H2O2 production under abiotic stress conditions was sufficient to induce PCD. To answer this, we applied salt stress to A-pao and S-pao cell suspension cultures to induce Spd exodus. A-pao plants retained lower ROS titers and PCD rates, compared with wild-type plants, in contrast with S-pao cells (Figure 5). When PAs were exogenously supplied rather than secreted, lower ROS and PCD levels were found in A-pao, in contrast with S-pao cell cultures (Figure 6). These observations reinforce the view that apoplastic ROS titers are produced via PA oxidation and that the size and rate of ROS accumulation determine PCD induction. Furthermore, apoplastic ROS could also exert parallel signaling effects, such as the expression of ROS stress–responsive genes. Interestingly, S-pao plants failed to accumulate mRNAs of stress-responsive genes, such as adc and samdc, as a response to the application of PAs in the apoplast, whereas in A-pao plants the corresponding mRNAs accumulated at a slower rate compared with wild-type plants (Figure 6). Taken together, these results suggest that apoplastic ROS generation induces either the expression of effector stress–responsive genes or the PCD syndrome, depending on a specific threshold.
Gene coregulation during stress is one of the most interesting topics of the ongoing research in plant biology, since the understanding of orchestrated responses gives evidence of the metabolic pathways and transduction signals important for stress compensation. To expand the previous results under stress conditions in vivo, we applied salt stress to S-pao and A-pao transgenic plants in order to induce Spd exodus and the concomitant responses. Depending on the stress magnitude, PA biosynthetic mRNAs, particularly those of spds, accumulated as an early response to treatment (Figure 7), as was the case in rice (Oryza sativa) (Kawasaki et al., 2001). A-pao plants responded to salt stress with a delay in increasing their corresponding PA biosynthetic genes. On the other hand, although in S-pao tobacco plants an increase in all biosynthetic PA mRNAs was evident under moderate salt stress treatments, no further response to more severe stress was found. This is consistent with the idea that, depending on the stress magnitude, cells induce defense genes or promote PCD, depending on the levels of apoplastic ROS.
To examine the role of the differential PA anabolism and/or ROS accumulation downstream of gene expression, we studied the PA biosynthetic rate. Under physiological conditions, the patterns of biosynthetic PA gene transcription were mirrored by their corresponding activities in A-pao and S-pao plants, with reduced PA anabolism in A-pao plants in contrast with S-pao plants (Figure 8). Under stress conditions, A-pao plants showed higher biosynthetic activity levels compared with wild-type and S-pao plants (Figure 8). In addition, S-pao plants were incapable of retaining high PA biosynthetic activities, suggesting that vulnerability could be correlated not only with higher ROS production but also with low biosynthetic activities, as well as high catabolic activities and vice versa. The fast increase in mRNAs of PA biosynthetic genes, especially in S-pao plants, could reflect an attempt to restore normal PA biosynthesis in a background with redistributed protein synthesis due to stress conditions. Thus, the early response involves the increase in transcript levels with no concomitant increase in their translation, but in a better stress-responsive background like A-pao plants, activity increases were more profound.
Since an increase of PA biosynthetic rate is considered important for stress-induced PA accumulation, we monitored PA titers under stress conditions (Figure 10). Indeed, higher PA levels were present under stress in A-pao plants, in contrast with S-pao plants. Both S-Put and S-Spm increased. On the other hand, the decrease of S-Spd could be, at least partly, the result of its secretion into the apoplastic compartment, where it is catabolized by PAO activity. To elucidate the role of inhibition of PA anabolism in intracellular PA titers and in PCD occurrence, we investigated the contribution of the PA anabolism-to-catabolism ratio to the occurrence of PCD in a normal catabolic background, this time using transgenic tobacco plants downregulating samdc via RNAi. SAMDC is considered the finest choice due to its ability to affect higher PA levels (Thu-Hang et al., 2002). Although these plants synthesized less Spd and Spm, they retained a normal PA catabolism (Figure 11). When they were exposed to salt stress (although retaining similar to wild-type Spd exodus and oxidation to the apoplast as well as similar to wild-type PA-derived apoplastic ROS production), they failed to respond and adapt, a result that reinforces the notion that increased PA anabolism is necessary for PCD inhibition/defense as a response to abiotic stress.
Taken together, the results reported here suggest that plants efficiently regulate the compartmentalization of PAs. Under physiological conditions, apoplastic PA titers and oxidation-derived ROS are below the threshold required to trigger stress responses, while stress induces the accumulation of apoplastic Spd. Plants may follow this strategy because the apoplast possesses low antioxidant potential and ROS-derived signals could be more efficiently transduced when produced in this compartment. Equal amounts of ROS generated within the cellular compartment could be scavenged through the highly efficient antioxidant machinery. However, when higher amounts of PAs are secreted from cells into the apoplast and catabolized by the high apoplastic PAO activity, ROS levels exceed a specific threshold and no longer signal the expression of defense genes but, instead, trigger PCD. This suggests that ROS, as in the case of Ca2+, could have specific apoplastic signatures, whereby a downstream array of events acts in concert to distinguish between the different signals. This promotes differential responses varying between two extremes, defense signaling and PCD. Overall, our data are in accordance with an emerging picture for apoplastic PA catabolism, which seems to contribute to abiotic stress responses/adaptation. We propose that PCD is strictly regulated by the ratio of PA anabolism to catabolism, while ROS generation/accumulation is a nodal point in cell fate decision. When PA anabolism predominates over PA catabolism, PCD fails to occur, whereas when the opposite occurs, PCD is induced.
METHODS
Plant Material and Growth Conditions
Tobacco plants (Nicotiana tabacum cv Xanthi) were grown in a growth chamber with irradiance of ∼100 μmol·m−2·s−1, temperature of 25 ± 2°C, 16-h/8-h photoperiod, and 75% RH. Calluses and cell suspension cultures were generated from leaf explants (Primikirios and Roubelakis-Angelakis, 2001).
pao and samdc Tobacco cDNA Cloning and RT-PCR
Total mRNA from petioles was extracted according to Iandolino et al. (2004) and was subjected to reverse transcription using the Omniscript reverse transcription kit according to the manufacturer's instructions (Invitrogen). To partially clone the pao described by Yoda et al. (2006), the primers FW (5′-GGCAGGATAAGGAAGGA-3′) and Rv (5′-GTCTGTTAGCATTGGTGTTCT-3′) were used, and the 621-bp product was cloned into pGEM-T easy vector (Promega) and sequenced. To clone the tobacco samdc cDNA, the primers Fw (5′-TCTAGATGGATTCGGCCTTG-3′) and Rv (5′-GGTACCCAGAGTAAACATGC-3′) were used, and the 1104-bp product was introduced into a pGEM-T easy vector (Promega) and directly sequenced. For quantitative reactions, total mRNA from leaves was extracted and treated with RNase-free DNase I for 45 min at 37°C. The samples were then subjected to RT-PCR using polyT as primer and the Super RT enzyme according to the manufacturer's instructions (HT Biotechnology). The samples were normalized according to actin using the primers Fw (5′-GATTTGCTGGTGATGATGC-3′) and Rv (5′-AAGGGTGCTTCAGTAAGTAG-3′). More specifically, samples were used for 30, 35, 37, and 40 cycles of PCR (1 min at 94°C initial denaturation, 35 s at 94°C cycling denaturation, 53°C primer annealing, a 30-s extension, and a 5-min final extension) and analyzed using gel electrophoresis to estimate whether the reaction was still in the logarithmic phase. The actin signal was radioactively detected following DNA gel blotting of the products, and this was shown to be quantitative over this range of amplification by estimating the product density using image analysis (see Supplemental Figure 4 online). The primers for pao were used for 30 to 35 cycles (logarithmic phase) of quantitative RT-PCR (2 min at 94°C initial denaturation, 40 s at 94°C cycling denaturation, 55°C primer annealing, a 1-min extension, and a 7-min final extension). Products were radioactively detected following DNA gel blotting of the products and quantified using image analysis (see Supplemental Figure 4 online). Reactions with samples treated with DNase I but not subjected to reverse transcription were also used as negative controls.
Vector Construction and Plant Transformation
The m pao cDNA (Tavladoraki et al., 1998) was introduced into a pART7 vector using EcoRI sites. PCR was performed on tobacco samdc cDNA using the primers Fw (5′-GCTCTAGATGGATTCGGCCTTG-3′) and Rv (5′-GGGGTACCCAGAGTAAACATGC-3′) to obtain a 460-bp fragment that was introduced in both the sense and antisense orientations (RNAi samdc), separated by a 600-bp intron, into the pART7 vector. The pART7 vectors in both cases were digested with NotI to give the pao cDNA and the RNAi samdc, downstream of the 35S cauliflower mosaic virus promoter. The excised fragments were introduced into the pART27 vector, which was used to transform Agrobacterium tumefaciens strain LBA4404 (Wi et al., 2006). Stable transformation was achieved using tobacco leaf discs, and transformants were selected against 150 mg/L kanamycin in MS medium (Murashige and Skoog, 1962). Preparation of transgenic tobacco plants overexpressing samdc and m pao was described previously (Wi et al., 2006; Moschou et al., 2008).
PA Analysis, Protein Extraction, and Enzyme Assays
S-, SH-, and PH-PA fractions were determined as described previously (Kotzabasis et al., 1993) using an HP 1100 high-performance liquid chromatograph (Hewlett-Packard). Apoplastic PAs were analyzed as described previously (Yoda et al., 2003, 2006), and the purity of apoplastic fluids was confirmed using malate dehydrogenase (EC 1.1.1.37) and glucose-6-phosphate dehydrogenase (EC 1.1.1.49) as cytoplasmic markers, as described previously (Boudart et al., 2005). Apoplastic fluids were further tested by measuring PAO activity and by protein gel blot against PAO.
Total proteins were extracted as described (Primikirios and Roubelakis-Angelakis, 2001). For PAO and DAO assays, a spectrophotometric method developed by Federico et al. (1985) was used with minor modifications. More specifically, K-phosphate buffer instead of Na-phosphate buffer was used, while for S-pao plants, 10 μL of extracts was used instead of 50 μL in wild-type and A-pao plants. A radiometric method was also used for PAO and DAO assays according to Paschalidis and Roubelakis-Angelakis (2005b). ADC, ODC, and SAMDC were assayed by measuring the release of 14CO2. l-[1-14C]Arg, l-[1-14C]Orn, and adenosyl-l-Met S-[carboxyl-14C], respectively (American Radiolabeled Chemicals), were used as radioactive substrates. Labeled CO2 was counted in an LS 6000SE (Beckman) scintillation counter (Paschalidis and Roubelakis-Angelakis 2005a). SPDS and SPMS were assayed by measuring the formation of Spd and Spm, respectively, according to Paschalidis and Roubelakis-Angelakis (2005a).
Protein Gel Blotting, RNA Extraction, and DNA and RNA Gel Blotting
Total protein extracts were electrophoretically resolved, transferred to membranes, and hybridized against an anti-PAO maize (Zea mays) polyclonal antibody (Angelini et al., 1995) and an anti-ADC grape (Vitis vinifera) polyclonal antibody (Primikirios and Roubelakis-Angelakis, 2001). DNA gel blotting was performed as described previously (Wi et al., 2006). For RNA gel blotting, total RNA was extracted with the optimized hot-phenol method according to Iandolino et al. (2004), transferred to a membrane, and hybridized to the corresponding 32P-labeled probe prepared using the RadPrime DNA labeling kit as described by the manufacturer (Invitrogen). Probes for adc, odc, samdc, spds, and m pao were prepared as described previously (Paschalidis and Roubelakis-Angelakis, 2005a, 2005b), while the probe for Nt pao was prepared using the partially cloned cDNA. X-ray films (Kodak) were used for visualization.
DNA Fragmentation and Epifluorescent ROS Detection
For the detection of nuclear DNA fragmentation, the method described by Skopelitis et al. (2006) was used. After a 24-h treatment, the cells were fixed in 4% (w/v) paraformaldehyde in PBS buffer (pH 7.4). Nuclei were stained with propidium iodide (1 μg/mL) and the free 3′-OH groups of fragmented DNA molecules were labeled by the TUNEL (Promega), as instructed by the manufacturer.
In situ localization of ROS was performed using the highly sensitive, cell-permeable probe 2′,7′-dichlorodihydrofluorescein diacetate (DCFMA; Molecular Probes). For PA treatments, the corresponding PA solution was supplied to the cell suspension culture. Cells were harvested, after centrifugation at 800g, and were incubated in 1 mL of buffer (20 mM K-phosphate, pH 6.0, supplemented with 50 μM DCFMA and 3 μg/mL horseradish peroxidase [Sigma-Aldrich]) for 10 min at 25°C in darkness. An aliquot of cells (0.1 mL) was removed, washed in the same buffer, and visualized immediately.
Photographs were taken using an epifluorescence microscope (Nikon Eclipse E800) with excitation/emission filter 450 to 490/520 nm using a SONY 655 DXC-950P camera. Positive controls for TUNEL assay with DNase I and menadione treatments, menadione for in situ ROS localization, and negative controls without terminal transferase for TUNEL assay and treatments with ascorbate and catalase for in situ ROS localization were also used.
Transmission Electron Microscopy for H2O2 Localization
H2O2 localization was also detected by the cerium chloride method, as described by Bestwick et al. (1997). In brief, small pieces (2 to 5 mm3) of tissues from the central laminae region of tobacco third leaves were incubated in freshly prepared 50 mM MOPS buffer, pH 7.2, containing 5 mM cerium chloride for 1 h. Subsequently, the samples were fixed in a mixture of 1.25% (v/v) glutaraldehyde and 1.25% (v/v) paraformaldehyde in 50 mM cacodylate buffer, pH 7.2, for 1 h. After washing in cacodylate buffer, samples were postfixed overnight in 1% osmium tetroxide (prepared in cacodylate buffer), dehydrated in a graded ethanol series, and embedded in London Resin White. Ultrathin sections were examined with a transmission electron microscope at 80 kV without poststaining. To confirm the specificity of cerium chloride staining for H2O2, samples were incubated for 20 min in 50 mM MOPS, pH 7.2, containing either 1 mM sodium azide (to inhibit peroxidase) or 25 mg/mL bovine liver catalase (to decompose H2O2). They were then transferred to cerium chloride solution, incubated for 1 h, and processed for transmission electron microscopy, as described above. Some samples were fixed without the cerium chloride treatment. After postfixation in OsO4, samples were processed for transmission electron microscopy, as outlined above.
Salt Stress Treatments and Inhibitor Applications
For all salt stress treatments, T2 generation S-pao, A-pao, and samdc RNAi transgenic tobacco plants were used. For the germination assessment, S-pao and A-pao seeds were surface-sterilized and placed on MS agar medium with 0, 200, and 300 mM NaCl. For growth assessment, seeds were first sown in MS medium; after 2 weeks, plants of the same size were transferred to 0, 100, 200, and 300 mM NaCl-containing MS medium for 1 month, and plant biomass was estimated as fresh weight. For the whole plant treatments, 1-month-old plants grown in pots were transferred to hydroponic cultures and supplemented with increasing concentrations of MS solution up to half-strength MS medium. After 2 d, the half-strength MS medium was supplemented with NaCl. For the cell suspension treatments, exponentially grown cell suspension cultures were supplemented with NaCl.
Inhibitor studies were performed in hydroponically grown wild-type plants. Concentrations were as follows: 0.1 mM DFMA, 1 mM DFMO, and 1 mM MGBG. Each inhibitor was sterilized using a 0.2-μm Supor Membrane PALL Acrodisc syringe filter (Pall).
Image and Statistical Analysis
Image and pixel analyses and mRNA quantification were performed with ImageJ 1.37v (rsb.info.nih.gov/ij), and statistical analysis was performed with SPSS 14v (www.spss.com).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: adc, AB110952; odc, AF321138; spds, DQ536198; samdc, AF321142; Nt pao, AB200262; m pao, AJ002204; actin, AB158612.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular and Biochemical Analysis of the A-pao Plants.
Supplemental Figure 2. Biomass of the Wild Type and A2, A6, and A8 Transgenic Lines Exposed to Salt Stress.
Supplemental Figure 3. Difference in Activity Levels of the PA Biosynthetic Enzymes and PA Accumulation in the Wild Type and S16-4, PS-124, and PS-144 Transgenic Lines.
Supplemental Figure 4. Quantitative RT-PCR Analysis of the actin and Nt pao Genes within the Exponential Phase.
Supplementary Material
Acknowledgments
We are grateful to P. Tavladoraki (University Roma Tre), to K. Samejima (Musashino University), and to K. Park (Sunchon National University) for the generous gifts of anti-MPAO polyclonal antibody and m Pao cDNA, decarboxylated S-adenosyl-l-methionine, and samdc-overexpressing plants, respectively. We also thank Eleftherios Zouros (University of Crete) for critically reading the manuscript and Alexandra Siakouli and Eva Papadogiorgaki for excellent technical assistance. This project was cofunded by national and European resources (EPEAEKII-Pythagoras-Herakleitos) and implemented in the frame of COST Actions 858 and 0605.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Kalliopi A. Roubelakis-Angelakis (poproube@biology.uoc.gr).
Online version contains Web-only data.
References
- Agalou, A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66 87–103. [DOI] [PubMed] [Google Scholar]
- Alcazar, R., Marco, F., Cuevas, J.C., Patron, M., Ferrando, A., Carrasco, P., Tiburcio, A.F., and Altabella, T. (2006). Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett. 28 1867–1876. [DOI] [PubMed] [Google Scholar]
- Angelini, R., Federico, R., and Bonfante, P. (1995). Maize polyamine oxidase: Antibody production and ultrastructural localization. J. Plant Physiol. 145 686–692. [Google Scholar]
- Bestwick, C.S., Brown, I.R., Bennet, M.H., and Mansfield, J.W. (1997). Localization of hydrogen peroxide accumulation during the hypersensitive reaction of lettuce cells to Pseudomonas syringae pv phaseoliocola. Plant Cell 9 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchereau, A., Aziz, A., Larher, F., and Martin-Tanguy, J. (1999). Polyamines and environmental challenges: Recent development. Plant Sci. 140 103–125. [Google Scholar]
- Boudart, G., Jamer, E., Rossignol, M., Lafitte, C., Borderies, G., Jauneau, A., Esquerré-Tugayé, M.T., and Pont-Lezica, R. (2005). Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: Identification by mass spectrometry and bioinformatics. Proteomics 5 212–221. [DOI] [PubMed] [Google Scholar]
- Capell, T., Bassie, L., and Christou, P. (2004). Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc. Natl. Acad. Sci. USA 101 9909–9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay, M.K., Gupta, S., Sengupta, D.N., and Ghosh, B. (1997). Expression of arginine decarboxylase in seedlings of indica rice (Oryza sativa L.) cultivars as affected by salinity stress. Plant Mol. Biol. 34 477–483. [DOI] [PubMed] [Google Scholar]
- Cona, A., Rea, G., Angelini, R., Federico, R., and Tavladoraki, P. (2006). Functions of amine oxidases in plant development and defense. Trends Plant Sci. 11 80–88. [DOI] [PubMed] [Google Scholar]
- Federico, R., Angelini, R., Cesta, A., and Pini, C. (1985). Determination of diamine oxidase in lentil seedlings by enzymic activity and immunoreactivity. Plant Physiol. 79 62–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iandolino, A.B., da Silva, F.G., Lim, H., Choi, H., Williams, L.E. and Cook, D.R. (2004). High-quality RNA, cDNA, and derived EST libraries from grapevine (Vitis vinifera L.). PMB Rep. 22 269–278. [Google Scholar]
- Kasinathan, V., and Wingler, A. (2004). Effect of reduced arginine decarboxylase activity on salt tolerance and on polyamine formation during salt stress in Arabidopsis thaliana. Physiol. Plant. 121 101–107. [DOI] [PubMed] [Google Scholar]
- Kasukabe, Y., He, L., Nada, K., Misawa, S., Ihara, I., and Tachibana, S. (2004). Overexpression of spermidine synthase enhances tolerance to multiple environmental stresses and up-regulates the expression of various stress-regulated genes in transgenic Arabidopsis thaliana. Plant Cell Physiol. 45 712–722. [DOI] [PubMed] [Google Scholar]
- Kawasaki, S., Borchert, C., Deyholos, M., Wang, H., Brazille, S., Kawai, K., Galbraith, D., and Bohnert, H.J. (2001). Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13 889–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzabasis, K., Christakis-Hampsas, M.D., and Roubelakis-Angelakis, K.A. (1993). A narrow-bore HPLC method for the identification and quantitation of free, conjugated and bound polyamines. Anal. Biochem. 214 484–489. [DOI] [PubMed] [Google Scholar]
- Kumria, R., and Rajam, M.V. (2002). Ornithine decarboxylase transgene in tobacco affects polyamines, in vitro-morphogenesis and response to salt stress. J. Plant Physiol. 159 983–990. [Google Scholar]
- Manavella, P.A., Arce, A.L., Dezar, C.A., Bitton, F., Renou, F.P., Crespi, M., and Chan, R.L. (2006). Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J. 48 125–137. [DOI] [PubMed] [Google Scholar]
- Moschou, P.N., Delis, I.D., Paschalidis, K.A., and Roubelakis-Angelakis, K.A. (2008). Transgenic tobacco plants overexpressing polyamine oxidase are not able to cope with oxidative burst generated by abiotic factors. Physiol. Plant. 133 140–156. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Paschalidis, K.A., and Roubelakis-Angelakis, K.A. (2005. a). Spatial and temporal distribution of polyamine levels and polyamine anabolism in different organs/tissues of the tobacco plant. Correlations with age, cell division/expansion, and differentiation. Plant Physiol. 138 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschalidis, K.A., and Roubelakis-Angelakis, K.A. (2005. b). Sites and regulation of polyamine catabolism in the tobacco plant. Correlations with cell division/expansion, cell cycle progression, and vascular development. Plant Physiol. 138 2174–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primikirios, N.I., and Roubelakis-Angelakis, K.A. (2001). Indications for posttranslational regulation of Vitis vinifera L. arginine decarboxylase. Plant Mol. Biol. 45 669–678. [DOI] [PubMed] [Google Scholar]
- Purnell, P.M., Skopelitis, D., Roubelakis-Angelakis, K.A., and Botella, J.R. (2005). Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222 167–180. [DOI] [PubMed] [Google Scholar]
- Rea, G., Concetta, P.M., Tavazza, R., Biondi, S., Gobbi, V., Ferrante, P., De Gara, L., Federico, R., Angelini, R., and Tavladoraki, P. (2004). Ectopic expression of maize polyamine oxidase and pea copper amine oxidase in the cell wall of tobacco plants. Plant Physiol. 134 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis, D.S., Paranychianakis, N.V., Paschalidis, K.A., Pliakonis, E.D., Delis, I.D., Yakoumakis, D.I., Kouvarakis, A., Papadakis, A.K., Stephanou, E.G., and Roubelakis-Angelakis, K.A. (2006). Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18 2767–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W., Newton, R.J., Li, C., and Charles, T.M. (2007). Enhanced stress tolerance in transgenic pine expressing the pepper CaPF1 gene is associated with the polyamine biosynthesis. Plant Cell Rep. 26 115–124. [DOI] [PubMed] [Google Scholar]
- Tavladoraki, P., Schinina, M.E., Cecconi, F., Di Agostino, S., Manera, F., Rea, G., Mariottini, P., Federico, R., and Angelini, R. (1998). Maize polyamine oxidase: Primary structure from protein and cDNA sequencing. FEBS Lett. 426 62–66. [DOI] [PubMed] [Google Scholar]
- Thu-Hang, P., Bassie, L., Safwat, G., Trung-Nghia, P., Christou, P., and Capell, T. (2002). Expression of a heterologous S-adenosylmethionine decarboxylase cDNA in plants demonstrates that changes in S-adenosyl-L-methionine decarboxylase activity determine levels of the higher polyamines spermidine and spermine. Plant Physiol. 129 1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waie, B., and Rajam, M.V. (2003). Effect of increased polyamine biosynthesis on stress responses in transgenic tobacco by introduction of human S-adenosylmethionine gene. Plant Sci. 164 727–734. [Google Scholar]
- Wi, S.J., Kim, W.T., and Park, K.Y. (2006). Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broad-spectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep. 25 1111–1121. [DOI] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K.S., and Zhu, J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (suppl.): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K., Takahashi, Y., Berberich, T., Imai, A., Miyazaki, A., Takahashi, T., Michael, A., and Kusano, T. (2006). The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Lett. 22 6783–6788. [DOI] [PubMed] [Google Scholar]
- Yoda, H., Hiroi, Y., and Sano, H. (2006). Polyamine oxidase is one of the key elements for oxidative burst to induce programmed cell death in tobacco cultured cells. Plant Physiol. 142 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda, H., Yamaguchi, Y., and Sano, H. (2003). Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol. 132 1973–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.