Protein production is essential for plant development, responses to environmental stimuli, and hormone signaling. Equally essential is the flip side of this process: removing key regulatory proteins so that the response ceases. In many cases, such as auxin responses and photomorphogenesis, this is accomplished by ubiquitin-mediated proteolysis. The ubiquitin pathway targets specific proteins for proteolysis by attaching the 76–amino acid protein ubiquitin to the targeted protein using three enzymes, an activating enzyme (E1), a conjugating enzyme (E2), and the ubiquitin ligase (E3) (reviewed in Stone and Callis, 2007). E3 ligases recruit substrate and thereby provide specificity; some ligases act as monomers, but some act as multisubunit complexes, thereby increasing substrate binding combinations. A large family of E3 ligases in plants contains a RING (for Really Interesting New Gene) domain; the Arabidopsis genome, for example, is predicted to encode 475 RING-type E3 ligases.
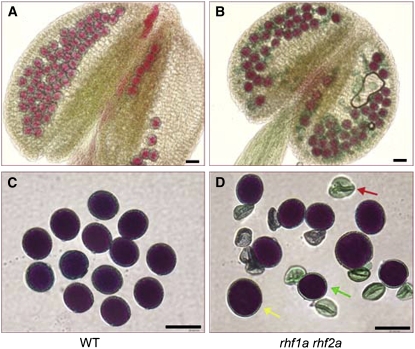
Two articles in this issue highlight the importance of shutting down protein production; indeed, the two very different processes studied use two different redundant pairs of RING-type E3 ligases to turn off the key regulator for that process. In the first case, the key regulator is a transcription factor that turns on the drought response. Qin et al. (pages 1693–1707) examine Arabidopsis DREB2A (for Drought Response Element Binding Protein 2A), which turns on the plant response to drought stress. Although DREB2A is induced by drought stress, it also has low-level expression in unstressed plants; this unneeded protein is removed by two RING E3 ligases, DRIP1 and DRIP2 (DREB2A Interacting Proteins). Failure to turn off this response is bad for the plant: although single mutants in either protein have no effect, the drip1 drip2 double mutant is stunted and also shows activation of drought response genes. In the second case, the key regulator is a cyclin-dependent kinase inhibitor that must be removed for the mitotic cell divisions that follow meiosis in Arabidopsis gametogenesis. Liu et al. (pages 1538–1554) investigate inhibitors of cyclin-dependent kinases known as Interactors of Cdc2 Kinase (ICKs) or Kip-related proteins (KRPs). They find that two RING-type E3 ligases are essential to remove ICK4/KRP6 and thus allow cells to proceed through the postmeiotic cell divisions in gametogenesis. Again, the single ligase mutants show no phenotype, but the double mutant has a strong phenotype, which is in this case, severe defects in both male and female gamete formation (see figure).
Figure 1.
Pollen development in wild-type ([A] and [C]) and double mutant plants for two key RING-type E3 ligases ([B] and [D]), showing shrunken pollen and grains of variable size (arrows). (Image reproduced from Liu et al. [2008].)
The phenotypic consequences of failure to degrade specific proteins show the advantages of redundant control of proteolysis. Interestingly, E3 ligase redundancy also shows the importance of reverse genetics in studying this process. Finding the targets of the hundreds of E3 ligases will inform studies of the regulation of multiple developmental processes.
References
- Liu, J., et al. (2008). Targeted degradation of cyclin-dependent kinase inhibitor ICK4/KRP6 by the RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20 1538–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, F., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress–responsive gene expression. Plant Cell 20 1693–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., and Callis, J. (2007). Ubiquitin ligases mediate growth and development by promoting protein death. Curr. Opin. Plant Biol. 10 624–632. [DOI] [PubMed] [Google Scholar]