Abstract
RNA interference (RNAi) acts constitutively to silence the innate immune response, and innate immunity genes are misregulated in Dicer-deficient Caenorhabditis elegans. Here, we show that inhibition of Dicer expression by RNAi in human cells up-regulates major histocompatibility complex class I–related molecules A and B (MICA and MICB). MICA and MICB are innate immune system ligands for the NKG2D receptor expressed by natural killer cells and activated CD8(+)T cells. We reveal that knockdown of Dicer elicits DNA damage. Up-regulation of MICA and MICB by Dicer knockdown is prevented by pharmacologic or genetic inhibition of DNA damage pathway components, including ataxia telangiectasia mutated (ATM) kinase, ATM- and Rad3-related kinase, or checkpoint kinase 1. Therefore we conclude that up-regulation of MICA and MICB is the result of DNA damage response activation caused by Dicer knockdown. Our results suggest that RNAi is indirectly linked to the human innate immune system via the DNA damage pathway.
Introduction
RNAi is important for regulation of heterochromatin structure and function (Bühler and Moazed, 2007; Locke and Martienssen, 2006). Loss of Dicer2, a key enzyme in the RNAi pathway, not only results in decondensation of heterochromatin but also leads to accumulation of extrachromosomal circular (ecc) repeated DNAs. Ligase IV, an essential regulator of nonhomologous end joining, and perhaps other DNA damage repair machinery, participate in ecc DNA formation (Peng and Karpen, 2007). This suggests that, in addition to increased accessibility of DNA repair and recombination proteins to repeated DNA caused by heterochromatin decondensation, activation of the DNA damage response may also contribute to the formation of ecc DNA in Dicer2 mutant cells. DNA replication timing is tightly regulated and correlates with chromatin state (Donaldson, 2005), and the timing of satellite DNA replication is misregulated in Dicer-deficient embryonic stem cells (Jørgensen et al., 2007). Stalled and collapsed replication forks elicit the DNA damage response (Sancar et al., 2004). In addition, RNAi is postulated to function as the genome's immune system that defends against molecular parasites such as transposons and viruses (Plasterk, 2002; Fire, 2005), and loss of Dicer may activate transposons, which in turn leads to DNA damage (Gasior et al., 2006). These observations collectively suggest that DNA damage response may be elicited in Dicer-deficient cells.
Innate immunity genes are misregulated in Caenorhabditis elegans deficient in Dicer, RDE-4, or RDE-1 (Welker et al., 2007). Mouse and human NKG2D ligands are up-regulated in nontumor cell lines by genotoxic stress and stalled DNA replication, conditions known to activate a major DNA damage checkpoint pathway initiated by ataxia telangiectasia mutated (ATM) or ATM- and Rad3-related (ATR) protein kinases (Gasser et al., 2005). Based on these observations, we hypothesize that loss of Dicer elicits a DNA damage response, which in turn induces expression of NKG2D ligands.
Results and discussion
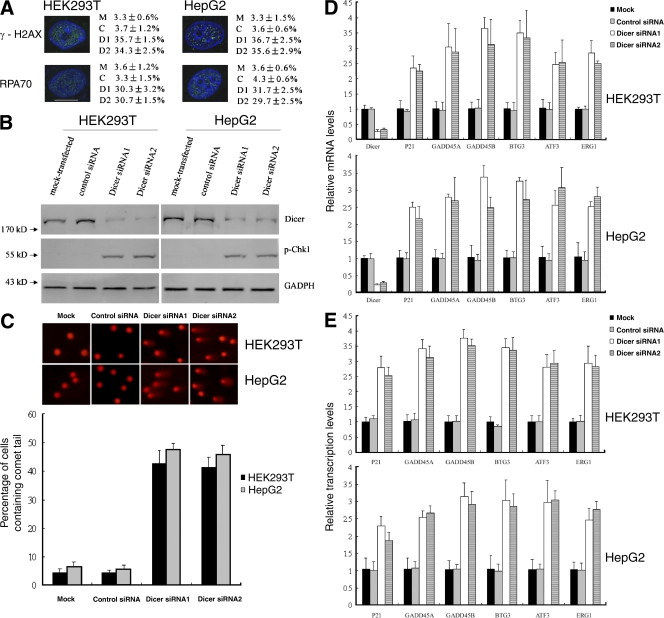
To test whether loss of Dicer causes DNA damage, we knocked down Dicer in HEK293T cells and human hepatoma HepG2 cells using two different siRNAs. The frequency of DNA double-strand breaks (DSBs) was measured by immunostaining for the phosphorylated form of histone H2AX (γ-H2AX), a widely used marker for DSBs (Foster and Downs, 2005). Compared with mock-transfected or control siRNA-transfected cells, intense γ-H2AX staining was observed in Dicer knockdown HEK293T cells. Quantification of the data revealed an ∼10-fold increase in the number of Dicer knockdown HEK293T cells positive for γ-H2AX compared with mock-transfected or control siRNA-transfected cells (Fig. 1 A). These results suggest that DSBs accumulate in the absence of Dicer. Similar results were obtained with HepG2 cells. To confirm these results, we used immunofluorescence in Dicer knockdown, mock-transfected, and control siRNA-transfected cells to examine the distribution of replication protein A 70 (RPA70), a protein involved in DNA replication, recombination, and repair, that becomes phosphorylated and forms intranuclear foci upon exposure of cells to DNA damage (Zou et al., 2006). Consistent with γ-H2AX staining, a much higher percentage of Dicer knockdown cells also displayed intense RPA foci, further indicating the accumulation of DNA damage (Fig. 1 A). Murchison et al. (2005) found that Dicer-deficient embryonic stem cells displayed only a slight increase in G1 and G0 cells and a corresponding decrease in cells in G2 and M phase. We also found that the cell cycle profile was not significantly affected by knocking down Dicer (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200801169/DC1), which excludes the possibility that increased γ-H2AX and RPA foci are simply the consequence of an increased percentage of cells in S phase. Furthermore, an increase in checkpoint kinase 1 (Chk1) phosphorylation on S345, an event associated with DNA damage, was found in Dicer knockdown cells (Fig. 1 B). As a more direct assessment of DNA damage, comet assay revealed that knockdown of Dicer resulted in accumulation of DNA breaks, as indicated by formation of a comet-like tail after single-cell gel electrophoresis (Fig. 1 C). Upon exposure of cells to DNA damage, growth arrest– and DNA damage–inducible gene α (GADD45A), growth arrest– and DNA damage–inducible gene β (GADD45B), p21, B cell translocation gene 3 (BTG3), activating transcription factor 3 (ATF3), and early growth response 1 (ERG1) are up-regulated (Fan et al., 2002; Thyss et al., 2005; Ou et al., 2007; Gévry et al., 2007). Real-time RT-PCR analysis indicated that the mRNA levels of these genes were increased in Dicer knockdown cells (Fig. 1 D). Loss of Dicer compromises miRNA biogenesis (Hutvágner et al., 2001; Murchison et al., 2005). miRNAs regulate the expression of target mRNAs by repressing translation and/or by promoting mRNA degradation (Behm-Ansmant et al., 2006), therefore, up-regulation of these genes may be the result of compromised miRNA biogenesis rather than the result of DNA damage. To test this possibility, we performed chromatin immunoprecipitation using anti-RNA polymerase II antibody (RNAPol-ChIP; Sandoval et al., 2004) to check transcription of these DNA damage–induced genes. Our results demonstrated that transcription of these genes was increased in Dicer knockdown cells (Fig. 1 E), which suggests that increased stability of these transcripts caused by reduction of miRNAs is not the only, if any, reason to explain their elevated mRNA levels. Real-time RT-PCR analysis revealed that the expression levels of IFN-β1, stat1, ISG15, IFIT1, IFIT2, OSA2, and UL16-binding protein (ULBP) 1, 2, and 3 were not up-regulated in Dicer knockdown cells compared with mock-transfected or control siRNA-transfected cells (Fig. S2 B, available at http://www.jcb.org/cgi/content/full/jcb.200801169/DC1; and not depicted), which excludes the possibility that heterochromatin decondensation induced by Dicer knockdown leads to general up-regulation of transcription.
Figure 1.
DNA damage accumulation in Dicer knockdown cells. (A) DNA damage assayed by immunostaining for γ-H2AX and RPA70. Mean ± SD indicates the fraction of γ-H2AX– or RPA-positive cells in mock-transfected (M), control siRNA-transfected (C), Dicer siRNA1-transfected (D1), and Dicer siRNA2-transfected (D2) cells. Bar, 10 μM. (B) Immunoblot analysis revealed an increase of Chk1 phosphorylation on S345 in Dicer knockdown cells. (C, top) Representative comet assay showing formation of DNA strand breaks (formation of a “comet tail”). (bottom) Mean ± SD indicates the fraction of cells containing a comet tail. (D) Levels of DNA damage–induced transcripts determined by real-time RT-PCR. (E) Transcription of DNA damage–induced genes assayed by RNAPol-ChIP. Data in D and E represent means ± SD from three independent experiments.
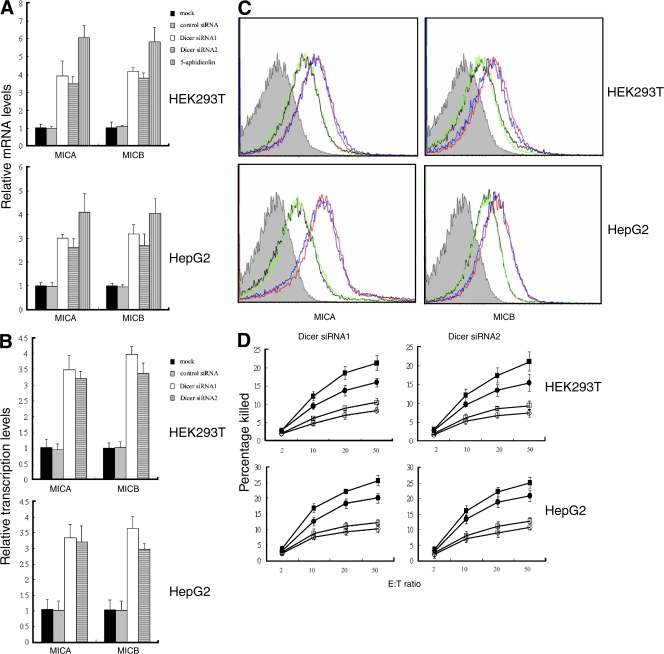
The DNA damage pathway regulates innate immune system ligands for the NKG2D receptor, and human NKG2D ligands are up-regulated by genotoxic stress and stalled DNA replication, conditions known to activate a major DNA damage checkpoint pathway (Gasser et al., 2005). Therefore, we tested whether NKG2D ligands were up-regulated in Dicer knockdown cells. Quantitative RT-PCR revealed markedly increased levels of major histocompatibility complex class I–related molecules A and B (MICA and MICB) mRNAs in Dicer knockdown cells compared with mock-transfected or control siRNA-transfected cells (Fig. 2 A), whereas the levels of ULBP1, -2, and -3 mRNAs were not significantly regulated by inhibiting the expression of Dicer (not depicted). Up-regulation of MICA and MICB was confirmed by flow cytometry (Fig. 2 C). In agreement with the up-regulation of MICA and MICB, Dicer knockdown cells exhibited greater sensitivity to lysis by NKL, a cell line derived from an aggressive form of human natural killer (NK) cell leukemia (Robertson et al., 1996). Lysis was partially inhibited by anti-NKG2D antibody, which indicated that up-regulated NKG2D ligands induce elevated lysis (Fig. 2 D). This result suggests that Dicer-deficient cells may be cleared by NK or other immune cells.
Figure 2.
Up-regulation of MICA and MICB in Dicer knockdown cells. (A) Up-regulation of MICA and MICB mRNAs determined by real-time RT-PCR. Cells treated with 4 μM aphidicolin for 16 h were used as positive control. (B) Transcription of MICA and MICB was monitored by RNAPol-ChIP. Data in A and B represent means ± SD from three independent experiments. (C) Cell surface expression of MICA and MICB measured by FACS. Dicer siRNA1-transfected (red) and Dicer siRNA2-transfected (blue) cells were compared with mock-transfected (black) and control siRNA-transfected (green) cells. Filled histograms indicate isotype control antibody staining. The x axis depicts staining intensity and the y axis depicts the relative number of cells. (D) Knockdown of Dicer sensitizes HepG2 and HEK293T cells to lysis by NKL cells. NKL cells were incubated with Dicer knockdown (squares) or control siRNA-transfected (circles) cells in the absence (closed) or presence (open) of anti-NKG2D antibody. The effector to target ratio (E:T) is indicated. Data represent means ± SD from three independent experiments.
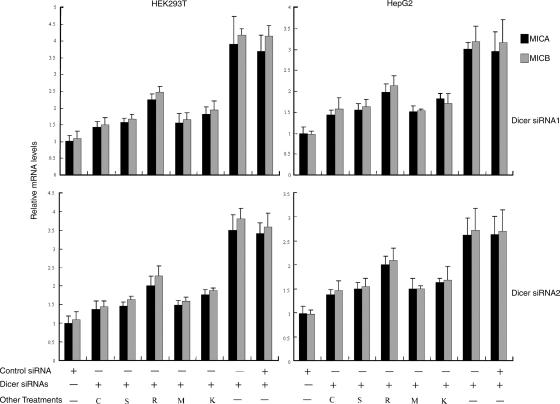
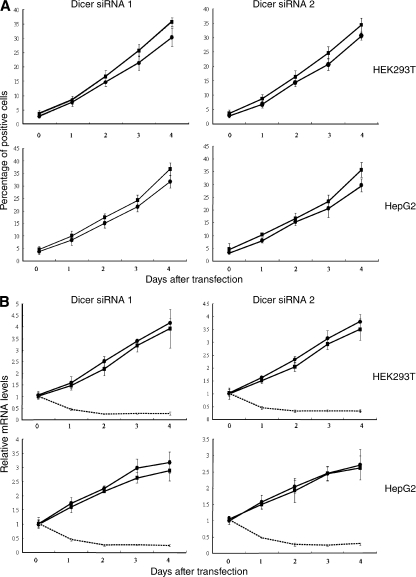
DNA damage–induced up-regulation of NKG2D ligands can be prevented by pharmacologic or genetic inhibition of ATR, ATM, or Chk1 (Gasser et al., 2005). The roles of ATR, ATM, and Chk1 in Dicer knockdown–induced MICA and MICB up-regulation were investigated. Up-regulation of MICA and MICB in response to Dicer knockdown was blocked by caffeine, an inhibitor of ATR and ATM, and by staurosporine, an inhibitor of Chk1 (Figs. 3 and S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200801169/DC1). As a more specific test, expression of ATM, ATR, and Chk1 was repressed using siRNAs (Fig. S3, E and F). Up-regulation of MICA and MICB induced by Dicer knockdown was inhibited by cotransfection of Dicer siRNAs with ATR siRNA (Figs. 3 and S3 B). In contrast, cotransfection with control siRNA did not significantly affect Dicer knockdown–induced up-regulation of MICA and MICB (Fig. 3). Similarly, inhibition of ATM and Chk1 expression by siRNAs also blocked MICA and MICB up-regulation in response to Dicer knockdown (Figs. 3 and S3, C and D). In addition, we demonstrated that DNA damage progressed in parallel with the kinetics of up-regulation of MICA and MICB in Dicer knockdown cells (Fig. 4). These data suggest that up-regulation of MICA and MICB is a consequence of DNA damage response activation. Compared with ATR suppression, ATM suppression had larger effects on MICA and MICB expression (Figs. 3 and S3, B and C), which suggests that MICA and MICB up-regulation is predominantly in response to DSBs.
Figure 3.
Inhibition of DNA damage response prevents MICA and MICB up-regulation in Dicer knockdown cells. Dicer knockdown-induced MICA and MICB up-regulation was prevented by incubation with caffeine (C) or staurosporine (S) and by cotransfection with ATR siRNA (R), ATM siRNA (M), or Chk1 siRNA (K). MICA and MICB mRNA levels were determined by real time RT-PCR, and were normalized to these in mock-transfected cells, which are defined as 1. Data represent means ± SD from three independent experiments.
Figure 4.
DNA damage progresses in parallel with the kinetics of up-regulation of MICA and MICB in Dicer knockdown cells. Cells were transfected with Dicer siRNAs. (A) γ-H2AX (▪) and RPA70 (•) staining was performed at the indicated time points. (B) Levels of MICA (▪), MICB (•), and Dicer (broken lines) transcripts were measured by real time RT-PCR. Means ± SD of three independent experiments are shown.
Some siRNAs are found to activate protein kinase R (PKR) and induce global up-regulation of IFN-stimulated genes (Sledz et al., 2003; Marques and Williams, 2005). This raises the question whether up-regulation of MICA and MICB is the consequence of nonspecific activation of the mammalian immune system by Dicer-specific siRNAs. To address this question, we checked the phosphorylation status of PKR. Western blotting demonstrated that there was no increase in PKR phosphorylation in Dicer knockdown cells compared with mock-transfected or control siRNA-transfected cells (Fig. S2 A). In addition, Dicer knockdown did not affect the expression of IFN-β1, stat1, ISG15, IFIT1, IFIT2, and OSA2 (Fig. S2 B), six genes nonspecifically induced by a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) siRNA due to activation of the IFN system (Sledz et al., 2003). Therefore, it is unlikely that up-regulation of MICA and MICB was an indirect consequence of nonspecific activation of the mammalian immune system by Dicer siRNAs. Although RNAPol-ChIP indicated that the transcription of MICA and MICB was increased in Dicer knockdown cells (Fig. 2 B), further studies are necessary to determine whether up-regulation of MICA and MICB is also the consequence of increased stability due to reduction of miRNAs.
Collectively, our results demonstrate that suppression of Dicer expression leads to cellular clearance via DNA damage, and the subsequent DNA damage response up-regulates the expression of MICA and MICB, which in turn induces the NK-mediated destruction. RNAi constitutes a key component of the innate immune response to viral infection in both plants and invertebrate animals (Cullen, 2006) and has been postulated to act constitutively to silence the innate immune genes (Welker et al., 2007). Our study suggests that RNAi is indirectly linked to the human innate immune system via the DNA damage pathway.
We propose the following model for how DNA damage is elicited in Dicer knockdown cells. Dicer is essential for heterochromatin formation and restricts the accessibility of DNA repair and recombination proteins to repeated DNA. Loss of Dicer leads to decondensation of heterochromatin and causes ecc DNA formation (Peng and Karpen, 2007), which may activate the DNA damage response. Furthermore, heterochromatin decondensation results in misregulation of DNA replication timing (Jørgensen et al., 2007) and causes DNA replication stress, which elicits DNA damage responses. In addition, heterochromatin decondensation may activate transposons and cause DNA damage (Gasior et al., 2006).
DNA damage activates cell cycle checkpoints that prevent progression through the cell cycle. However, Murchison et al. (2005) and our study found that the cell cycle profile was not significantly changed in Dicer-deficient cells (Fig. S1). This conflict can be reconciled by the fact that Ago1 binds to 14-3-3 proteins and inhibits nuclear import of the mitosis-inducing phosphatase Cdc25 (Stoica et al., 2006). Because Ago1 interacts strongly with Dicer (Doi et al., 2003), the latter may also interact with 14-3-3 proteins and repress nuclear import of Cdc25. Therefore, knockdown of Dicer, on one hand, induces DNA damage and activates Chk1, which in turn phosphorylates Cdc25. 14-3-3 proteins bind to phosphorylated Cdc25 and induce export of this phosphatase from the nucleus (Lopez-Girona et al., 1999). Nuclear exclusion of Cdc25 correlates with an inability to enter mitosis because the phosphatase no longer has access to its substrate, the Cdc13–Cdc2 complex, which is located in the nucleus (Lopez-Girona et al., 1999). On the other hand, knockdown of Dicer may facilitate nuclear import of Cdc25 and induce cells to enter mitosis. This scenario may also explain why dcr1 mutant Schizosaccharomyces pombe cells are defective for the DNA damage–induced cell cycle checkpoint (Carmichael et al., 2004).
Materials and methods
Cell culture
HepG2 and HEK293T cells were cultured in RPMI 1640 medium supplemented with 10% FBS. All cultures were maintained at 37°C in a moist atmosphere containing 5% CO2 in air. The NKL cell line (provided by M.J. Robertson, Indiana University School of Medicine, Indianapolis, IN), was grown in RPMI 1640 with 15% FBS and 200 U/ml interleukin 2.
RNAi
siRNAs were prepared by in vitro transcription with the Silencer siRNA Construction kit (Ambion). Target sequences of siRNAs were as follows: Dicer siRNA1 (AAGGCTTACCTTCTCCAGGCT), Dicer siRNA2 (AATTGGCTTCCTCCTGGTTAT), ATM siRNA (AACATACTACTCAAAGACATT), ATR siRNA (AACCTCCGTGATGTTGCTTGA), Chk1 siRNA (AAGCGTGCCGTAGACTGTCCA), and control siRNA (AATTCTCCGAACGTGTCACGT).
Transfection was performed using siPORT NeoFX (Ambion) according to the manufacturer's instructions.To maximally inhibit Dicer expression and keep the cells at optimal density, cells were transfected with two rounds of siRNAs. In brief, cells were split 2 d after the first round of Dicer siRNAs treatment and then treated again for another 2 d. To pharmacologically inhibit ATM, ATR, or Chk1, cells were treated with 2 mM caffeine (Alfa Aesar) or 8 nM staurosporine (Sigma-Aldrich) 24 h after the second round of Dicer siRNAs transfection. To genetically inhibit the DNA damage response, Dicer siRNAs were cotransfected with ATM, ATR, or Chk1 siRNAs, respectively.
Flow cytometry
Cell surface MICA and MICB were analyzed using mouse anti–human MICA and anti-MICB. In brief, cells were incubated with anti-MICA (MAB1300; R&D Systems), anti-MICB (MAB1599; R&D Systems), or the isotype control (MAB0041; R&D Systems) for 1 h at 4°C. Then cells were washed three times with PBS and incubated with phycoerythrin-conjugated goat anti–mouse IgG (F0102; R&D Systems) for 30 min at 4°C. Finally, the fluorescence was detected with FACSAria (BD Biosciences) and analyzed with WinMDI software (http://facs.scripps.edu/software.html). For cell cycle analysis, cells were fixed in cold ethanol, stained with 50 μg/ml propidium iodide, and analyzed by FACSAria.
Cellular cytotoxicity assay
HEK293T cells express several other activating ligands besides NKG2DL that are recognized by the various activating receptors on polyclonal NK cells (Welte, et al., 2003). Therefore, we used the NKL cell line, which has been well characterized for its NKG2D expression and NKG2D-mediated effector functions, to study NKG2D-mediated recognition of Dicer knockdown cells. Target cells were labeled with 100 μCi Na2 51CrO4 (GE Healthcare) for 2 h at 37°C and washed three times. In blocking experiments, anti-NKG2D (MAB139; R&D Systems) were added at 10 μg/ml to the target cells during the labeling procedure. Cells were washed, and effector cells were titrated on the target cells and incubated for 4 h at 37°C. Maximum release was determined from target cells lysed in 1% Triton X-100. The percentage of lysis was calculated as follows: 100 × (experimental release – spontaneous release)/(maximum release – spontaneous release). Experiments were performed in triplicate.
Western blotting
Cells were lysed in RIPA buffer. Total protein was measured using the Bradford protein assay. Samples were denatured at 100°C for 5 min. Equal amounts of total protein were loaded to each well for electrophoresis in SDS-polyacrylamide gels and then transferred to polyvinylidene fluoride microporous membranes (Millipore). Membranes were then incubated with primary antibody followed by incubation with horseradish peroxidase–linked secondary antibodies. The primary antibodies included anti-Dicer (ab14601; Abcam), anti-phospho-PKR (Thr 451; Cell Signaling Technology), anti-phospho-Chk1 (Ser345) antibody (Cell Signaling Technology), anti-Chk1 (Cell Signaling Technology), and anti-GAPDH (KC-5G4; Kangchen Biotech).
Immunofluorescence microscopy
Cells were fixed and stained with anti–phospho-histone H2AX (S139) antibody (R&D Systems) and anti-RPA70 (Cell Signaling Technology). Images were acquired with identical exposure conditions and analyzed with a confocal microscope (TCS-SP5 AOBS) using a 63× HCX PL APO 1.4 NA lens and LAS AF software (all from Leica). 100–300 cells were screened for each sample.
Quantitative real-time RT-PCR
Total RNA was prepared using TRIZOL (Invitrogen) and incubated with RNase-free Dnase I (Fermentas) for 30 min. The DNA-free RNA was reverse transcribed using MMLV reverse transcription (TOYOBO Bio-Technology) according to the manufacturer's instructions. Samples prepared without reverse transcription served as negative control templates. SYBR green PCR was performed in triplicate using the ABI PRISM 7300 Sequence Detection system (Applied Biosystems). All samples were normalized to the signal generated from GAPDH or 18S rRNA. Primer sequences (forward and reverse) were as follows: Dicer, TCCACGAGTCACAATCAACACGG and GGGTTCTGCATTTAGGAGCTAGATGAG; ATM, TGGATCCAGCTATTTGGTTTGA and CCAAGTATGTAACCAACAATAGAAGAAGTAG; ATR, TGTCTGTACTCTTCACGGCATGTT and AAGAGGTCCACATGTCCGTGTT; Chk1, GGTGAATATAGTGCTGCTATGTTGACA and TTGGATAAACAGGGAAGTGAACAC; MICA, CTTGGCCATGAACGTCAGG and CCTCTGAGGCCTCRCTGCG; MICB, ACCTTGGCTATGAACGTCACA, CCCTCTGAGACCTCGCTGCA; ULBP1, GTACTGGGAACAAATGCTGGAT and AACTCTCCTCATCTGCCAGCT; ULBP2, TTACTTCTCAATGGGAGACTGT and TGTGCCTGAGGACATGGCGA; ULBP3, CCTGATGCACAGGAAGAAGAG and TATGGCTTTGGGTTGAGCTAAG; P21, CCTCATCCCGTGTTCTCCTTT and GTACCACCCAGCGGACAAGT; GADD45B, GGGTGTACGAGTCGGCCAA and TGGCCAAGAGGCAGAGGA; GADD45A, TCAGCGCACGATCACTGTC and CCAGCAGGCACAACACCAC; ATF3, AAGAACGAGAAGCAGCATTTGAT and TTCTGAGCCCGGACAATACAC; ERG1, GCCTGCGACATCTGTGGAA and CGCAAGTGGATCTTGGTATGC; BTG3, ATATCGCCCAATTCCAGTGA and TCTCAACATGACACCAACACAA; OAS2, TCAGAAGAGAAGCCAACGTGA and CGGAGACAGCGAGGGTAAAT; Stat1, CCATCCTTTGGTACAACATGC and TGCACATGGTGGAGTCAGG; IFIT1, TCATCAGGTCAAGGATAGTCTG and GGTGTTTCACATAGGCTAGTAG; IFIT2, ACTGCAACCATGAGTGAGAAC and GCCTCGTTTTGCCCTTTGAG; IFNβ1, CAGCAATTTTCAGTGTCAGAAGC and TCATCCTGTCCTTGAGGCAGT; ISG15, TCATCCTGTCCTTGAGGCAGT and ACTCATCTTTGCCAGTACAGGAG; GAPDH, ATGACATCAAGAAGGTGGTG and CATACCAGGAAATGAGCTTG; and 18S rRNA, CGGCTACCACATCCAAGGAA and GCTGGAATTACCGCGGCT.
RNAPol-ChIP
Chromatin immunoprecipitation was performed as described by Sandoval et al. (2004). RNA pol II antibody (sc-899; Santa Cruz Biotechnology, Inc.) immunoprecipitated DNA was amplified with primer pairs in triplicate, and amplification was monitored using SYBR green chemistry on the ABI PRISM 7300 Sequence Detection system. Samples prepared without antibody served as negative controls. All samples were normalized to the signal generated from GAPDH or β-actin. Primers of GAPDH, ATF3, ERG1, BTG3, and P21 for real-time RT-PCR were also used for RNAPol-ChIP; other primer sequences (forward and reverse) were as follows: GADD45A, GGATCCTGCCTTAAGTCAACTTATTT and AAAACTTCAGTGCAATTTGGTTCA; GADD45B, TGAACTTGGTTGGTCCTTGTC and TATGCTTCCCATCTCGCTCT; MICA, GTCCTGGATCAACACCCAGT and AAGAGGGAAAGTGCTCGTGA; MICB, ACTTTCCCTCTGTTTCCTGAC and AGCAGTCGTGAGTTTGCC; and ACTB, TTCCTGGGCATGGAGTC and CAGGTCTTTGCGGATGTC.
Comet assay
Cells were harvested 4 d after transfection and resuspended in PBS. A low–melting point agarose (0.625%) suspension at 37°C was then added to the cell suspension at a ratio of 4:1 and immediately transferred to a slide precoated with 0.8% regular agarose. The cells on the slides were lysed with ice-cold high-salt lysis buffer (2.5 mol/liter NaCl, 100 mmol/liter EDTA, 10 mmol/liter Tris, pH 10, 1% Triton X-100, and 10% DMSO) for 1 h in dark at 4°C. Cells were then covered in alkali solution (1 mmol/liter EDTA, 300 mmol/liter sodium hydroxide, pH > 13, and 10 μg/ml proteinase K) for 20 min followed by electrophoresis (300 mmol/liter NaCl, and 100 mmol/liter Tris, pH 9) in the dark at 25 V, 300 mA (1 V/cm) for 15 min. The slides were neutralized using 400 mmol/liter Tris, pH 7.5 and washed with double-distilled H2O twice before being staining with 20 μg/ml EB. Comet images were visualized by using a fluorescence microscope (Eclipse TE 2000-S; Nikon) and NIS-Elements 2.3 software (Nikon).
Online supplemental material
Fig. S1 shows that Dicer knockdown does not affect the cell cycle profile. Fig. S2 shows that up-regulation of MICA and MICB is not the consequence of nonspecific activation of the mammalian immune system by Dicer-specific siRNAs. Fig. S3 shows that inhibition of the DNA damage response prevents MICA and MICB up-regulation in Dicer knockdown cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801169/DC1.
Supplementary Material
Acknowledgments
We thank Wei Sun, Liting Wang (Third Military Medical University), and Hua Zhao (Chongqing Medical University) for help with confocal analysis. We also wish to thank International Science Editing, Compuscript Ltd. for editing the manuscript.
This work was supported by the National Natural Science Foundation of China (grant Nos. 3070078 and 30400532), the Natural Science Foundation Project of the Chongqing Science and Technology Commission (grant No. 2007BB5307), and the Medical Science Foundation Project of Health Bureau of Chongqing Province (grant No. 06-2-083).
Abbreviations used in this paper: ATM, ataxia telangiectasia mutated; ATR, ATM- and Rad3-related; BTG3, B cell translocation gene 3; Chk1, checkpoint kinase 1; DSB, DNA double-strand break; ecc, extrachromosomal circular; ERG1, early growth response 1; GADD45, growth arrest– and DNA damage–inducible gene; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MICA, major histocompatibility complex class I–related gene A; MICB, major histocompatibility complex class I–related gene B; NK, natural killer; PKR, protein kinase R; RPA70, replication protein A 70; ULBP, UL16-binding protein.
References
- Behm-Ansmant, I., J. Rehwinkel, T. Doerks, A. Stark, P. Bork, and E. Izaurralde. 2006. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20:1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühler, M., and D. Moazed. 2007. Transcription and RNAi in heterochromatic gene silencing. Nat. Struct. Mol. Biol. 14:1041–1048. [DOI] [PubMed] [Google Scholar]
- Carmichael, J.B., P. Provost, K. Ekwall, and T.C. Hobman. 2004. ago1 and dcr1, two core components of the RNA interference pathway, functionally diverge from rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol. Biol. Cell. 15:1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. 2006. Is RNA interference involved in intrinsic antiviral immunity in mammals? Nat. Immunol. 7:563–567. [DOI] [PubMed] [Google Scholar]
- Doi, N., S. Zenno, R. Ueda, H. Ohki-Hamazaki, K. Ui-Tei, and K. Saigo. 2003. Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr. Biol. 13:41–46. [DOI] [PubMed] [Google Scholar]
- Donaldson, A.D. 2005. Shaping time: chromatin structure and the DNA replication programme. Trends Genet. 21:444–449. [DOI] [PubMed] [Google Scholar]
- Fan, F., S. Jin, S.A. Amundson, T. Tong, W. Fan, H. Zhao, X. Zhu, L. Mazzacurati, X. Li, K.L. Petrik, et al. 2002. ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene. 21:7488–7496. [DOI] [PubMed] [Google Scholar]
- Fire, A. 2005. Nucleic acid structure and intracellular immunity: some recent ideas from the world of RNAi. Q. Rev. Biophys. 38:303–309. [DOI] [PubMed] [Google Scholar]
- Foster, E.R., and J.A. Downs. 2005. Histone H2A phosphorylation in DNA double-strand break repair. FEBS J. 272:3231–3240. [DOI] [PubMed] [Google Scholar]
- Gasior, S.L., T.P. Wakeman, B. Xu, and P.L. Deininger. 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357:1383–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, S., S. Orsulic, E.J. Brown, and D.H. Raulet. 2005. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 436:1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévry, N., H.M. Chan, L. Laflamme, D.M. Livingston, and L. Gaudreau. 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21:1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner, G., J. McLachlan, A.E. Pasquinelli, E. Bálint, T. Tuschl, and P.D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 293:834–838. [DOI] [PubMed] [Google Scholar]
- Jørgensen, H.F., V. Azuara, S. Amoils, M. Spivakov, A. Terry, T. Nesterova, B.S. Cobb, B. Ramsahoye, M. Merkenschlager, and A.G. Fisher. 2007. The impact of chromatin modifiers on the timing of locus replication in mouse embryonic stem cells. Genome Biol. 8:R169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, S.M., and R.A. Martienssen. 2006. Slicing and spreading of heterochromatic silencing by RNA interference. Cold Spring Harb. Symp. Quant. Biol. 71:497–503. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature. 397:172–175. [DOI] [PubMed] [Google Scholar]
- Marques, J.T., and B.R. Williams. 2005. Activation of the mammalian immune system by siRNAs. Nat. Biotechnol. 23:1399–1405. [DOI] [PubMed] [Google Scholar]
- Murchison, E.P., J.F. Partridge, O.H. Tam, S. Cheloufi, and G.J. Hannon. 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 102:12135–12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, Y.H., P.H. Chung, F.F. Hsu, T.P. Sun, W.Y. Chang, and S.Y. Shieh. 2007. The candidate tumor suppressor BTG3 is a transcriptional target of p53 that inhibits E2F1. EMBO J. 26:3968–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J.C., and G.H. Karpen. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9:25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk, R.H. 2002. RNA silencing: the genome's immune system. Science. 296:1263–1265. [DOI] [PubMed] [Google Scholar]
- Robertson, M.J., K.J. Cochran, C. Cameron, J.M. Le, R. Tantravahi, and J. Ritz. 1996. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp. Hematol. 24:406–415. [PubMed] [Google Scholar]
- Sancar, A., L.A. Lindsey-Boltz, K. Unsal-Kaccmaz, and S. Linn. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73:39–85. [DOI] [PubMed] [Google Scholar]
- Sandoval, J., J.L. Rodríguez, G. Tur, G. Serviddio, J. Pereda, A. Boukaba, J. Sastre, L. Torres, L. Franco, and G. López-Rodas. 2004. RNAPol-ChIP: a novel application of chromatin immunoprecipitation to the analysis of real-time gene transcription. Nucleic Acids Res. 32:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz, C.A., M. Holko, M. J.de Veer, R.H. Silverman, and B.R. Williams. 2003. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 5:834–839. [DOI] [PubMed] [Google Scholar]
- Stoica, C., J.B. Carmichael, H. Parker, J. Pare, and T.C. Hobman. 2006. Interactions between the RNA interference effector protein Ago1 and 14-3-3 proteins: consequences for cell cycle progression. J. Biol. Chem. 281:37646–37651. [DOI] [PubMed] [Google Scholar]
- Thyss, R., V. Virolle, V. Imbert, J.F. Peyron, D. Aberdam, and T. Virolle. 2005. NF-kappaB/Egr-1/Gadd45 are sequentially activated upon UVB irradiation to mediate epidermal cell death. EMBO J. 24:128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker, N.C., J.W. Habig, and B.L. Bass. 2007. Genes misregulated in C. elegans deficient in Dicer, RDE-4, or RDE-1 are enriched for innate immunity genes. RNA. 13:1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, S.A., C. Sinzger, S.Z. Lutz, H. Singh-Jasuja, K.L. Sampaio, U. Eknigk, H.G. Rammensee, and A. Steinle. 2003. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 33:194–203. [DOI] [PubMed] [Google Scholar]
- Zou, Y., Y. Liu, X. Wu, and S.M. Shell. 2006. Functions of human replication protein A (RPA): from DNA replication to DNA damage and stress responses. J. Cell. Physiol. 208:267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.