Abstract
Subunit oligomerization of many proteins is mediated by coiled-coil domains. Although the basic features contributing to the thermodynamic stability of coiled coils are well understood, the mechanistic details of their assembly have not yet been dissected. Here we report a 13-residue sequence pattern that occurs with limited sequence variations in many two-stranded coiled coils and that is absolutely required for the assembly of the Dictyostelium discoideum actin-bundling protein cortexillin I and the yeast transcriptional activator GCN4. The functional relationship between coiled-coil “trigger” sequences was manifested by replacing the intrinsic trigger motif of GCN4 with the related sequence from cortexillin I. We demonstrate that these trigger sequences represent autonomous helical folding units that, in contrast to arbitrarily chosen heptad repeats, can mediate coiled-coil formation. Aside from being of general interest for protein folding, trigger motifs should be of particular importance in the protein de novo design.
The parallel two-stranded α-helical coiled coil is the most frequently encountered subunit oligomerization motif in intracellular proteins (1–4). This structural motif appears to offer the simplest system for studying both the intra- and intermolecular interactions that govern the folding and stability of multisubunit proteins (5–8). The coiled-coil structure that was first postulated by Crick in 1953 (9) consists of two right-handed amphipathic α-helices coiled around one another in a left-handed manner (9, 10). The sequences of α-helical coiled coils are characterized by a heptad repeat of seven residues denoted a to g with a 3,4-hydrophobic repeat of mostly apolar amino acids at positions a and d (11, 12). Two-stranded coiled-coil structures are stabilized by the hydrophobic interface between the α-helices, which is formed by residues at positions a and d, and to a lesser extent by e and g (10).
A rather puzzling and frequently made observation is that relatively long heptad-repeat-containing polypeptide chain fragments derived from stable coiled-coil domains fail to associate into coiled-coil structures. This failure cannot be simply explained by instability caused by the type of residues occupying the a and d positions of the heptad repeats, or by electrostatic repulsion of the two chains. This observation raises the question of whether distinct sites exist within heptad-repeat-containing amino acid sequences that are necessary to mediate coiled-coil formation. We recently have addressed this question in detail by using the two-stranded parallel coiled-coil oligomerization domain of the actin-bundling protein Dictyostelium discoideum cortexillin I, which consists of 18 continuous heptad repeats (13). We have documented that a distinct 14-residue “trigger” sequence exists within the cortexillin I oligomerization domain that is absolutely required to mediate proper assembly of the domain into a parallel homodimeric coiled coil (6). This result suggests that the presence of heptad repeats, per se, is not sufficient for stable coiled-coil formation.
Here we have generalized our concept on the existence of coiled-coil trigger sequences. We report a 13-residue pattern that occurs with limited sequence variations in many two-stranded coiled coils, including myosin, kinesin, tropomyosin, and the leucine zipper domain of the yeast transcriptional activator GCN4. The functional relationship between coiled-coil trigger sequences is manifested by replacing the intrinsic trigger motif of GCN4 with the related sequence from cortexillin I. Furthermore, by using 16-residue-long synthetic peptides, we demonstrate that these trigger sequences represent autonomous folding units. We found that trigger sequences, in contrast to arbitrarily chosen heptad repeats, can mediate coiled-coil formation.
MATERIALS AND METHODS
Recombinant GCN4 Fragments.
The recombinant fragments GCN4p-wt, GCN4p-Cort, GCN4p-Cort T/L, GCN4p-c1, and GCN4p-c2 were expressed in Escherichia coli strain JM109(DE3) (Novagen). Synthetic genes were prepared with optimal codon usage for E. coli (14) and ligated into the BamHI/EcoRI site of pPEP-T (15). DNA manipulations for cloning were performed according to standard protocols (16). Recombinant insert DNA was verified by Sanger dideoxy DNA sequencing.
Production and purification of 6×His-tagged fusion proteins by affinity chromatography on Ni2+-Sepharose (Novagen) was performed under denaturing conditions as described in the manufacturer’s instructions. Separation of recombinant GCN4 fragments from the 6×His-tagged carrier protein was carried out as described by Kammerer et al. (17). If not stated otherwise, recombinant fragments were analyzed at 5°C in 5 mM sodium phosphate buffer (pH 7.4) supplemented with 150 mM sodium chloride.
Synthetic Peptides.
The 16-residue peptides GCN4p2–17, GCN4p16–31, and cI-p were purchased from MedProbe (Oslo, Norway). Purity of the peptides, which was >95%, had been verified by qualitative amino acid and mass spectral analysis. Exact concentrations of peptide solutions were determined by quantitative amino acid analysis.
Analytical Ultracentrifugation (AUC).
AUC was performed on a Optima XL-A analytical ultracentrifuge (Beckman Instruments, Palo Alto, CA) equipped with a 12-mm Epon double-sector cells in an An-60 Ti rotor. Sedimentation equilibrium runs were performed at 5°C at rotor speeds of 40,000 and 48,000 rpm. Chain concentrations (monomer) were in the range of 0.1–0.5 mg/ml for the recombinant proteins and 0.1, 0.5, and 1 mg/ml for the synthetic peptides. Average molecular masses were evaluated by using a floating baseline computer program that adjusts the baseline absorbance to obtain the best linear fit of ln(absorbance) versus radial distance square (18). A partial specific volume of 0.73 ml/g was used for all calculations.
CD Spectroscopy.
CD analysis of the synthetic 16-residue peptides and the recombinant GCN4 fragments was performed as described (6, 17).
RESULTS AND DISCUSSION
We recently have identified a 14-residue trigger sequence that is indispensable for the formation of the two-stranded parallel coiled-coil oligomerization domain in the D. discoideum actin-bundling protein known as cortexillin I (6, 13). Interestingly, analysis of a number of well-characterized two-stranded coiled coils has revealed related putative trigger sequences (Fig. 1). Furthermore, the functional significance of these distinct sequences is supported by published data. Trybus et al. (19), for example, have reported that smooth muscle myosin fragments require between 25 and 37 heptad repeats of the coiled-coil domain to achieve full dimer stability. We have identified a putative trigger sequence within heptad repeats 28 and 29 (Fig. 1). Recently, Tripet et al. (20) established a stable two-stranded coiled-coil structure for the neck region in kinesin. Although N-terminal deletion mutants of the neck region still formed stable coiled-coil structures, removal of the C-terminal part containing the identified trigger sequence (Fig. 1) abolished coiled-coil formation. Likewise, mutational analysis of the GCN4 leucine zipper (denoted GCN4p1) indicated that removal of N-terminal residues did not affect dimerization, whereas coiled-coil formation was completely abolished in fragments where part of the identified trigger sequence (Fig. 1) residing in the C-terminal half of the polypeptide had been removed (21). Hu et al. (22) analyzed the importance of the amino acid side chains at eight positions that form the hydrophobic interface of the GCN4 leucine zipper dimer. Those authors demonstrated in single randomization experiments that among the individual a and d positions Leu-267, Val-271, and Leu-274 were the most sensitive to mutations in terms of dimerization propensities. Notably, all three residues comprise hydrophobic a and d positions of the trigger sequence of the leucine zipper coiled coil (Fig. 1). In addition, heptad repeats similar to the trigger sequences have been used in a de novo-designed tropomyosin-derived synthetic dimeric coiled coil (Fig. 1) (23). A comparison of the above trigger sequences is illustrated in the sequence alignment of Fig. 1. By using two derived sequence patterns, we identified related sites in other two-stranded coiled coils (see Fig. 1 for some selected examples).
Figure 1.
Alignment of related trigger sequences from two-stranded coiled coils. The coiled-coil trigger sequences from D. discoideum cortexillin I, yeast transcriptional activator GCN4, human kinesin, chicken gizzard smooth muscle myosin II heavy chain, and a tropomyosin-derived synthetic de novo coiled coil were identified based on their functional importance for coiled-coil formation reported in the literature. Additional sequences of known two-stranded coiled coils were localized by using the GCG Wisconsin sequence analysis software package. A sequence pattern search against swiss-prot with LEX(R, K)(L, V, I, A)X(R, E, K, D)XE and LEXE(L, V, I, A)X(R, E, K, D)X(R, K) has been applied, and zero mismatches were allowed. Human skeletal muscle tropomyosin β-chain, human tpr, and human Clip170 represent some selected examples. For each protein analyzed only one match with the derived sequence patterns was found. Heptad positions are indicated by lowercase letters. Residues that match the search pattern are in bold. Conserved potential interhelical attractive electrostatic interactions between ionizable side chains at positions e and g (residue i in chain 1 to residue i′+5 in chain 2; g to e′) are indicated in the derived consensus sequence. Moreover, with the exception of kinesin and tropomyosin all selected examples contain a possible i, i+8 intrahelical ionic interaction that exists in the N-terminal helix of the crystal structure of RNase A (24). Numbers refer to the amino acid positions within the native proteins. x, any residue; h, hydrophobic residue; c, charged residue.
The characteristic feature of all of these trigger sequences is the distinct pattern of hydrophobic and hydrophilic residues, where the latter may be involved in several possible intra- and interchain electrostatic interactions. In addition to a conserved LeuGlu di-peptide sequence, the conservation of a possible g to e′ interchain attractive electrostatic interaction (residue i in one chain to residue i′+5 in the neighboring chain) is particularly striking. This interhelical electrostatic interaction is, for example, observed in the crystal structure of the GCN4 leucine zipper (between Glu-270 and Lys-275′; see Fig. 1) (10). With the exception of kinesin and tropomyosin all selected examples contain a possible i, i+8 intrahelical ionic interaction, which is, for example, present in the N-terminal helix of the crystal structure of RNase A (between Glu-2 and Arg-10; see figure 1 in ref. 24). Our derived consensus sequence, xxLExc-hxcxccx (see Fig. 1), appears predominantly in two-stranded coiled coils. Screening known three-stranded coiled-coil proteins with the two sequence patterns (see Fig. 1 legend) yielded only one match (mouse and rat laminin γ1 chain). Moreover, the sequence pattern could not be identified in coiled coils containing four and five α-helices. Because this distinct sequence pattern could not be found in all known two-stranded coiled coils suggests the existence of different types of functionally equivalent coiled-coil trigger sequences in these proteins. It also should be noted that by screening protein sequences using our rather restrictive search patterns (zero mismatches were allowed) additional potential coiled-coil trigger sequences (even in the same heptad-repeat-containing segment) may go undetected.
The functional relationship between different (but closely related) coiled-coil trigger sequences was manifested by replacing the trigger residing within the GCN4 leucine zipper by the cortexillin I homologue (i.e., GCN4p-Cort; Fig. 2A). The leucine zipper from GCN4 was selected because this particular coiled coil has been studied in great detail both structurally and functionally. In addition, the following recombinant control peptides were produced: GCN4p-wt served as a positive control and corresponds to the wild-type GCN4 leucine zipper sequence. GCN4p-c1 is a chimeric peptide in which the trigger sequence in GCN4p-wt has been replaced by an arbitrarily chosen two-heptad repeat segment (Asp-270 to Glu-282; ref. 13) from cortexillin I (Fig. 2A). These two heptad repeats meet the criteria for coiled-coil formation in the sense that they contain Leu at the a and the d positions (1). Finally, GCN4p-c2 differs from GCN4p-wt in that its two N-terminal heptad repeats (which do not include the trigger sequence) have been exchanged for the two arbitrarily chosen cortexillin I heptad repeats. Notably, Asn-16, which is known to favor the parallel two-stranded coiled-coil structure of the GCN4 leucine zipper (25), was not replaced in any of the engineered fragments.
Figure 2.
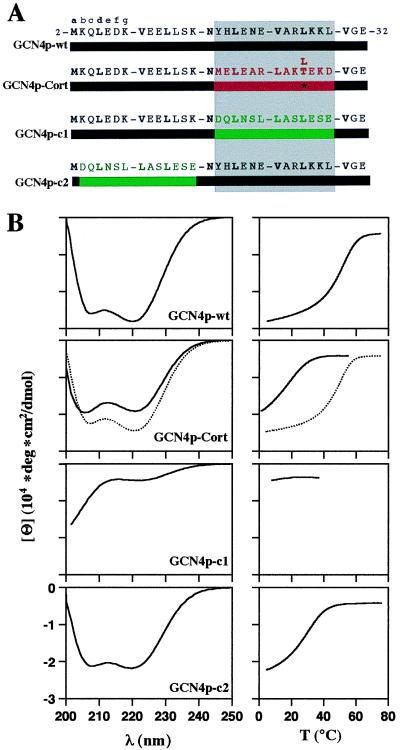
The coiled-coil trigger sequences of GCN4 and cortexillin I are functionally equivalent. (A) Sequences of the recombinant GCN4 polypeptide chain fragments used in this study. GCN4p-wt corresponds to the GCN4 leucine zipper. GCN4p-Cort is a chimeric protein in which the GCN4 trigger sequence has been replaced by the related trigger sequence of cortexillin I (Met-312 to Asp-324) (6, 13). GCN4p-c1 and GCN4p-c2 represent control peptides in which either the GCN4 trigger sequence or the two N-terminal heptad repeats have been replaced by an arbitrarily chosen two-heptad repeat segment from cortexillin I (Asp-270 to Glu-282). Heptad repeats are represented as blocks, and heptad positions are indicated by lowercase letters. The 3,4-hydrophobic repeat with mostly hydrophobic amino acid residues at heptad positions a and d is indicated in bold. Numbers refer to the amino acid positions within GCN4p1 (10). GCN4 sequences are represented in black. The trigger sequence of cortexillin I is shown in red, and the two arbitrarily chosen cortexillin I control heptad repeats are in green. The trigger site critical for coiled-coil formation is indicated by the gray-shaded box. (B) CD spectra (Left) and thermal unfolding profiles (Right) of wild-type and chimeric GCN4 fragments. CD spectra and thermal unfolding profiles of GCN4p-Cort and GCN4p-Cort T/L are represented as solid and dashed lines, respectively. Thermal stability of the proteins was monitored by the CD signal change at 222 nm, [Θ]222. Polypeptide chain concentrations were 75 μM in 5 mM sodium phosphate buffer, pH 7.4, containing 150 mM sodium chloride.
The secondary structures and assembly products of the recombinant GCN4 peptides displayed in Fig. 2A were analyzed by CD spectroscopy (Fig. 2B) and AUC (Table 1). Consistent with the values reported in the literature (21), GCN4p-wt formed a dimer with an α-helical spectrum and a thermal unfolding profile exhibiting a sigmoid shape. GCN4p-Cort also folded into a dimer with a similarly high α-helix content. The thermal stability of the chimeric coiled coil, however, was significantly reduced compared with the wild-type peptide. This decrease in thermal stability of the chimera could be attributed to a Thr residue occupying a heptad d position in the cortexillin I trigger sequence (Fig. 2A). When changing this residue to Leu (GCN4p-Cort T/L), which is the corresponding amino acid in the trigger sequence of GCN4p-wt, the formation of a trimer was observed whose transition temperature of thermal unfolding was significantly increased. Moreover, replacement of the N-terminal two heptad repeats by the two arbitrarily chosen cortexillin I heptad repeats (GCN4p-c2) did not abolish the peptide’s ability to fold into a dimeric coiled-coil structure. In contrast, replacement of the GCN4 coiled-coil trigger sequence by the two arbitrary cortexillin I heptad repeats (GCN4p-c1) abolished chain association and resulted in a CD spectrum characteristic of randomly coiled proteins.
Table 1.
Mean molar ellipticities at 222 nm, melting temperatures at the midpoint of transition, and molecular masses of the GCN4 leucine zipper fragments and peptides and cortexillin I-derived peptides
| Peptides | [Θ]222,a deg⋅cm2⋅dmol−1 | Tm,c °C | Molecular mass,d Da |
|---|---|---|---|
| GCN4p-wt | −27.9 × 103 | 53 | 7600 (3828) |
| GCN4p-Cort | −18.1 × 103 | 19 | 5700 (3749) |
| GCN4p-Cort T/L | −24.6 × 103 | 50 | 11700 (3761) |
| GCN4p-cl | −4.5 × 103 | n.d. | 4300 (3634) |
| GCN4p-c2 | −22.1 × 103 | 29 | 7400 (3688) |
| GCN4p16-31 | −12.2 × 103b | n.d. | 1900 (1882) |
| GCN4p2-17 | −1.9 × 103b | n.d. | 2200 (2008) |
| cI-p | −2.5 × 103b | n.d. | 1600 (1692) |
All fragments were analyzed in 5 mM sodium phosphate buffer (pH 7.4) containing 150 mM sodium chloride, except GCN4p16-31 and GCN4p2-17, which were analyzed in 1 mM sodium phosphate buffer (pH 7.4). The corresponding amino acid sequences of the fragments are shown in Figs. 2 and 3. n.d.: not determined.
[Θ]222 measured at 5°C at peptide concentrations (monomer) of 75 μM.
Determined at 3°C and at peptide concentrations of 30 μM.
Tm determined at chain concentrations (monomer) of 75 μM.
Average molecular masses were determined at 5°C. The sequence predicted molecular masses are in parentheses.
To characterize further the coiled-coil trigger sequence of the GCN4 leucine zipper, we analyzed a 16-residue peptide, denoted GCN4p16–31 (Fig. 3A), by CD spectroscopy (Fig. 3B) and AUC (Table 1). At 3°C, pH 7.4, and at low ionic strength the peptide revealed a CD spectrum with well-defined minima at 205 and 222 nm (Fig. 3B), indicating 40–50% helicity (26). Helix formation by GCN4p16–31 is a monomolecular process and is not the result of aggregation as judged from the lack of concentration dependence of [Θ]222 (data not shown) and AUC at different peptide concentrations (Table 1).
Figure 3.
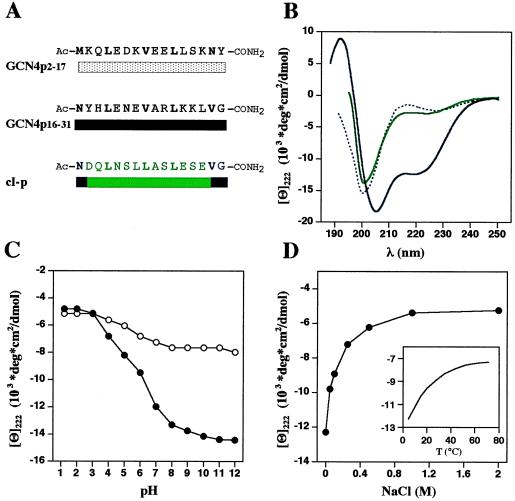
The GCN4 coiled-coil trigger sequence is an autonomous helical folding unit. (A) Synthetic peptide sequences used in this study. GCN4p2–17 and GCN4p16–31 represent N- and C-terminal fragments of the GCN4 leucine zipper. cI-p mainly corresponds to the two arbitrarily chosen cortexillin I control heptad repeats (see Fig. 2). (B) CD spectra of GCN4p16–31 (black solid line), GCN4p2–17 (black dashed line), and cI-p (green line) at 3°C. Peptide concentrations were 30 μM in 1 mM sodium phosphate buffer (pH 7.4). (C) pH dependence of [Θ]222 of GCN4p16–31 at 3°C (•) and 50°C (○) in the absence of sodium chloride. Peptide concentration was 30 μM in 1 mM sodium citrate, 1 mM sodium phosphate, 1 mM sodium borate buffer. (D) The effect of ionic strength (at 3°C) and temperature (without sodium chloride, Inset) on [Θ]222 of peptide GCN4p16–31. Peptide concentration was 30 μM in 1 mM sodium phosphate buffer (pH 7.4). Throughout the pH range and salt concentrations evaluated, helix formation by GCN4p16–31 remained strongly temperature dependent and occurred as a monomolecular reaction.
The helix content of GCN4p16–31 also has been evaluated as a function of pH and ionic strength (Fig. 3 C and D). Accordingly, the pH titration data presented in Fig. 3C indicate that one or both of the negatively charged Glu (residues 20 and 22) and the uncharged form of His-18 are required for stable helix formation. Consistent with this finding, an intrahelical ion pair between Glu-22 and Arg-25 (i, i+3) is apparent in the crystal structure of the GCN4 leucine zipper (10). GCN4p16–31 is most stable at basic pH, suggesting a repulsive intrachain electrostatic interaction between Arg-25 and Lys-28 (i, i+3). In addition, another potential attractive electrostatic interaction exists between Glu-20 and Lys-28 (i, i+8; observed in the N-terminal helix of the crystal structure of RNase A, ref. 24). However, it is difficult to interpret the peptide’s pH-dependence profile in terms of interacting residues, because in addition to potential ion-pair and salt-bridge (H-bonded ion-pair) interactions other pH-sensitive factors exist that may affect helix stability (27). Nevertheless, the strong ionic strength dependence of GCN4p16–31 supports the importance of side-chain interactions in helix stability (Fig. 3D). Screening of the possible charge-charge interactions by sodium chloride demonstrates that the net interaction between side chains is attractive and helix stabilizing.
Generally, peptides prepared from helix-containing segments of proteins are only marginally helical in aqueous solution (28). To assess whether peptides originating from coiled-coil oligomerization domains form significant monomer helices, we designed two 16-residue control peptides (Fig. 3A): GCN4p2–17 corresponds to the first two N-terminal heptad repeats of the GCN4 leucine zipper, and cI-p includes the arbitrarily chosen two-heptad repeat segment from cortexillin I (see Fig. 2A). As illustrated in Fig. 3B, at 3°C, pH 7.4, and at low ionic strength the CD spectra of both peptides were characteristic of randomly coiled proteins. Both spectra were neither significantly dependent on concentration nor sensitive to temperature and ionic strength (data not shown). In agreement with these results, AUC yielded molecular masses consistent with a monomeric structure of the two peptides (Table 1).
In conclusion, our findings (see also ref. 6) demonstrate that the trigger sequences of cortexillin I and GCN4 are functionally related with regard to their absolute necessity for coiled-coil formation. For the GCN4 leucine zipper, it has been established that the apolar residues occupying the a and d positions are mainly responsible for the stability of the coiled-coil structure (5). In this context, it is interesting to note that coiled-coil trigger sequences contain different types of amino acids at the d position of the second heptad repeat, including polar and charged residues (Fig. 1). Accordingly, the decreased thermal stability of the chimera GCN4p-Cort could be attributed to the Thr residue at the d position of the fourth heptad repeat (Fig. 2). However, the lack of coiled-coil formation of GCN4p-c1 is rather surprising because the trigger sequence has been replaced by an arbitrarily chosen two-heptad repeat segment from cortexillin I that is optimal in the sense that both heptads contain Leu residues at their a and d positions (Fig. 2). Hence, the failure of chain association of GCN4p-c1 cannot be explained by destabilizing residues at the hydrophobic core positions of the leucine zipper. Conversely, a stability of GCN4p-c1 comparable with GCN4p-wt is expected because Leu was found to be the most stabilizing aliphatic amino acid residue in the heptad d position of dimeric leucine zipper coiled coils (29). The finding of the existence of coiled-coil trigger sequences explains the frequent observation that even long stretches of heptad repeats, per se, are often not sufficient to mediate stable coiled-coil formation (6, 19, 30). This finding further indicates that coiled-coil domains must be considered as sequence-dependent cooperative units, a view that is supported by the two-state nature of the conformational transitions observed for many coiled coils. Hence, the 4.5 times larger size of the cortexillin I coiled-coil compared with the GCN4 leucine zipper also may explain why its trigger sequence tolerates a Thr residue at a heptad d position, which clearly destabilizes the coiled coil.
Our data on the 16-residue peptides comprising the functionally related coiled-coil trigger sequences of cortexillin I (6) and GCN4 (Fig. 3) are consistent with representing autonomous helical folding units (31). In contrast, two synthetic peptides comprising arbitrarily chosen heptad repeats revealed no secondary structure (Fig. 3B). Notably, trigger sequences, per se, do not dimerize, thus emphasizing that additional heptad repeats are required for stable coiled-coil dimer formation. This finding is consistent with recent studies reporting the minimum length required for the formation of stable coiled coils to be in the range of 21–23 residues (21, 32, 33). Indeed, Lumb et al. (21) observed that the GCN4p11–33 fragment, which is only slightly longer than our peptide GCN4p16–31 comprising mainly the trigger sequence, can associate into a helical dimer at 1 mM peptide concentration. Importantly, the attractive electrostatic intrachain interactions, which are most likely responsible for the observed stability of the monomer helices of GCN4p16–31 and cI-t (6), also are observed in the high-resolution structures of GCN4p1 (10) and cortexillin I (P. Burkhard, M.O.S., and R.A.K., unpublished work). What is less clear, however, is how exactly the helicity of the trigger sequences contributes to the formation and stability of the coiled coils.
Together with the kinetic data reported on coiled coils in the literature it appears reasonable to speculate from our peptide data that under physiological conditions some partial folding of the monomer species occurs before dimerization. Because it is known that helix nucleation is typically 10 times faster than the fastest loop closure reaction (34), it follows that incipient helices are already present when formation of the tertiary structure begins. Accordingly, as illustrated in Fig. 4i, our findings with GCN4p2–17 and GCN4p16–31 (Fig. 3) indicate the occurrence of a monomeric helical segment within the C-terminal half of the GCN4 leucine zipper. This proposal is consistent with recent work demonstrating two-state kinetic folding and unfolding of GCN4p1 and indicating that widespread helix is not formed within the monomer (35, 36). Furthermore, a recent NMR study on the thermal unfolding of GCN4p1 (37) revealed transient chemical shifts of Val-23 and Ala-24 within the trigger sequence of the unfolded monomer species in the range expected for residues being in a helical conformation. Notably, the presence of a helical segment within the monomer significantly limits the number of possible chain conformations and hence would provide an effective structural framework for the interaction of critical core residues. We therefore may speculate that stable dimer formation of GCN4 involves the interaction of two helical trigger sites at some stage in the folding pathway. As illustrated schematically in Fig. 4ii, such a mechanism ideally would align the dimer in parallel register. The interhelical g to e′ attractive ionic interaction between Glu-270 and Lys-275′ (10) might be important in this chain recognition and alignment process. Such a mechanism is supported by computer simulations on possible folding pathways of GCN4p1, for example, suggesting that dimer formation starts from the collision of short helical stretches, typically near or at the chain ends (38). However, the collision of monomers does not necessarily involve the coiled-coil trigger site because Sosnick et al. (36) reported a heterogeneous population of transition states for GCN4p1, suggesting several possible pathways for the folding of the leucine zipper dimer. Nevertheless, it has been demonstrated that if helices in proteins can be stabilized rapidly, then helix formation is expected to dominate the folding kinetics of helical proteins (39, 40). Finally, interacting helices then “zip up” along the molecule to form the stable coiled-coil structure (Fig. 4iii).
Figure 4.
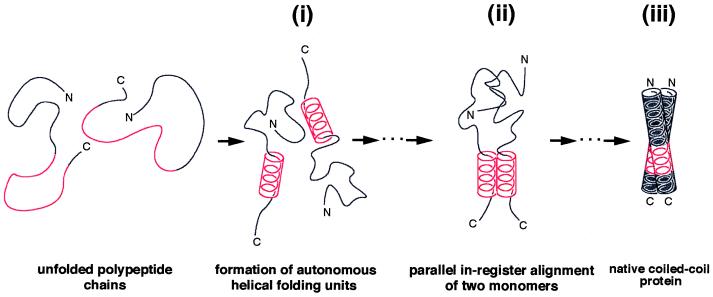
Proposed mechanism of coiled-coil formation in GCN4. Our peptide data (Fig. 3) indicate the presence of a short helical segment within the monomer (i). The fluctuating amphipathic helical stretch corresponds to the autonomous folding unit within the C-terminal half of the GCN4 leucine zipper monomer. Notably, the presence of a helical segment within the monomer significantly limits the number of possible chain conformations and would provide an ideal scaffold for the interaction of critical core residues. Hence, stable dimer formation may involve the interaction of two helical trigger sites at some stage in the folding pathway (ii). Interacting helices then “zip up” along the dimer to finally form the stable coiled-coil structure (iii). It should be noted that such a mechanism ideally would arrange the two-stranded coiled coil in parallel register.
Taken together, our findings on cortexillin I and GCN4 (Fig. 2) demonstrate that distinct trigger sequences within coiled coils represent autonomous helical folding units, which, in contrast to arbitrarily chosen heptad repeats, control chain assembly. Clearly, kinetic folding and unfolding measurements of rationally designed GCN4 leucine zipper mutants now will be necessary to more profoundly elucidate the role of the trigger sequence in coiled-coil formation and stabilization. Aside from being of general interest for protein folding, a detailed understanding of the structural and functional properties of trigger sequences should be of particular interest in the de novo design of proteins, as this area has become very important in recent years (7, 8).
Acknowledgments
We are indebted to Dr. Thomas Kiefhaber for helpful discussions. We thank Dr. David A. D. Parry for critical reading of the manuscript. We are grateful to Mr. Robert Wyss for figure preparation. This work was supported by grants from the Swiss National Science Foundation, the Maurice E. Müller Foundation of Switzerland, and the Canton Basel-Stadt.
ABBREVIATION
- AUC
analytical ultracentrifugation
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Cohen C, Parry D A D. Proteins. 1990;7:1–15. doi: 10.1002/prot.340070102. [DOI] [PubMed] [Google Scholar]
- 2.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 3.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 4.Kohn W D, Mant C T, Hodges R S. J Biol Chem. 1997;272:2583–2586. doi: 10.1074/jbc.272.5.2583. [DOI] [PubMed] [Google Scholar]
- 5.Harbury P B, Zhang T, Kim P S, Alber T. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 6.Steinmetz M O, Stock A, Schulthess T, Landwehr R, Lustig A, Faix J, Gerisch G, Aebi U, Kammerer R A. EMBO J. 1998;17:1883–1891. doi: 10.1093/emboj/17.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeGrado W F, Wasserman Z R, Lear J D. Science. 1989;243:622–628. doi: 10.1126/science.2464850. [DOI] [PubMed] [Google Scholar]
- 8.Hodges R S. Biochem Cell Biol. 1996;74:133–154. doi: 10.1139/o96-015. [DOI] [PubMed] [Google Scholar]
- 9.Crick F H C. Acta Crystallogr. 1953;6:689–697. [Google Scholar]
- 10.O’Shea E K, Klemm J D, Kim P S, Alber T. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 11.Sodek J, Hodges R S, Smillie L B, Jurasek L. Proc Natl Acad Sci USA. 1972;69:3800–3804. doi: 10.1073/pnas.69.12.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLachlan A D, Stewart M. J Mol Biol. 1975;98:293–304. doi: 10.1016/s0022-2836(75)80119-7. [DOI] [PubMed] [Google Scholar]
- 13.Faix J, Steinmetz M O, Boves H, Kammerer R A, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerisch G. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- 14.Dong H, Nilsson L, Kurland C G. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 15.Brandenberger R, Kammerer R A, Engel J, Chiquet M. J Cell Biol. 1996;135:1583–1592. doi: 10.1083/jcb.135.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 17.Kammerer R A, Schulthess T, Landwehr R, Lustig A, Fischer D, Engel J. J Biol Chem. 1998;273:10602–10608. doi: 10.1074/jbc.273.17.10602. [DOI] [PubMed] [Google Scholar]
- 18.van Holde K E. Physical Biochemistry. 2nd Ed. Englewood Cliffs, NJ: Prentice Hall; 1985. pp. 93–136. [Google Scholar]
- 19.Trybus K M, Freyzon Y, Faust L Z, Sweeney H L. Proc Natl Acad Sci USA. 1997;94:48–52. doi: 10.1073/pnas.94.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripet B, Vale R D, Hodges R S. J Biol Chem. 1997;272:8946–8956. doi: 10.1074/jbc.272.14.8946. [DOI] [PubMed] [Google Scholar]
- 21.Lumb K J, Carr C M, Kim P S. Biochemistry. 1994;33:7361–7367. doi: 10.1021/bi00189a042. [DOI] [PubMed] [Google Scholar]
- 22.Hu J C, O’Shea E K, Kim P S, Sauer R T. Science. 1990;250:1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- 23.Hodges R S, Semchuk P D, Taneja A K, Kay C M, Parker J M, Mant C T. Pept Res. 1988;1:19–30. [PubMed] [Google Scholar]
- 24.Wlodawer A, Sjölin L. Biochemistry. 1983;22:2720–2728. doi: 10.1021/bi00280a021. [DOI] [PubMed] [Google Scholar]
- 25.Gonzalez L, Jr, Woolfson D N, Alber T. Nat Struct Biol. 1996;3:1011–1018. doi: 10.1038/nsb1296-1011. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y-H, Yang J T, Chau K H. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- 27.Marqusee S, Baldwin R L. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muñoz V, Serrano L. Nat Struct Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 29.Moitra J, Szilák L, Krylov D, Vinson C. Biochemistry. 1997;36:12567–12573. doi: 10.1021/bi971424h. [DOI] [PubMed] [Google Scholar]
- 30.Holtzer M E, Crimmins D L, Holtzer A. Biopolymers. 1995;35:125–136. doi: 10.1002/bip.360350113. [DOI] [PubMed] [Google Scholar]
- 31.Shoemaker K R, Fairman R, Kim P S, York E J, Stewart J M, Baldwin R L. Cold Spring Harbor Symp Quant Biol. 1987;52:391–398. doi: 10.1101/sqb.1987.052.01.045. [DOI] [PubMed] [Google Scholar]
- 32.Fairman R, Chao H-G, Mueller L, Lavoie T B, Shen L, Novotny Y, Matsueda G R. Protein Sci. 1995;4:1457–1469. doi: 10.1002/pro.5560040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su J Y, Hodges R S, Kay C M. Biochemistry. 1994;33:15501–15510. doi: 10.1021/bi00255a032. [DOI] [PubMed] [Google Scholar]
- 34.Eaton W A, Muñoz V, Thompson P A, Chan C-K, Hofrichter J. Curr Opin Struct Biol. 1997;7:10–14. doi: 10.1016/s0959-440x(97)80003-6. [DOI] [PubMed] [Google Scholar]
- 35.Zitzewitz J A, Bilsel O, Luo J, Jones B E, Matthews C R. Biochemistry. 1995;34:12812–12819. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 36.Sosnick T R, Jackson S, Wilk R R, Englander S W, DeGrado W F. Proteins. 1996;24:427–432. doi: 10.1002/(SICI)1097-0134(199604)24:4<427::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 37.Holtzer M E, Lovett E G, d’Avignon D A, Holtzer A. Biophys J. 1997;73:1031–1041. doi: 10.1016/S0006-3495(97)78136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vieth M, Kolinski A, Brooks III C L, Skolnick J. J Mol Biol. 1994;237:361–367. doi: 10.1006/jmbi.1994.1239. [DOI] [PubMed] [Google Scholar]
- 39.Burton R E, Huang G S, Daugherty M A, Calderone T L, Oas T G. Nat Struct Biol. 1997;4:305–310. doi: 10.1038/nsb0497-305. [DOI] [PubMed] [Google Scholar]
- 40.Viguera A R, Villegas V, Aviles F X, Serrano L. Folding Design. 1997;2:23–33. doi: 10.1016/S1359-0278(97)00003-5. [DOI] [PubMed] [Google Scholar]



