Abstract
Turnover of actin filaments in cells requires rapid actin disassembly in a cytoplasmic environment that thermodynamically favors assembly because of high concentrations of polymerizable monomers. We here image the disassembly of single actin filaments by cofilin, coronin, and actin-interacting protein 1, a purified protein system that reconstitutes rapid, monomer-insensitive disassembly (Brieher, W.M., H.Y. Kueh, B.A. Ballif, and T.J. Mitchison. 2006. J. Cell Biol. 175:315–324). In this three-component system, filaments disassemble in abrupt bursts that initiate preferentially, but not exclusively, from both filament ends. Bursting disassembly generates unstable reaction intermediates with lowered affinity for CapZ at barbed ends. CapZ and cytochalasin D (CytoD), a barbed-end capping drug, strongly inhibit bursting disassembly. CytoD also inhibits actin disassembly in mammalian cells, whereas latrunculin B, a monomer sequestering drug, does not. We propose that bursts of disassembly arise from cooperative separation of the two filament strands near an end. The differential effects of drugs in cells argue for physiological relevance of this new disassembly pathway and potentially explain discordant results previously found with these drugs.
Introduction
Actin filaments turn over rapidly in cells, and their assembly through barbed end growth is thought to generate protrusive force. Rapid assembly must be balanced by equally rapid disassembly to replenish the monomer pool and allow remodeling of cell shape. Disassembly is arguably the least understood step in the actin dynamics cycle (Pollard and Borisy, 2003). We need to elucidate its mechanism to understand the morphogenesis of dynamic actin assemblies and answer the fundamental question of how polymerization dynamics transduce the energy of ATP hydrolysis into mechanical work. Physiological disassembly mechanisms must fulfill two requirements: (1) fast kinetics and (2) disassembly in a cytoplasm that contains high concentrations of polymerizable monomer (∼5–20 μM, estimated from Dabiri et al., 1990; Pollard, 1986). This monomer will tend to make exposed filament ends grow rather than shrink. Hydrolysis and phosphate release of polymer-bound nucleotide is thought to provide the thermodynamic drive for actin filament disassembly (Mitchison, 1992; Pollard and Borisy, 2003), but the mechanism by which this chemical energy is transduced into protein dynamics is not known. One widely discussed disassembly mechanism involves subunit dissociation from slow-growing pointed ends of filaments. This mechanism gives rise to treadmilling of individual filaments (Wegner, 1976). Another widely discussed mechanism involves severing (Pollard and Borisy, 2003), which increases the number of filament ends where subunit dissociation can potentially occur. A third plausible mechanism, subunit dissociation from fast-growing barbed ends, has been seldom discussed because barbed ends are thought to either grow or be capped (Marchand et al., 1995; Pantaloni et al., 2001).
We recently reconstituted a purified protein system consisting of cofilin, coronin, and actin-interacting protein 1 (Aip1) that catalyzes the disassembly of Listeria monocytogenes comet tails (Brieher et al., 2006), dense actin assemblies that structurally and biochemically resemble the leading edge of motile cells (Cameron et al., 2001). These three purified components together reconstitute disassembly that is both fast (τ < 100 s) and insensitive to polymerizable monomer (∼10 μM ATP-actin monomer). Cofilin belongs to a family of widely conserved, essential proteins that promote actin disassembly in many, perhaps all, cell types and structures (Maciver and Hussey, 2002). It preferentially binds ADP-actin filaments and presumably helps to couple the energy of ATP hydrolysis to actin turnover (Carlier et al., 1997; Blanchoin and Pollard, 1999). Paradoxically, cofilin can also promote actin assembly, presumably by providing new barbed ends to seed polymerization (Ichetovkin et al., 2002; Ghosh et al., 2004). Coronin binds actin filaments and also plays a role in physiological actin turnover (de Hostos et al., 1991; Foger et al., 2006; Cai et al., 2007b). Its mechanism of action is still controversial and may be isoform dependent (Cai et al., 2007a). We used coronin 1a, which was purified to homogeneity from calf thymus (Brieher et al., 2006). Aip1 works with cofilin to promote actin disassembly in a variety of organisms (Ono, 2001; Rogers et al., 2003; Ketelaar et al., 2004; Okada et al., 2006). It appears to interact with filament barbed ends (Okada et al., 2002; Balcer et al., 2003) and confer monomer sensitivity to the three-component disassembly reaction (Brieher et al., 2006). Although our previous work began to define roles for each factor in the purified protein system (Brieher et al., 2006), it did not determine how the full purified system promotes disassembly on a single-filament level. We address this question here.
Results
Actin filaments disassemble in bursts
We imaged the disassembly of single, fluorescently labeled actin filaments catalyzed by the three-component purified protein system consisting of cofilin, coronin, and Aip1. Concentrations of individual components were chosen to be those sufficient for full rate enhancement in L. monocytogenes comet tail disassembly (Brieher et al., 2006). All reactions also contained 5 μM of unlabeled actin monomer and 2 mM ATP, which reflects the physiological requirement that disassembly must occur in substantial concentrations of polymerizable monomer. We polymerized fluorescent-labeled actin filaments in a perfusion chamber using the actin cross-linker filamin to immobilize filaments to the coverslip surface. In all assays, filaments were polymerized for <1 min before initiation of disassembly. Filament aging leads to kinetic changes we will detail elsewhere (unpublished data). Newly assembled filaments were then disassembled by perfusion of the three-component system. Immediately after perfusion, a streaming video of actin filaments (acquisition time per image = 400 ms) was acquired by wide-field fluorescence microscopy (Fig. 1 A and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1).
Figure 1.
Actin filaments disassemble in bursts in cofilin, coronin, and Aip1. (A) Time-lapse wide-field epifluorescence images of fluorescently labeled actin filaments in the presence of 2 μM cofilin, 1 μM coronin, 200 nM Aip1, 5 μM of actin monomer, and 2 mM ATP. Filaments shorten and disappear from the field of view. Bar, 3 μm. (B) Successive time-lapse images showing a single actin filament (f1) over time, along with kymographs drawn along the contours of representative filaments (f1–f4). The red lines on the image of f1 at t = 0 denote the contour on which the kymograph was drawn. Time is given on the x axis of the kymograph, whereas the position along the filament contour is given on the y axis. Mean integration time for a single image was 400 ms for f1–f3 and 16 ms for f4. Triangles denote endwise bursting (f1–f3); yellow triangles denote initial burst (f1–f3), red triangles denote successive proximal bursts (f1 and f2), and green triangle denotes a successive distal burst (f3). Same-side bursts occurred more frequently (78%) than opposite-side bursts (22%; P < 0.001, one-tailed z test). The square denotes internal disassembly event counted as a severing event (f3). Bar, 1 μm. (C) Histogram of filament burst size. The mean burst size was 260 subunits. (D) Histogram of waiting times between successive bursts (red), fit to a single exponential (black). Single exponential fit gave characteristic decay time of τ = 14 s.
In the presence of cofilin, coronin, and Aip1, filaments shortened and disappeared from the field of view within the first 100 s (Fig. 1, A and B; and Video 1). We tracked the locations of filament ends using kymographs drawn along the contours of individual actin filaments (Fig. 1, B and C, filaments f1–f4). Kymographs showed that filaments did not shrink smoothly from an end; instead, they disassembled in infrequent bursts, with filaments abruptly losing mass from an end (Fig. 1 B, triangles). Most disassembly events occurred at filament ends, but events were occasionally initiated in the middle of the filament; these were scored as severing events (Fig. 1 B, blue square; see Fig. 2 for a detailed comparison of this reaction to a cofilin-mediated severing reaction). In an attempt to resolve molecular events within an endwise burst, the acquisition time per image was reduced to 16 ms using a high-speed camera (Fig. 1 B, filament f4). At this frame rate, kymographs were noisy, but disassembly still appeared primarily as bursts at ends. The abruptness of the burst argues against mechanisms involving successive dissociation of individual subunits from an end; the instantaneous off rate for such a mechanism would have to be ∼104 subunits/s.
Figure 2.
Filament bursting is distinct from cofilin-mediated severing. (A) Time-lapse images of two actin filaments disassembling in 10 μM cofilin, 5 μM of actin monomer, and 2 mM ATP. Long filaments were chosen to illustrate the occurrence of multiple severing events within a single filament. Bar, 1 μm. (B) Bar graph showing the fraction of disassembly events scored as bursting (red) or severing (green), either in the full depolymerizing system (left) or in cofilin alone (right). In the full system, bursting occurred with significantly higher frequency than severing (χ2 = 190, df = 1, P < 0.01). However, in the presence of high concentrations of cofilin, severing occurred with significantly higher frequency than bursting (χ2 = 55, df = 1, P < 0.01) and was the predominant disassembly mechanism observed under these conditions. We note that mean initial filament length did not differ significantly between different experiments and was not the cause of the differences observed.
Many filaments underwent multiple bursts of disassembly. Successive endwise bursts primarily occurred from the same end where the initial burst occurred (Fig. 1 B, filaments f1 and f2, red triangles), with a small fraction of bursts occurring from the opposite end as well (Fig. 1 B, filament f3, green triangle). The significantly higher frequency of same-side bursts (78%) versus opposite-side bursts (22%; n = 158, P < 0.001, 1-tailed z test) suggests either a favored end for disassembly or vectorial propagation of an instability inward from either end. Endwise bursts had a mean size of 260 subunits, and the time between successive bursts from the same end was exponentially distributed with a characteristic decay time of 14 s (Fig. 1, C and D).
The abrupt bursts of filament disassembly catalyzed by cofilin, coronin, and Aip1 appeared quite different from a classical cofilin-mediated severing reaction. To directly compare these mechanisms, we repeated the imaging experiments using cofilin alone at 10 μM, 10-fold higher than the concentration of cofilin used in the three-component system. Previous studies have also found that cofilin efficiently severed filaments at this concentration (Maciver et al., 1991). We found that a cofilin-alone severing reaction showed no preference for filament ends and generated two detectable daughter filaments after each event (Fig. 2 A). To quantitatively compare the disassembly reaction catalyzed by a three-component system to the cofilin-alone reaction, we scored disassembly events under both conditions and counted them as either bursting or severing events. For scoring purposes, a severing event was one in which both daughter filaments remained attached to the surface of the coverslip for at least two frames after breakage. Such an analysis revealed that, although filaments in the three-component system underwent occasional internal disassembly events that were scored as severing events (Fig. 1 B, f3, square), bursting from ends was the predominant mode of disassembly (Fig. 2 B). In contrast, severing was the predominant mode of disassembly catalyzed by cofilin alone (Fig. 2 B), which is consistent with previous studies (Maciver et al., 1991; Andrianantoandro and Pollard, 2006). Our cofilin-alone data reinforce our conclusion that bursting disassembly catalyzed by cofilin, coronin, and Aip1 is a novel mechanism of filament disassembly.
Actin filaments are structurally polarized, so we next tested whether the bursts of disassembly occur preferentially from a particular end of the filament. To separately probe disassembly near barbed ends and pointed ends, we imaged disassembly of filaments elongated from fragments of Limulus polyphemus,acrosomal processes, which are highly bundled arrays of filaments all oriented in the same direction (Bonder and Mooseker, 1983). Filaments grown off the barbed end of acrosomal bundles have free barbed ends but pointed ends that are connected to the acrosome. The reverse applies at the pointed end. We polymerized bundles of filaments off the barbed ends and pointed ends of L. polyphemus acrosomes using 5 μM of fluorescently labeled actin monomer and 2 mM ATP and then perfused them with cofilin, coronin, and Aip1, as well as unlabeled actin monomer and ATP. To minimize filament aging effects, filaments were perfused within 2 min after initiation of polymerization. Interestingly, filament bundles grown from either barbed ends (Fig. 3 A, b) or pointed ends (Fig. 3 A, p) of acrosomal processes disassembled at comparable rates (Fig. 3 A). Most of the polymer mass in the filament bundles disappeared within the first 2 min; the remainder, which manifest as a short stub of filaments at the acrosomal barbed end, disassembled with considerably slower kinetics (Fig. S1 and Video 2, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1). To quantify the fast component of disassembly, we first subtracted out the slowly varying component of the decay curve (see Materials and methods). Bundles with either exposed barbed ends or exposed pointed ends exhibited similar polymer mass decay curves and inferred decay rate constants (Fig. 3, B and C). These results suggest that bursting does not occur preferentially from a particular end of the filament but can proceed vectorially from either end. We also note that in our single-filament assay, internal filament disassembly events catalyzed by cofilin, coronin, and Aip1 frequently gave rise to two new filament ends that both underwent bursting disassembly (unpublished data), which is consistent with both ends undergoing bursting disassembly at similar rates.
Figure 3.
Actin filaments disassemble with similar kinetics from both ends. (A) Time-lapse images of actin filament bundles grown off fragments of L. polyphemus acrosomal processes. Filaments in the long bundle have exposed barbed ends (b), whereas filaments in the short bundle have exposed pointed ends (p). The two bundles shown were elongated from opposite ends of the same L. polyphemus acrosomal fragment. The brightness of the shorter bundle was increased relative to the longer one for ease of visualization. At 0 s, filament bundles were perfused with 2 μM cofilin, 1 μM coronin, 300 nM Aip1, 5 μM of actin monomer, and 2 mM ATP. Bar, 1 μm. (B) Total actin polymer mass in the filament bundles as a function of time measured for all bundles (red), bundles with exposed barbed ends (green), and bundles with exposed pointed ends (blue). To compare the rates of rapid disassembly, the slowly varying component of each decay curve was removed (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1; see Materials and methods). (C) Bar graph comparing decay rates for bundles with exposed barbed ends (b) with those with exposed pointed ends (p). Data represent mean and standard deviation of three independent experiments.
Bursting disassembly generates barbed ends not recognized by CapZ
The disassembly pathway catalyzed by cofilin, coronin, and Aip1 may involve reaction intermediates with structurally altered filament ends. To test this hypothesis, we probed the barbed ends of filaments using fluorescently labeled CapZ, a protein that caps standard filament barbed ends with fast kinetics and high affinity (Caldwell et al., 1989; Schafer et al., 1996). We polymerized filament bundles from L. polyphemus acrosomal fragments, treated them under different depolymerizing conditions, and then perfused them with 3 μM Alexa 488 CapZ for 10 s. This CapZ incubation was sufficient to completely cap all standard barbed ends (unpublished data), which is consistent with previous kinetic studies (kon ∼3 μM−1 s−1 from Schafer et al., 1996). When filament bundles were incubated with CapZ in the absence of disassembly, they recruited CapZ at their tips, which is consistent with all filaments recruiting CapZ to their barbed ends (Fig. 4 A, middle). When filament bundles were partially disassembled with cofilin alone before CapZ incubation, their fluorescence intensity tapered toward the end of the bundle, which reflects shortening of filaments due to severing (Fig. 4 B, left). Cofilin-treated filament bundles recruited CapZ not only at their tips but also along their lengths (Fig. 4 B, middle), which is consistent with binding of CapZ to barbed ends of severed filaments. To quantify the fraction of capped filaments before and after cofilin treatment, we measured for individual filament bundles the CapZ/filament ratio, which reports on the proportion of capped filaments (see Materials and methods). This analysis showed that cofilin-treated filament bundles showed no significant decrease in the fraction of capped filaments compared with untreated bundles (Fig. 4, A and B, right; 1-tailed t test, t = 1.6, degrees of freedom [df] = 14, P > 0.01). We conclude that filaments severed by cofilin retain the ability to bind CapZ at their barbed ends, as expected.
Figure 4.
Bursting disassembly generates barbed ends not recognized by CapZ. Fluorescence images show Alexa 647 actin (left) and Alexa 488 CapZ (right) in filament bundles. Barbed ends are oriented toward to the right. The bar graph shows the CapZ/actin filament ratio, a measure of the fraction of capped barbed ends in the bundle. Data represent mean and standard deviation of eight filament bundles. (A) Filament bundles polymerized from L. polyphemus acrosomal fragments were incubated directly with Alexa 488 CapZ. (B) Filament bundles were partially disassembled with 8 μM cofilin for 90 s then incubated with CapZ. (C) Filament bundles were partially disassembled with 2.5 μM cofilin, 1.5 μM coronin, and 50 nM Aip1 for 25 s, then incubated with CapZ. (D) Filament bundles were partially disassembled with cofilin, coronin, and Aip1 as in C, incubated in buffer for 10 s, then incubated with CapZ. Bar, 1 μm.
When filament bundles were partially disassembled with cofilin, coronin, and Aip1 before CapZ incubation, their fluorescence also tapered due to disassembly (Fig. 4 C, left). However, unlike cofilin-treated filament bundles, these bundles recruited almost no CapZ (Fig. 4 C, middle). The CapZ/filament ratio for bundles treated with cofilin, coronin, and Aip1 was much lower than that for untreated bundles (Fig. 4, A and C, right; 1-tailed t test, t = 11.1, df = 22, P < 0.01) and not significantly different from zero (Fig. 4 C, right; t test, t = 2.3, df = 15, P > 0.01). A similar decrease in CapZ recruitment was also seen when CapZ was incubated concurrently with cofilin, coronin, and Aip1 (unpublished data). These results suggest that, unlike a pure severing reaction, bursting disassembly by the three-component system generates as reaction intermediates barbed ends that are not recognized by CapZ. To determine whether these barbed ends were stable in buffer, we partially disassembled filament bundles with cofilin, coronin, and Aip1, then incubated them in buffer for 10 s before incubation with CapZ. These filament bundles recruited significantly more CapZ than bundles treated directly with CapZ (Fig. 4, C and D; 1-tailed t test, t = 4.8, df = 22, P < 10−4), which suggests that, in the absence of disassembly factors, nonstandard barbed ends rapidly revert to a standard CapZ-recognizable form. Consistent with this interpretation, others have observed that cofilin and Aip1 do not inhibit filament elongation from barbed ends when washed out (Ono et al., 2004). These data suggest that bursting disassembly generates unstable filament intermediates with nonstandard barbed ends not recognized by CapZ.
Barbed end–capping agents inhibit actin disassembly in vitro
Our experiments with fluorescent CapZ argue against a disassembly mechanism where depolymerization factors form a stable cap with filament barbed ends to prevent monomer addition and filament annealing. They instead suggest that bursting disassembly may involve active disruption of filament internal structure, leading to ragged barbed ends that cannot recruit CapZ. To test these ideas further, we imaged single filaments after perfusion with cofilin, coronin, and actin monomer with or without Aip1, and in the presence of CapZ or cytochalasin D (CytoD), a well-characterized barbed end–capping drug.
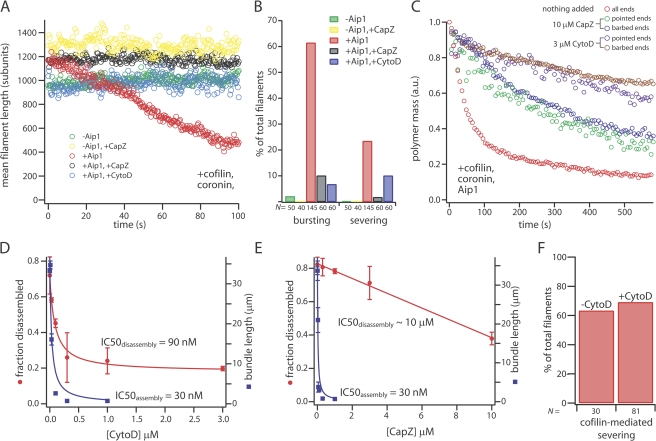
Omission of Aip1 from the three-component system stopped disassembly (Fig. 5 A, green) and greatly reduced the frequency of both bursts and scored severing events (Fig. 5 B, green). Omission of either cofilin or coronin had the same effect (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1), which is consistent with a requirement for all three factors for rapid monomer-insensitive disassembly in L. monocytogenes comet tails (Brieher et al., 2006). CapZ could not substitute for Aip1 in the three-component reaction (Fig. 5, A and B, yellow), which suggests a role for Aip1 in disassembly that is distinct from barbed-end capping. When added to the full three-component system, both CytoD and CapZ strongly inhibited filament disassembly (Fig. 5 A, black and blue), blocking both endwise bursting and internal severing modes of filament disassembly (Fig. 5 B, black and blue). To separately test the effect of CytoD and CapZ on disassembly from filament barbed ends and pointed ends, we imaged acrosomal filament bundles in the presence of the full three-component system and either CytoD or CapZ (Fig. 5 C). Remarkably, disassembly of filaments grown off barbed ends and pointed ends were inhibited with equal efficacy, which suggests that filament segments near both ends disassemble through the same CytoD/CapZ-sensitive mechanism (Fig. 5 C). CytoD inhibited filament bundle disassembly with an IC50 of 90 nM (measured using total polymer mass from all filament bundles in a field of view), which is similar to its IC50 for inhibition of polymerization off acrosomal fragments (30 nM; Fig. 5 D). In contrast, although CapZ also inhibited polymerization off acrosomal fragments with a low IC50 (30 nM; Fig. 5 E), its inhibitory effects on disassembly did not saturate at 10 μM (Fig. 5 E). The difference in IC50s between CytoD- and CapZ-mediated inhibition of disassembly suggests differences in the detailed mechanism between these two barbed end–capping agents. For example, barbed ends generated during disassembly may have a conformation that is recognized by CytoD but not by CapZ. Regardless of the detailed mechanism, their common inhibitory effects suggest an important role for barbed end–capping agents in controlling filament stability in the three-component system, even when disassembly occurs by bursting from the pointed end. In contrast, CytoD did not inhibit filament severing in high concentrations of cofilin alone (Fig. 5 F), which is consistent with others' results (Ono et al., 2004).
Figure 5.
Barbed end–capping factors inhibit actin disassembly by cofilin, coronin, and Aip1. (A) Mean filament length over time under the following conditions: 2 μM cofilin, 1 μM coronin, 5 μM of actin monomer, and 2 mM ATP, as well as 200 nM Aip1 (red), no Aip1 (green), 5 μM CapZ (yellow), 200 nM Aip1 + 1 μM CytoD (black), or 200 nM Aip1 + 5 μM CapZ (blue). Fast monomer-insensitive disassembly required Aip1 (red vs. green, yellow). CytoD and CapZ inhibited disassembly in the full system (black, blue vs. red). (B) Bar graph showing the percentage of filaments that underwent endwise bursting or severing. The number of filaments analyzed is given below the bars. The percentages of filaments that either underwent endwise bursting or severing increased significantly in the presence of Aip1 (red vs. green, yellow) and decreased significantly when the barbed-end cappers CytoD and CapZ were added to the full system (black, blue vs. red; severing frequency of −CapZ vs. +CapZ, χ2 = 4.9, df = 1, P < 0.05; all other pairwise comparisons, χ2 > 40, df = 1, P < 0.001). (C) Polymer mass decay for bundles with either exposed barbed ends or pointed ends in 3 μM CytoD (brown and purple) or 10 μM CapZ (blue and green), as well as polymer mass decay for all filament bundles in the absence of barbed-end cappers (red). Disassembly of bundles with either exposed barbed ends or pointed ends were inhibited by CytoD/CapZ with equal efficacy. (D) Fraction of polymer mass disassembled as a function of CytoD concentration (red circles). Best fit of the data to a hyperbola (red curve) shows an IC50 of 90 nM for inhibition of disassembly. Length of filament bundles polymerized for a fixed period of time in the presence of varying concentrations of CytoD is also shown (blue squares). Hyperbolic best fit (blue curve) shows an IC50 of 30 nM for inhibition of polymerization. (E) Fraction of polymer disassembled as a function of CapZ concentration (red squares). The titration curve did not reach saturation; the red curve denotes best fit of the data to a straight line. The length of filament actin bundle polymerized in the presence of varying concentrations of CapZ is also shown (blue circles). Polymerization conditions were identical to those in D. Hyperbolic best fit (blue curve) gave an IC50 of 30 nM for inhibition of polymerization. Error bars indicate SD. (F) CytoD does not inhibit cofilin-mediated severing. The bar graph shows the percentage of filaments that severed during a 400 s video either in the absence of CytoD (left) or in the presence of 1 μM CytoD (right). Conditions: 8 μM cofilin and 5 μM of actin monomer in assay buffer.
Barbed end–capping agents inhibit actin disassembly in cells
To determine whether physiological actin disassembly also occurs by a mechanism that is inhibited by barbed end–capping agents, we treated mammalian tissue culture cells with CytoD and measured disassembly by several different assays. As a control, we used latrunculin B (LatB), a drug that also inhibits polymerization but does so by sequestering a free monomer. We first used a simple bulk assay for total cellular F-actin. Human HeLa S3 cells grown in suspension were treated with either 10 μM CytoD or LatB, fixed at various time points after treatment, and then stained with TRITC-phalloidin to quantify bulk F-actin levels. Before drug treatment, F-actin was present throughout the entire cell and was enriched at the cell cortex (Fig. 6 A, top). Treatment of the cells with CytoD did not reduce the amount of F-actin per cell (Fig. 6 A, middle; and Fig. 6 B, blue). The TRITC-phalloidin signal remained high for >30 min after CytoD treatment. The drug was active in these cells as it induced extensive blebs after a few minutes (unpublished data). In contrast, treatment of cells with LatB led to a rapid drop in the amount of F-actin per cell (Fig. 6 A, bottom; and Fig. 6 B, red). The TRITC-phalloidin signal dropped by nearly threefold over the first 100 s, which implies a timescale of disassembly consistent with fluorescence recovery after photobleaching studies of cortical actin turnover (Murthy and Wadsworth, 2005) and our data with the purified protein system. The differential responses of cellular F-actin levels to CytoD and LatB treatment provides evidence that CytoD also actively inhibits actin filament disassembly in vivo, as it also does in our purified protein system in vitro.
Figure 6.
Barbed end–capping drugs inhibit actin disassembly in mammalian tissue-culture cells. (A) Images of suspension HeLa S3 cells showing F-actin stained with TRITC-phalloidin (top) and DNA stained with Hoechst (bottom). Images show untreated cells (left), cells treated with 10 μM CytoD for 480 s (middle), and cells treated with 10 μM LatB for 480 s (right). All images used identical acquisition settings and contrast levels. Bar, 10 μm. (B) The level of F-actin per cell at different times after treatment with either CytoD (blue) or LatB (red). F-actin levels remained high after CytoD treatment. In contrast, F-actin levels fell rapidly upon LatB treatment. Each point represents the mean and SD of at least 10 fields of cells. (C) Images of actin comet tails in L. monocytogenes–infected BSC-1 cells taken at successive times after drug treatment. In the absence of any drugs (control), comet tails disassembled within 50 s. Upon LatB treatment, comet tails also disassembled within 50 s, which suggests that there is no effect of LatB on comet tail disassembly kinetics. In contrast, comet tails did not disassemble in CytoD- and KabC-treated cells for at least 50 s after drug addition, which implies that CytoD and KabC inhibited comet tail disassembly. All drugs were added at time 0. Bar, 5 μm. (D) Curves showing decay of actin polymer mass from L. monocytogenes comet tails. In the absence of drugs, actin disassembled at an initial rate of kcontrol = (2.3 ± 0.2) × 10−2s−1. LatB did not change the disassembly rate, with kLatB = (2.6 ± 0.2) × 10−2. In contrast, CytoD reduced the L. monocytogenes comet tail decay rate to kCytoD = (1.9 ± 6) × 10−4s−1. Data represent the mean of polymer mass in multiple tails (control, 27 tails; CytoD treatment, 40 tails; LatB treatment, 27 tails). Disassembly rates were obtained by fitting the initial slope of the decay curve to a straight line.
To probe disassembly on a well-characterized actin array in vivo, we imaged actin comet tails formed by the intracellular pathogen L. monocytogenes (Sanger et al., 1992; Theriot et al., 1992) while perturbing cells with CytoD or LatB. To control for secondary effects of CytoD, we also used in this assay kabiramide C (KabC), a barbed end–capping drug structurally unrelated to CytoD (Tanaka et al., 2003). We recorded time-lapse fluorescence videos of L. monocytogenes actin comet tails inside mammalian tissue culture cells expressing GFP-actin. Before drug addition, actin comet tail assembly propelled L. monocytogenes forward in the cytoplasm. Comet tail actin disassembled within a minute after assembly (Fig. 6 C, control; and Fig. 6 D, green; Theriot et al., 1992), causing a decrease in filament density away from the bacterium. Both CytoD and KabC stopped comet tail assembly and L. monocytogenes motility within a few seconds after addition (Fig. 6 C), which is consistent with the drugs capping barbed ends to inhibit actin polymerization. Both CytoD and KabC also inhibited disassembly of comet tails, which showed no visible decrease in fluorescence for up to a minute after drug addition (Fig. 6, C and D, blue; and Videos 3 and 4, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1). Cell rounding induced by both drugs hindered longer observation; however, tails were still visible in rounded-up cells up to 10 min after CytoD addition, which implies a reduction in the disassembly rate by at least 20-fold. Although LatB also rapidly stopped tail assembly and L. monocytogenes movement, it did not inhibit disassembly. Comet tails disappeared after drug addition at a rate similar to that in untreated cells (Fig. 6, C and D, red; and Video 5). Others have previously noted that comet tails are stable in CytoD-treated cells (Sanger et al., 1992) but did not attribute this observation to inhibition of disassembly.
Lamellipodia and other dynamic cellular actin assemblies contain dense dendritic arrays resembling those seen in L. monocytogenes comet tails (Sechi et al., 1997; Cameron et al., 2001). To test if these arrays show similar sensitivities to CytoD versus LatB with respect to disassembly, we imaged the ruffling edge of BSC-1 cells stably expressing a fusion of actin to monomeric RFP and photoactivatable GFP (mRFP-PAGFP-actin; Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1). PAGFP allows localized illumination of specific actin subpopulations, and the in-tandem mRFP permits simultaneous viewing of the bulk actin population. Like GFP-actin, this probe localized to actin-rich cellular structures like the ruffling cell edge and stress fibers (Fig. 7 A). During the experiments, we imaged both red and green fluorescence and compared actin dynamics before and after drug treatment both in bulk and in photoactivated subpopulations. Before drug treatment, a dense actin assembly was present at the ruffling cell edge (Fig. 7, A and D, red). Consistent with existing turnover data (Wang, 1985; Watanabe and Mitchison, 2002; Ponti et al., 2004), this assembly was highly dynamic; photoactivated subpopulations disappeared within 50 s after photoactivation (Fig. 7, A and D, green; and Video 6). After LatB treatment, the bulk actin assembly disappeared within 50 s (Figs. 7 B and 6 E, red). A subpopulation of the assembly photoactivated immediately after drug addition decayed with kinetics similar to those in bulk (Fig. 7, B and E, green). These results are consistent with LatB's function as a monomer-sequestering agent that stops polymerization without affecting depolymerization. In contrast, CytoD stabilized the bulk actin assembly at the cell edge, stopping ruffling and rendering the assembly much less dynamic (Fig. 7 C and F, red; and Video 6). The bulk assemblies remained static for >100 s, which reflects simultaneous inhibition of polymerization and depolymerization. Subpopulations of the assemblies photoactivated after CytoD treatment persisted for >100 s (Fig. 7, C and F, green), which suggests that CytoD actively inhibits disassembly and does not simply promote local nucleation or accumulation of actin. The similar inhibitory effect of barbed end–capping factors on observed cellular actin assemblies and on single filaments in vitro suggests that the three-component system reconstitutes the physiological mechanism of actin disassembly in L. monocytogenes comet tails, the leading edge, and the bulk cortex of round cells.
Figure 7.
Barbed-end cappers inhibit disassembly of dynamic actin at the ruffling cell edge. (A–C) Time-lapse images of ruffling edges in BSC-1 cells expressing mRFP-PAGFP-actin taken before CytoD addition (A), after LatB addition (B), or after CytoD addition (C). The red channel shows bulk actin and the green channel shows photoactivated fluorescence. Images were taken before activation (left), immediately after activation (middle), and 100 s after activation (right). (D–F) Kymographs taken along the lines shown in A, B, and C, respectively. The color kymograph shows bulk actin in red and photoactivated actin in green. The black and white kymograph shows photoactivated actin alone. Arrows show the time of CytoD addition (D and F) or LatB addition (E). (A and D) Before drug treatment, photoactivated subpopulations of actin decayed within 100 s. (B and E) LatB did not inhibit actin disassembly. Both bulk and photoactivated subpopulations of actin disappeared within 100 s. (C and F) CytoD inhibited disassembly of dynamic actin assemblies at the ruffling edge. Photoactivated actin subpopulations remained visible for >100 s, and bulk actin fluorescence persisted at the ruffling cell edge. Bar, 5 μm.
Discussion
This study aimed to elucidate the mechanism of rapid monomer-insensitive actin disassembly by the three-component system consisting of cofilin, coronin, and Aip1. By directly imaging filament disassembly, we found that this three-component system disassembled filaments in abrupt bursts, where 100-subunit-long segments near both filament ends were lost very rapidly. Bursting disassembly generated filament barbed ends not recognized by CapZ, a protein that binds tightly to standard barbed ends. When added to the three-component system, CapZ and CytoD inhibited bursting disassembly, and did so from both ends with equal efficacy. CytoD and KabC, a structurally unrelated barbed-end capper, also inhibited actin disassembly in mammalian cells, which suggests that the three-component system reconstitutes a prevalent physiological disassembly pathway.
Mechanism of actin filament bursting disassembly
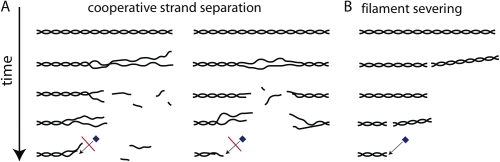
What is the mechanism underlying bursting filament disassembly? We propose the following model (Fig. 8 A): during a burst of disassembly, cooperative weakening of lateral interactions between the two strands of the actin filament causes the strands to transiently separate near an end (Fig. 8 A, left). Cofilin has been shown to disrupt lateral contacts between subunits in adjacent strands (Bobkov et al., 2004) as well as induce splaying or unwinding of strands observed by electron microscopy (Bremer et al., 1991; McGough et al., 1997). Aip1, which is believed to bind filaments at barbed ends and along their lengths (Okada et al., 2002), may also catalyze cofilin-induced strand separation. Consistent with this idea, Aip1 interacts with the F-actin subunit near surfaces that form lateral interactions with adjacent filament strands in the Holmes model (Holmes et al., 1990; Amberg et al., 1995; Rodal et al., 1999; Clark and Amberg, 2007). As separated strands lack stabilizing lateral interactions, they are highly unstable and rapidly disassemble into constituent subunits. Rapid disassembly of separated filament strands is plausible, as single-stranded polymers are not limited by dissociation of individual subunits from ends (Oosawa and Asakura, 1975). This burst leaves behind a nonstandard filament end (Fig. 4), which can then initiate another burst at a significantly faster rate, giving rise to a successive series of bursts from the same filament end (Fig. 1 B, f1 and f2). Occasionally, strand separation also occurs in the middle of the filament, giving rise to internal bursts of disassembly scored as severing events in our assay (Fig. 1 B, f3; and Fig. 8 A, right). However, most disassembly occurs as bursts from either filament end (Fig. 2 B), as strands separate more easily near ends.
Figure 8.
Candidate mechanisms for actin filament disassembly by cofilin, coronin, and Aip1. (A) Cooperative strand separation. (B) Filament severing. Black lines denote filament strands. The blue box denotes CapZ. The red bar denotes inhibition of CapZ binding. Successive illustrations depict reaction intermediates in each disassembly pathway.
An alternative interpretation of our observations is that bursting disassembly simply involves filament severing, where a spatially localized disruption of the filament causes it to break at the site of disruption (Fig. 8 B). Both severed daughter filaments are completely intact but one daughter filament immediately diffuses away and cannot be detected. However, we favor cooperative strand separation over severing for several reasons: (1) strand separation accounts for preferential bursting from an end that has already undergone a burst (Fig. 1), whereas severing does not; (2) strand separation accounts for preferential initiation of disassembly near filament ends (Fig. 1), whereas severing does not (moreover, the classical severing we observed using cofilin alone was not biased toward ends [Fig. 2], which argues that nothing in our experimental setup, such as filament immobilization, is causing an artificial bias toward ends); (3) we were able to score severing by single filament imaging (Fig. 2), which argues that we would have seen severing if it contributed to bursting; (4) strand separation accounts for disassembly intermediates with lowered affinity for CapZ (Fig. 4; it is also possible that barbed ends are structurally intact but capped by Aip1 after a burst and thus unable to bind CapZ; however, this putative Aip1–barbed end interaction would be quite unstable [τ < 10 s; Fig. 4 D]); and (5) strand separation plausibly accounts for inhibition of disassembly by capping factors (5–7), whereas severing does not (Fig. 5 F). In our view, severing better describes the behavior of filaments treated with high concentrations of cofilin alone (Fig. 2). Future efforts to distinguish between these alternative models will require electron microscopy of intermediates or development of spectroscopic methods that report on filament internal structure.
The mechanism of inhibition of disassembly by barbed end–capping agents presents a paradox we have yet to fully resolve. Mutual antagonism between barbed end capping and bursting disassembly suggests a possible means of inhibition. Bursting disassembly may inhibit binding of CytoD/CapZ by separating filament strands; conversely, CytoD/CapZ may inhibit bursting disassembly by keeping strands together near barbed ends. However, filaments do not disassemble preferentially from barbed ends but instead disassemble near both ends with similar kinetics by a similar capping-sensitive mechanism. We considered several possible explanations. One possibility is that barbed end factors control the stability of the whole filament by binding only to the free barbed end. However, the effect of barbed-end binding would have to propagate through the L. polyphemus acrosomal bundle, which itself is highly stabilized. Alternatively, barbed end factors may act in solution as opposed to on the polymer to inhibit disassembly. For instance, CytoD may interact with an actin monomer to perturb the activity of the purified system (Goddette and Frieden, 1986). However, we failed to detect any possible indirect effects of CytoD in solution (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1). A third possibility is that barbed end factors may bind along the filament length to inhibit strand separation. Consistent with this idea, in vitro studies point to low-affinity cytochalasin B binding sites along the filament length (Hartwig and Stossel, 1979).
Implications for actin dynamics in cells
What are the implications of this new disassembly reaction for dynamic organization and morphogenesis of actin assemblies in cells, assuming that it is physiologically relevant? Actin cytoskeletal networks in cells must disassemble through a mechanism that is both fast and insensitive to monomers. Monomer insensitivity is a basic thermodynamic requirement for nonequilibrium polymer turnover in the cell, and speed is important to allow fast remodeling of the actin cytoskeleton. The three-component reaction may achieve speed by disassembling entire filaments in cells in a single burst, as many actin filaments do not exceed hundreds of subunits in length in cells (Sechi et al., 1997; Cameron et al., 2001). Moreover, the three-component reaction may achieve monomer insensitivity by generating nonstandard filament ends (Fig. 4 C) that cannot readily add subunits, which is akin to the low affinity of shrinking microtubule (+) ends for tubulin. We have not visualized these nonstandard ends and speculate that the two filament strands may be unequal in length or partially separated (Fig. 8 A).
In contrast to bursting disassembly by the three-component system, pure severing by cofilin alone nucleates assembly by generating filament ends that can grow (Ichetovkin et al., 2002; Andrianantoandro and Pollard, 2006). The opposite effects of these two reactions may help resolve the paradoxical role of cofilin in cells, where it promotes disassembly in some cases (Kiuchi et al., 2007; Okreglak and Drubin, 2007) and assembly in others (Ichetovkin et al., 2002; Ghosh et al., 2004). Local activity of Aip1 and/or coronin may be important in determining whether cofilin catalyzes assembly or disassembly in cells.
The inhibitory effects of barbed-end cappers in our experiments suggest a role for barbed end–associated factors such as CapZ, formin, or gelsolin in modulating not only filament assembly but also disassembly. Conversely, bursting disassembly may antagonize barbed end–capping agents in cells, possibly accounting for the cofilin-dependent fast dissociation of CapZ from dynamic actin assemblies in cells (Miyoshi et al., 2006). Similarly, the side-binding factors (Siripala and Welch, 2007) may also modulate filament stability in cells. We note that the rate of bursting disassembly in our in vitro assays decreased when filaments were immobilized with N-ethylmaleimide–inactivated myosin instead of filamin (unpublished data). It will be a challenge to examine how filaments disassemble in the full repertoire of factors that interact with actin filaments in cells.
Differential drug effects
We finally note that CytoD and LatB are widely used research tools and are often used interchangeably. Previous studies demonstrated, but did not explain, differences between these drugs with respect to actin polymerization (Higashida et al., 2004), gross morphology (Spector et al., 1989), cell mechanical properties (Wakatsuki et al., 2001), and other cellular processes (Forer and Pickett-Heaps, 1998; Morales et al., 2000; Omata et al., 2000). The dramatic difference in their effects on disassembly (Figs. 6 and 7) has not been recognized before and may explain these different effects on cell physiology. In the future, differential effects of these two drugs can be used to test the importance of disassembly in different actin-dependent cellular processes, a question that has largely been overlooked.
Materials and methods
Protein purification
Actin was prepared from rabbit skeletal muscle according to the method of Pardee and Spudich (1982). The F-actin was then depolymerized by dialysis into G buffer (2 mM Tris-Cl, 0.2 mM CaCl2, 0.2 mM ATP, 0.1% β-mercaptoethanol, and 0.005% NaN3, pH 8.0) and further purified by an additional cycle of polymerization/depolymerization as described by Pardee and Spudich (1982). Actin was then repolymerized and labeled on lysines using approximately stoichiometric amounts of Alexa 488 and Alexa 647 N-hydroxysuccinimide–ester (Invitrogen) according to the procedure of Kellogg et al. (1988). The labeled actin was subject to two additional cycles of polymerization and depolymerization and then frozen in aliquots at −80°C. Cofilin, coronin, and Aip1 were purified as described by Brieher et al. (2006).
Construction of perfusion chambers
Perfusion chambers were assembled using glass coverslips (48366–227; VWR International), glass slides (3050; Gold Seal), and thin parafilm strips. Glass coverslips were cleaned for imaging by sonication for 30 min in 1% Micro-90 detergent (99999–01; Cole-Parmer), 60 min in 2 M KOH, 30 min in acetone, and 30 min in ethanol. During each solution change, coverslips were rinsed extensively using 10 or more changes of distilled water. Cleaned coverslips were then stored in ethanol until the day of the experiment.
To assemble the perfusion chamber, two parafilm strips were placed on surface of the glass slide. The coverslips were then removed from ethanol, dried using filtered air, and pressed onto the parafilm strips to create a fluid channel. The chamber was then baked at 85°C for 20 s to create a seal between the parafilm and the glass. Assembled chambers were stored in a box and used within 2 d.
Imaging of single actin filaments
Alexa 488– or Alexa 647–labeled actin was treated with 1 mM EGTA and 0.2 mM MgCl2 on ice for 5 min to convert calcium-ATP-actin to magnesium-ATP-actin. To remove residual filaments, the actin was then diluted in G buffer (0.2 mM CaCl2, 0.2 mM ATP, and 2 mM Tris-Cl, pH 8), left on ice for 20 min, and centrifuged at 436,000 g with a k-factor of 7 for 20 min. The supernatant, which contained actin monomer, was retained and kept on ice. Filamin, an actin cross-linking protein, was diluted into assay buffer (50 mM KCl, 2 mM MgCl2, 2 mM ATP, and 100 mM K+-Hepes, pH 7.8) to a final concentration of 10–50 μg/ml and adsorbed onto coverslips of perfusion chambers using a 10-min incubation. Chambers were then washed with blocking solution (5 mg/ml casein, 0.2% Tween 20, and 0.1% Pluronic F-127 in assay buffer) for 5 min. Immediately before polymerization, Alexa 488 and Alexa 647 actin monomer were mixed to a final concentration of 10 μM, 30% Alexa 647–labeled and 5% Alexa 488–labeled. 10× polymerization buffer (500 mM KCl, 20 mM MgCl2, 20 mM ATP, and 1 M K+-Hepes, pH7.8) was then added, and the mixture was allowed to polymerize in the perfusion chamber for 30–60 s. The chamber was then perfused with 6–7 chamber volumes of assay buffer supplemented with an oxygen scavenging system (4.5 mg/ml glucose, 0.2 mg/ml glucose oxidase, and 35 μg/ml catalase) and 10% blocking solution for imaging. Actin filaments in the perfusion chamber were imaged immediately at room temperature using an upright wide-field epifluorescence microscope (E90i) with a 60× 1.4 NA oil objective (both from Nikon). To distinguish newly polymerized filaments from filaments already present before salt addition, an image of filaments in the field of view was first taken in both the far-red channel (Alexa 647) and the green channel (Alexa 488). The disassembly factors were diluted into imaging buffer and then perfused into the chamber, and a streaming video of filament disassembly was taken in the far-red channel using either an intensifying charge-coupled device (CCD) camera (ORCA; Hamamatsu) or an electron microscopy CCD camera (iXon; Andor Technology PLC) with image acquisition software (Metamorph 6.0; Invitrogen).
Segmentation of single filaments by automated image analysis
Single actin filaments in the time-lapse images were segmented using a custom image analysis algorithm written in MATLAB (Mathworks). For each image in a time-lapse sequence, background correction was performed by subtracting a grayscale morphological opening of the image from its parent. The contrast of the image was then adjusted. The low contrast value was set to: Clow = Imode + n Iwidth, where Imode is the grayscale value at the peak of the image intensity histogram, Iwidth is the width of the peak of the intensity histogram, and n is an adjustable parameter. The high contrast value was set to: Chigh = Imode + f(Imax − Imode), where Imax is the highest grayscale value in the image and 0 < f < 1 is the degree of saturation. The image was then segmented and the segmentation was iteratively refined using the following procedure: the contrast-adjusted image was passed through a Canny edge detector, which generated a binary image with closed contours around the actin filaments. Single-pixel gaps in the contours in the binary images were bridged, and the regions enclosed in the contours were filled. Unfilled open contours were removed by morphological opening, and the filled objects were thinned to lines. The shape of each object was automatically inspected, and only objects without branch points (corresponding to linear segments) were retained. The grayscale values from pixels corresponding to objects in the segmentation were than averaged to obtain mean filament intensity Ifil from this estimate, and the new high contrast value of the image was then set to: (2) Chigh = Imode + f(Ifil − Imode).
The procedure was then repeated until Ifil converged to a constant value. A typical image would require approximately three or four iterations before convergence. This iterative procedure achieves automated segmentation robust to fluctuations in fluorescence intensity, variations in filament density, and the presence of bright outliers. It also does not require input parameters that explicitly depend on microscope acquisition settings.
Once segmentation was achieved, lengths of individual linear contours were estimated by the number of pixels occupied by the contours in the binary image. The lengths from all linear objects were then averaged to give the mean filament length.
Imaging of actin filament bundles grown off L. polyphemus acrosomal processes
L. polyphemus acrosomal actin bundles (a gift from G. Waller and P. Matsudaira, Massachusetts Institute of Technology, Cambridge, MA) were absorbed to coverslips of perfusion chambers for 5 min. Perfusion chambers were then incubated with blocking solution (5 mg/ml casein, 0.2% Tween 20, and 0.1% Pluronic F-127 in assay buffer) for 5 min. Filaments were elongated off acrosomal bundles by incubation with 3 μM Alexa 647–labeled actin monomer in assay buffer for 2 min and then washed. For time-lapse imaging experiments, filament bundles were perfused with disassembly factors and 100 μg/ml filamin, which held bundles together for imaging. For experiments with Alexa 488 CapZ, filament bundles were treated under different depolymerizing conditions, incubated with 3 μM Alexa 488 CapZ for 10 s, washed again, bundled with 200 μg/ml filamin, and then imaged. Unless otherwise stated, all perfusion steps were performed using assay buffer supplemented with oxygen scavenging system and 10% blocking solution. Imaging was performed using the fluorescence microscope described for single filaments.
Measurement of polymer mass decay in acrosome filament bundles
To quantify polymer mass in fluorescent filament bundles, we wrote an automated image analysis routine in MATLAB (Mathworks). The routine measures integrated intensity of a bright object over a dark background. The grayscale value for the background in each image was estimated by finding the mode of the image histogram, assuming that the objects of interest were small in the field of view. The pixel intensities were background-subtracted using the obtained value and then summed to obtain the integrated fluorescence intensity. This procedure was repeated for all images in the time-lapse sequence. The polymer mass decay curves of all filament bundles were obtained by running this routine for the entire field of view (Fig. S1 A, left). Polymer mass decays for long bundles and short bundles were obtained by running the routine on the appropriate regions of interest (Fig. S1 A, middle and right). The polymer mass exhibited a rapidly decaying component followed by a decay with significantly slower kinetics. We then subtracted from the polymer mass decay curve the slow-varying component, which was obtained by fitting the last segment of the curve to a straight line and extrapolating to time 0 (Fig. S1 B, red and orange lines). The resultant decay curve was then normalized to unity at time 0, and the characteristic rate of polymer mass decay was then obtained by fitting the first few points of the normalized decay curve to a straight line (Fig. S1 C).
For experiments involving Alexa 488 CapZ (capping protein was a generous gift of A. Lebensohn, Harvard Medical School, Boston, MA), the integrated CapZ fluorescence in a single filament bundle was measured in a region of interest surrounding the filament bundle. Background was measured in a neighbouring region of interest where no filament bundles were present. The number of actin filaments in the bundle was estimated by taking the maximal fluorescence intensity along the filament bundle length. The integrated CapZ fluorescence was divided by estimated actin filament number to obtain the CapZ/actin filament ratio (Fig. 3 B), which measures the proportion of CapZ-bound filaments in the filament bundle.
Drug treatment of suspension cells
HeLa S3 cells were cultured in suspension in shaking Fernbach flasks in CO2 independent medium (Invitrogen) plus 10% fetal calf serum. 10 ml of log phase culture (∼105 cells/ml) were removed, disaggregated by passing twice through a 27-gauge needle, and kept in a water bath at 37° with occasional agitation. At time 0, CytoD or LatB was added from a 2,000× stock in DMSO to a final concentration of 10 μM. 0.5 ml aliquots of cells were removed at each time point and fixed by dilution into 1 ml of PBS (155 mM NaCl, 1 mM potassium phosphate, 3 mM sodium phosphate, 1 mM MgCl2, and 1 mM CaCl2, pH 7.4) + 2% formaldehyde. After 10–20 min at 25°C, the fixed cells were collected by centrifugation (1,000 g for 5 min) and resuspended in 100 μl of PBS + 0.1% Triton X-100 + 0.5 μM TRITC-phalloidin + 1 μM Hoechst dye. This was a saturating amount of TRITC-phalloidin, as judged by the presence of excess unbound reagent in solution after the incubation. After 30 min at 25°C with occasional agitation, the cells were diluted with 1 ml of PBS + 0.1% Triton X-100, collected by centrifugation, resuspended in 50 μl of PBS + 60% glycerol, and mounted between a slide and coverslip. Random fields were imaged with a 10× objective. Integrated intensity of the TRITC-phalloidin signal in each field of view was measured using the image analysis routine described for acrosome filament bundles. The total intensity in the field of view was then divided by the number of cells present to obtain the integrated intensity per cell.
Infection of tissue culture cells with L. monocytogenes
Before infection, BSC-1 cells were cultured in DME media on polylysine-coated glass coverslips. Cells were then infected with an adenovirus expressing GFP-actin and incubated overnight. These GFP-actin–expressing cells were then infected with L. monocytogenes for 3 h, washed with DME containing 50 μg/ml gentamicin, and incubated at 37°C for up to 24 h before live imaging.
Live imaging of L. monocytogenes comet tails in tissue culture cells
Imaging was performed using a 100× 1.4 NA oil objective on an epifluorescence microscope (TE300; Nikon). BSC-1 cells expressing GFP-actin were imaged in DME containing 50 μg/ml gentamicin and 20 mM Hepes at pH 7.4 and were maintained at 37°C by a heated metal stage. Infected cells containing visible GFP-actin comet tails were located for time-lapse image acquisition. Time-lapse movies of GFP-actin fluorescence in comet tails were then acquired with a cooled-CCD camera (ORCA-ER; Hammamatsu) using image acquisition software (MetaMorph; MDS Analytical Technologies). For experiments involving drug treatment, the imaging media was exchanged for media containing the drug of interest during the course of image acquisition. Drug concentrations used: 6 μM LatB, 5 μM CytoD, and 0.5 μM KabC (provided by G. Marriot, University of Wisconsin, Madison, WI). In all cases, the concentration of DMSO was <0.5%.
Measurement of polymer mass decay in L. monocytogenes comet tails in cells
Polymer mass in L. monocytogenes actin comet tails were obtained using the same image processing routine detailed for analysis of L. polyphemus acrosomal filament bundles. Polymer mass decay curves were obtained in regions of interest around the part of the comet tail that had polymerized immediately before drug addition. Decay curves from multiple cells were then normalized and averaged together.
Preparation of mRFP-PAGFP-actin in a retroviral vector
The mRFP-PAGFP-actin plasmid was based on an EGFP-tagged human β-actin from Clontech Laboratories, Inc. First, EGFP was excised using an NheI–BglII digest and replaced by mRFP (a gift of R. Tsien, University of California, San Diego, La Jolla, CA). Second, PAGFP (a gift from R. Wedlich-Soldner, Max Planck Institute, Martinsried, Germany) was PCR amplified and ligated in the mRFP-actin between XhoI and BglII. In addition, to enhance flexibility of the linking regions, glycine residues were inserted between mRFP and PAGFP (one added) and between PAGFP and actin (three added; Fig. S4). The plasmid sequence was verified by sequencing. For reliable expression inside cells, a retrovirus encoding mRFP-PAGFP-actin was generated. mRFP-PAGFP-actin was PCRed and ligated into pLNCX2 (Clontech Laboratories, Inc.) between HindIII and NotI, and this construct was transfected into a GPG-293 packaging cell line for production of retrovirus (Ory et al., 1996).
Preparation of a cell line stably expressing mRFP-PAGFP actin
BSC-1 cells were infected with DME media containing retrovirus with the mRFP-PAGFP construct and a G418 selectable marker. 3 d later, cells were split into media containing 500 μg/ml G418 (Invitrogen). Upon confluence, cells were then resplit in selective media at very low density (1:1,000, 1:5,000, and 1:10,000). 3 wk later, individual colonies were picked and regrown to confluence. Individual clones were then selected based on cell vitality and morphology, as well as actin morphology and mRFP-PAGFP-actin fluorescence intensity, and cultured on glass coverslips for imaging.
Imaging of photoactivated fluorescent actin in tissue culture cells
Imaging was performed using a 100×1.4 NA oil objective on an inverted microscope (TE2000U; Nikon) with an Ultraview spinning disk confocal unit (PerkinElmer). BSC-1 cells expressing PAGFP-mRFP-actin were imaged in CO2-independent medium (18045–088; Invitrogen) supplemented with l-glutamine and were maintained at 37°C by a heated metal stage. Cells were excited with 488- or 568-nm light from an argon-krypton laser (Melles Griot), and images were taken with a cooled-CCD camera (ORCA-ER) using image acquisition software (MetaMorph). During the time lapse, media containing the drug was applied to the cells, and the PAGFP-mRFP-actin was photoactivated using a nitrogen pulse laser tuned to 405 nm (Photonic Instruments, Inc.). Both LatB and CytoD were diluted in CO2-independent media to a final concentration of 5 μM in 0.5% DMSO.
Online supplemental materials
Fig. S1 shows decay rates of L. polyphemus actin bundles with exposed barbed versus pointed ends. Fig. S2 shows that monomer-insensitive disassembly of single filaments requires cofilin, coronin, and Aip1. Fig. S3 tests the effects of CytoD acting in solution on actin disassembly. Fig. S4 is a diagram of the mRFP-PAGFP-actin construct used for the photoactivation experiments in cells. Video 1 shows disassembly of single actin filaments in the presence of cofilin, coronin, Aip1, and G-actin. Video 2 shows disassembly of actin grown off L. polyphemus bundles under the same conditions. Videos 3, 4, and 5 show decay of L. monocytogenes actin comet tails in the presence of CytoD, KabC, or LatB, respectively. Video 6 shows actin turnover in ruffling BSC-1 cells as a function of CytoD. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801027/DC1.
Acknowledgments
We thank Gerard Marriot for his generous gift of KabC, the Randy King Laboratory for GFP-actin adenovirus, Andres Lebensohn for capping protein, and Guichy Waller and Paul Matsudaira for L. polyphemus acrosomal sperm processes. We also thank Roger Tsien and Roland Wedlich-Soldner for plasmids. The photoactivation experiments were performed using the spinning disk confocal microscope in the Nikon Imaging Center at the Harvard Medical School. In addition, we also thank members of the Mitchison Laboratory for feedback, support, and encouragement; and Jue Shi for help with statistics.
This work was supported by National Institutes of Health grant GM 23928. H.Y. Kueh is a Howard Hughes Medical Institute predoctoral fellow. G.T. Charras is a Royal Society University Research Fellow.
Abbreviations used in this paper: Aip1, actin-interacting protein 1; CCD, charge-coupled device; CytoD, cytochalasin D; df, degrees of freedom; KabC, kabiramide C; LatB, latrunculin B; PAGFP, photoactivatable GFP; mRFP, monomeric red fluorescence protein.
References
- Amberg, D.C., E. Basart, and D. Botstein. 1995. Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2:28–35. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro, E., and T.D. Pollard. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell. 24:13–23. [DOI] [PubMed] [Google Scholar]
- Balcer, H.I., A.L. Goodman, A.A. Rodal, E. Smith, J. Kugler, J.E. Heuser, and B.L. Goode. 2003. Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13:2159–2169. [DOI] [PubMed] [Google Scholar]
- Blanchoin, L., and T.D. Pollard. 1999. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274:15538–15546. [DOI] [PubMed] [Google Scholar]
- Bobkov, A.A., A. Muhlrad, A. Shvetsov, S. Benchaar, D. Scoville, S.C. Almo, and E. Reisler. 2004. Cofilin (ADF) affects lateral contacts in F-actin. J. Mol. Biol. 337:93–104. [DOI] [PubMed] [Google Scholar]
- Bonder, E.M., and M.S. Mooseker. 1983. Direct electron microscopic visualization of barbed end capping and filament cutting by intestinal microvillar 95-kdalton protein (villin): a new actin assembly assay using the Limulus acrosomal process. J. Cell Biol. 96:1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer, A., R.C. Millonig, R. Sutterlin, A. Engel, T.D. Pollard, and U. Aebi. 1991. The structural basis for the intrinsic disorder of the actin filament: the “lateral slipping” model. J. Cell Biol. 115:689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieher, W.M., H.Y. Kueh, B.A. Ballif, and T.J. Mitchison. 2006. Rapid actin monomer-insensitive depolymerization of Listeria actin comet tails by cofilin, coronin, and Aip1. J. Cell Biol. 175:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai, L., A.M. Makhov, and J.E. Bear. 2007. a. F-actin binding is essential for coronin 1B function in vivo. J. Cell Sci. 120:1779–1790. [DOI] [PubMed] [Google Scholar]
- Cai, L., T.W. Marshall, A.C. Uetrecht, D.A. Schafer, and J.E. Bear. 2007. b. Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell. 128:915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell, J.E., S.G. Heiss, V. Mermall, and J.A. Cooper. 1989. Effects of CapZ, an actin capping protein of muscle, on the polymerization of actin. Biochemistry. 28:8506–8514. [DOI] [PubMed] [Google Scholar]
- Cameron, L.A., T.M. Svitkina, D. Vignjevic, J.A. Theriot, and G.G. Borisy. 2001. Dendritic organization of actin comet tails. Curr. Biol. 11:130–135. [DOI] [PubMed] [Google Scholar]
- Carlier, M.F., V. Laurent, J. Santolini, R. Melki, D. Didry, G.X. Xia, Y. Hong, N.H. Chua, and D. Pantaloni. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, M.G., and D.C. Amberg. 2007. Biochemical and genetic analyses provide insight into the structural and mechanistic properties of actin filament disassembly by the Aip1p cofilin complex in Saccharomyces cerevisiae. Genetics. 176:1527–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabiri, G.A., J.M. Sanger, D.A. Portnoy, and F.S. Southwick. 1990. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc. Natl. Acad. Sci. USA. 87:6068–6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos, E.L., B. Bradtke, F. Lottspeich, R. Guggenheim, and G. Gerisch. 1991. Coronin, an actin binding protein of Dictyostelium discoideum localized to cell surface projections, has sequence similarities to G protein beta subunits. EMBO J. 10:4097–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foger, N., L. Rangell, D.M. Danilenko, and A.C. Chan. 2006. Requirement for coronin 1 in T lymphocyte trafficking and cellular homeostasis. Science. 313:839–842. [DOI] [PubMed] [Google Scholar]
- Forer, A., and J.D. Pickett-Heaps. 1998. Cytochalasin D and latrunculin affect chromosome behaviour during meiosis in crane-fly spermatocytes. Chromosome Res. 6:533–549. [DOI] [PubMed] [Google Scholar]
- Ghosh, M., X. Song, G. Mouneimne, M. Sidani, D.S. Lawrence, and J.S. Condeelis. 2004. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 304:743–746. [DOI] [PubMed] [Google Scholar]
- Goddette, D.W., and C. Frieden. 1986. The kinetics of cytochalasin D binding to monomeric actin. J. Biol. Chem. 261:15970–15973. [PubMed] [Google Scholar]
- Hartwig, J.H., and T.P. Stossel. 1979. Cytochalasin B and the structure of actin gels. J. Mol. Biol. 134:539–553. [DOI] [PubMed] [Google Scholar]
- Higashida, C., T. Miyoshi, A. Fujita, F. Oceguera-Yanez, J. Monypenny, Y. Andou, S. Narumiya, and N. Watanabe. 2004. Actin polymerization-driven molecular movement of mDia1 in living cells. Science. 303:2007–2010. [DOI] [PubMed] [Google Scholar]
- Holmes, K.C., D. Popp, W. Gebhard, and W. Kabsch. 1990. Atomic model of the actin filament. Nature. 347:44–49. [DOI] [PubMed] [Google Scholar]
- Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., T.J. Mitchison, and B.M. Alberts. 1988. Behaviour of microtubules and actin filaments in living Drosophila embryos. Development. 103:675–686. [DOI] [PubMed] [Google Scholar]
- Ketelaar, T., E.G. Allwood, R. Anthony, B. Voigt, D. Menzel, and P.J. Hussey. 2004. The actin-interacting protein AIP1 is essential for actin organization and plant development. Curr. Biol. 14:145–149. [DOI] [PubMed] [Google Scholar]
- Kiuchi, T., K. Ohashi, S. Kurita, and K. Mizuno. 2007. Cofilin promotes stimulus-induced lamellipodium formation by generating an abundant supply of actin monomers. J. Cell Biol. 177:465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver, S.K., and P.J. Hussey. 2002. The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 3:reviews3007.1–3007.12 [DOI] [PMC free article] [PubMed]
- Maciver, S.K., H.G. Zot, and T.D. Pollard. 1991. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J. Cell Biol. 115:1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand, J.B., P. Moreau, A. Paoletti, P. Cossart, M.F. Carlier, and D. Pantaloni. 1995. Actin-based movement of Listeria monocytogenes: actin assembly results from the local maintenance of uncapped filament barbed ends at the bacterium surface. J. Cell Biol. 130:331–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough, A., B. Pope, W. Chiu, and A. Weeds. 1997. Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 138:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J. 1992. Compare and contrast actin filaments and microtubules. Mol. Biol. Cell. 3:1309–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, T., T. Tsuji, C. Higashida, M. Hertzog, A. Fujita, S. Narumiya, G. Scita, and N. Watanabe. 2006. Actin turnover-dependent fast dissociation of capping protein in the dendritic nucleation actin network: evidence of frequent filament severing. J. Cell Biol. 175:947–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales, M., M.A. Colicos, and Y. Goda. 2000. Actin-dependent regulation of neurotransmitter release at central synapses. Neuron. 27:539–550. [DOI] [PubMed] [Google Scholar]
- Murthy, K., and P. Wadsworth. 2005. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 15:724–731. [DOI] [PubMed] [Google Scholar]
- Okada, K., L. Blanchoin, H. Abe, H. Chen, T.D. Pollard, and J.R. Bamburg. 2002. Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 277:43011–43016. [DOI] [PubMed] [Google Scholar]
- Okada, K., H. Ravi, E.M. Smith, and B.L. Goode. 2006. Aip1 and cofilin promote rapid turnover of yeast actin patches and cables: a coordinated mechanism for severing and capping filaments. Mol. Biol. Cell. 17:2855–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okreglak, V., and D.G. Drubin. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata, W., H. Shibata, L. Li, K. Takata, and I. Kojima. 2000. Actin filaments play a critical role in insulin-induced exocytotic recruitment but not in endocytosis of GLUT4 in isolated rat adipocytes. Biochem. J. 346(Pt 2):321–328. [PMC free article] [PubMed] [Google Scholar]
- Ono, S. 2001. The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152:1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono, S., K. Mohri, and K. Ono. 2004. Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279:14207–14212. [DOI] [PubMed] [Google Scholar]
- Oosawa, F., and S. Asakura. 1975. Thermodynamics of the Polymerization of Proteins. Academic Press, New York. 204 pp.
- Ory, D.S., B.A. Neugeboren, and R.C. Mulligan. 1996. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. USA. 93:11400–11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni, D., C. Le Clainche, and M.F. Carlier. 2001. Mechanism of actin-based motility. Science. 292:1502–1506. [DOI] [PubMed] [Google Scholar]
- Pardee, J.D., and J.A. Spudich. 1982. Purification of muscle actin. Methods Enzymol. 85 Pt B:164–181. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D. 1986. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103:2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T.D., and G.G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- Ponti, A., M. Machacek, S.L. Gupton, C.M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. [DOI] [PubMed] [Google Scholar]
- Rodal, A.A., J.W. Tetreault, P. Lappalainen, D.G. Drubin, and D.C. Amberg. 1999. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., U. Wiedemann, N. Stuurman, and R.D. Vale. 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol. 162:1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger, J.M., J.W. Sanger, and F.S. Southwick. 1992. Host cell actin assembly is necessary and likely to provide the propulsive force for intracellular movement of Listeria monocytogenes. Infect. Immun. 60:3609–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, D.A., P.B. Jennings, and J.A. Cooper. 1996. Dynamics of capping protein and actin assembly in vitro: uncapping barbed ends by polyphosphoinositides. J. Cell Biol. 135:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi, A.S., J. Wehland, and J.V. Small. 1997. The isolated comet tail pseudopodium of Listeria monocytogenes: a tail of two actin filament populations, long and axial and short and random. J. Cell Biol. 137:155–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siripala, A.D., and M.D. Welch. 2007. SnapShot: actin regulators I. Cell. 128:626. [DOI] [PubMed] [Google Scholar]
- Spector, I., N.R. Shochet, D. Blasberger, and Y. Kashman. 1989. Latrunculins–novel marine macrolides that disrupt microfilament organization and affect cell growth: I. Comparison with cytochalasin D. Cell Motil. Cytoskeleton. 13:127–144. [DOI] [PubMed] [Google Scholar]
- Tanaka, J., Y. Yan, J. Choi, J. Bai, V.A. Klenchin, I. Rayment, and G. Marriott. 2003. Biomolecular mimicry in the actin cytoskeleton: mechanisms underlying the cytotoxicity of kabiramide C and related macrolides. Proc. Natl. Acad. Sci. USA. 100:13851–13856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot, J.A., T.J. Mitchison, L.G. Tilney, and D.A. Portnoy. 1992. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature. 357:257–260. [DOI] [PubMed] [Google Scholar]
- Wakatsuki, T., B. Schwab, N.C. Thompson, and E.L. Elson. 2001. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 114:1025–1036. [DOI] [PubMed] [Google Scholar]
- Wang, Y.L. 1985. Exchange of actin subunits at the leading edge of living fibroblasts: possible role of treadmilling. J. Cell Biol. 101:597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, N., and T.J. Mitchison. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086. [DOI] [PubMed] [Google Scholar]
- Wegner, A. 1976. Head to tail polymerization of actin. J. Mol. Biol. 108:139–150. [DOI] [PubMed] [Google Scholar]