Abstract
The structure of a 29-nucleotide RNA containing the sarcin/ricin loop (SRL) of rat 28 S rRNA has been determined at 2.1 Å resolution. Recognition of the SRL by elongation factors and by the ribotoxins, sarcin and ricin, requires a nearly universal dodecamer sequence that folds into a G-bulged cross-strand A stack and a GAGA tetraloop. The juxtaposition of these two motifs forms a distorted hairpin structure that allows direct recognition of bases in both grooves as well as recognition of nonhelical backbone geometry and two 5′-unstacked purines. Comparisons with other RNA crystal structures establish the cross-strand A stack and the GNRA tetraloop as defined and modular RNA structural elements. The conserved region at the top is connected to the base of the domain by a region presumed to be flexible because of the sparsity of stabilizing contacts. Although the conformation of the SRL RNA previously determined by NMR spectroscopy is similar to the structure determined by x-ray crystallography, significant differences are observed in the “flexible” region and to a lesser extent in the G-bulged cross-strand A stack.
The highly conserved sarcin/ricin loop (SRL), a component of 23–28 S rRNA, is essential for protein synthesis because it participates in the binding of elongation factors (EFs) to the ribosome. The function of this domain was illucidated by studying ribosomes treated with its namesake ribotoxins, sarcin and ricin (1, 2). The ribotoxins cleave a single covalent bond in the SRL RNA, which kill cells by inactivating ribosomes. Sarcin is a ribonuclease that cleaves the phosphodiester backbone on the 3′-side of G4325 (rat 28 S rRNA numbering is used throughout) (1, 2). Ricin depurinates the 5′-adjacent A4324 (3). EF-dependent functions are specifically impaired when ribosomes are treated with either toxin, whereas other ribosomal functions including EF-G-independent translocation are unaffected (1). Further indications of the domain’s importance are that 12 of its 17 nucleotides are near universal and that chemical footprinting (4) marks it as the only rRNA region protected by both EFs.
An SRL RNA can mimic the SRL in the ribosome because binding of EF-G to an Escherichia coli or rat SRL RNA is only ≈10-fold weaker than binding to intact E. coli ribosomes (5). Further, only the GTP-bound and apoprotein forms of EF-G bind the E. coli SRL RNA, whereas GDP-bound EF-G does not (5). EF-G binding is most affected by mutation of G4319 (5). Both toxins recognize and cleave a single bond in the SRL oligonucleotide (Fig. 1). Comprehensive mutational analysis of the SRL established that sarcin recognition and cleavage are largely dependent on a single base, the bulged G4319 (7). In contrast, each of the 4 nt of the GAGA tetraloop is necessary, and a GAGA tetraloop is sufficient for ricin recognition and depurination (6).
Figure 1.
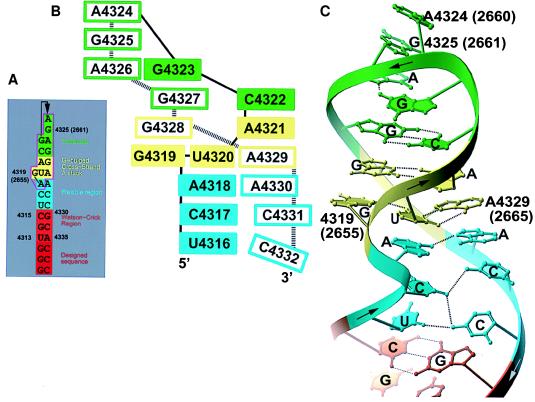
(A) The SRL sequence from the rat 28 S rRNA (with corresponding numbering for the E. coli 23 S rRNA in parenthesis) drawn with base pairs represented as adjacent nucleotide. The extended stem of the hairpin is red; the Watson–Crick region is orange; the flexible region is cyan; the G-bulged cross-strand stack is yellow; and the tetraloop is green. Bases within the magenta box are nearly universal. Figs. 2–5 use the same color scheme. (B) A base-stacking diagram of the SRL. (C) A Ribbon drawing of the SRL structure. To show the cross-strand stacks more clearly, the bases located on the 5′-half of the loop are filled while those bases on the 3′-half are not in Figs. 2–5.
The perception of RNA loop structure was altered by the determination of the conformation of the rat SRL by NMR spectroscopy (8, 9). Because the rules that govern formation of non-Watson–Crick base pairs were and remain largely unknown, regions devoid of canonical pairs had been depicted as single stranded loops. However, the 17 nt SRL RNA actually folds into a compact structure dominated by non-Watson–Crick pairs (8). Since 1993, a steady stream of RNA structures has demonstrated that canonical helical structure is regularly interrupted by compact noncanonical motifs (10–12).
We have determined the structure of the SRL RNA by x-ray crystallography, which provides an opportunity to compare the structure of the same RNA molecule solved by x-ray crystallography and NMR spectroscopy. NMR spectroscopy is a less powerful method for determination of the structure of RNA than it is for proteins: the number of distance constraints is much smaller in RNA than in proteins because RNA has fewer protons than proteins. Comparison of the cross-strand purine stacks and the GNRA tetraloops in this x-ray structure with the same motifs in the crystal structures of other RNA molecules shows that the structure of these RNA-building blocks are nearly identical in different molecular and crystalline environments. Finally, the SRL structure provides insights into how elongation factors and ribotoxins recognize this essential rRNA region.
MATERIALS AND METHODS
Purification and Crystallization.
Variants of the SRL 29-mer r(GGGUGCUCAGUACGAGAGGAACCGCACCC) were synthesized at the Yale Keck Microchemical facility on Applied Biosystems 394 or 3948 synthesizers by using β-cyanoethyl chemistry. Nucleotides 1–3 and 27–29 (Fig. 1) were designed to pair and create a blunt end, whereas nucleotides 4–26 corresponds to rat 28S nucleotides 4313–4335. Each variant contained a single modified nucleotide, either a deoxyribo-5-bromo-U or a deoxyribo-5-bromo-C, at position 4, 6, 25, 27, 28, or 29. The RNAs were deprotected with triethylamine trihydrofluoride (13) and purified by gel electrophoresis. Before crystallization, the SRL RNA was annealed at a concentration of ≈5 mg/ml in 5 mM MgCl2 and 50 mM Mops (pH 7.0) by heating to 55°C for 10 min and then slowly cooled to 25°C. Crystals grew in 3–5 days by vapor equilibration of 6 μl of drops with a well solution at 20°C. Drops were prepared by mixing 4 μl of annealed RNA with 2 μl of a well solution that contained 3.0–3.2 M (NH4)2SO4 and buffer X, which contained 50 mM Mops (pH 7.0), 20 mM MgCl2, 1 mM spermine, and 2 mM CoCl2. Crystals were stabilized in 3.3 M (NH4)2SO4 and buffer X. They were transferred to a cryo-stabilizing solution with 15% (wt/vol) xylitol added 3–5 min before their freezing in propane. Only the unmodified SRL RNA or the SRL RNA brominated at positions 27 and 29 crystallized.
Data Collection and Structure Determination.
Complete anomalous diffraction data from SRL crystals brominated at position 27 were measured at the bromine absorption edge, λ1 (determined by fluorescent energy scans on a brominated SRL crystal), at the absorption maximum, λ2, and at a remote wavelength, λ3 (space group P6122: a = 56.54 Å, c = 107.28 Å). In addition, complete anomalous data were collected at the absorption maximum, λ2, from SRL crystals with a bromine at position 29 (data not shown). Both data sets were collected on beamline F2 at the Cornell High Energy Synchrotron Source by using the Princeton 1K charge-coupled device (CCD) (see Table 1). Intensity data for each wavelength were merged separately and scaled by denzo and scalepack (14). Local scaling by using the program newlsc written by A. M. Friedman (Purdue University, West Lafayette, IN) led to a moderate improvement in the data quality. Phases were improved by cycling between heavy atom parameter refinement by mlphare (15) and solvent flattening of the maps by dm (15) using the multi-wavelength anomalous dispersion data collected on the SRL crystal with a bromine at position 27. Data collected at λ2 on the SRL crystal with bromine at position 29 did not improve the experimental maps, so only the phases derived from the bromine 27 crystal were used. The program o (16) was used for model building and crystallographic and NMR system was used for coordinate refinement (17); the register of the RNA chain in the map was established by the bromine positions on bases 27 and 29. Initial refinement used simulated annealing with a maximum likelihood target in which calculated phases were restrained by experimental phases (18). The final refinement used a 2.1-Å resolution data set on beamline F1 at Cornell High Energy Synchrotron Source by using the 4K ADSC charge-coupled device (CCD) detector (space group P6122: a = 56.81 Å, c = 107.92 Å) using SRL crystals containing a bromine at position 27. Intensities for this data set were merged and scaled with mosflm, scala, and agrovata (15), and refinement proceeded with CNS by using a maximum likelihood target function (Table 1). Although the electron density was best in the Watson–Crick base paired stem region, it also was clear in the conserved region. In contrast, the weak electron density for C23 corresponding to rat C4332 shows it is not well ordered.
Table 1.
Data collection, phasing, and refinement statistics
| Phasing Resolution | Observations (unique) 20-2.5 Å | Completeness overall, % | Bijvoet Rsym,* % acentric (centric) | Phasing power, acentric† 20-5.5 Å; 5.5-2.5 Å | 〈m〉‡ 20-5.5 Å; 5.5-2.5 | |||
|---|---|---|---|---|---|---|---|---|
| λ1 = 0.9199 Å | 18472 (5873) | 88.8 | 0.65 | 0.28 | ||||
| λ2 = 0.9194 Å | 23314 (6052) | 91.1 | 0.62 | .12 | ||||
| λ3 = 0.90499 Å | 17097 (5734) | 84.7 | 5.2 (3.6) | 1.71 | 0.56 | |||
| Refinement Resolution, Å | Observations (unique) | Completeness overall, % | Rsym,* % | Number of atoms (nt) | Waters, Mg2+, Br | R (Rfree) | rms Deviations bond, Å | Angle, ° |
| λ4 = 0.9188 Å 20-2.1 | 22518 (6036) | 92.7 | 4.3 | 621 (29) | 10, 9, 1 | 0.280 (0.330) | 0.009 | 1.4 |
Rsym = Σ|I − 〈I〉|/ΣI.
Phasing power is defined as rms 〈FH〉/rms closure error reported for acentric data.
〈m〉 is the figure of merit, defined as cos〈σ(ΔΦ)〉.
Figures were generated with ribbons (19), except for the van der Waal surfaces, which were rendered with grasp (20). All atoms were used for the comparisons between SRL structures determined by x-ray and NMR; other comparisons used only the backbone atoms.
RESULTS AND DISCUSSION
The purine-rich SRL RNA folds into a hairpin whose structure is distorted from A-form by three regions dominated by non-Watson–Crick base pairs (Fig. 1). At the base of the SRL hairpin, there is a Watson–Crick paired stem that stacks with nucleotides that we call the flexible region due to the paucity of stabilizing inter base pair hydrogen bonds. This flexible region in turn stacks with a G-bulged cross-strand A stack, whose bulged G4319 forms a G4319⋅U4320⋅A4329 base triple with the reversed Hoogsteen pair of the cross-strand stack. At the top of the hairpin, there is a closing Watson–Crick C4322·G4327 pair that is shared with the cross-strand stack and a GAGA (4323–4326) tetraloop.
The Flexible Region.
The flexible region consists of two pyrimidine·pyrimidine pairs and an A4318·A4330 pair and lies between the Watson–Crick stem and the cross-strand stack (Figs. 1 and 2). Unlike typical non-Watson–Crick base pairs that are stabilized by either two direct (21) or one direct and one water-mediated inter-base hydrogen bond (12, 22, 23), these pyrimidine·pyrimidine pairs have only one direct hydrogen bond. The conformation of these pyrimidine·pyrimidine pairs was not determined by the original structure determination by NMR spectroscopy (8, 9). Because the U4316·C4332 and the C4317·C4331 pairs are sheared, they present a portion of their Watson–Crick face either to the minor groove in the case of the 5′-side pyrimidines or to the major groove in the case of the 3′-side pyrimidines. The 5′-side of C4332 is unstacked, which may further destabilize this region.
Figure 2.
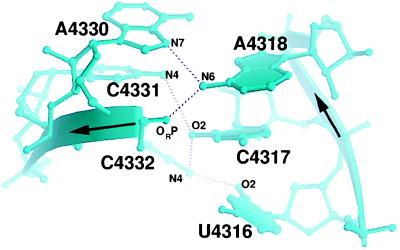
The presumed flexible region. C4331 and C4317 are not co-planar, and each base pair has only one direct hydrogen bond.
The G-Bulged Cross-Strand A Stack.
Like other reversed Hoogsteen cross-strand A stacks (12), this motif has a Watson–Crick C4322·G4327 pair followed by a sheared A4321·G4328 pair and a reversed Hoogsteen U4320·A4329 pair (Fig. 3). The module has characteristic cross-strand stacking of the adenosines and the backbone kink of the A4329 in the reversed Hoogsteen pair as well as stabilizing intra- and inter-strand contacts involving the 2′-hydroxyl of G4328. The SRL cross-strand A stack superimposes on each of the two in E. coli 5S rRNA (12) with a 0.6 Å rms difference. In contrast to standard reversed Hoogsteen cross-strand stacks (12), the SRL has an extra bulged G4319 that pairs with U4320 of the reversed Hoogsteen pair to form a base triple. The bulged-G4319 is unstacked on its 5′-side whereas on its 3′-side it stacks with the opposite strand G arising from the sheared A4321⋅G4328 pair resulting in a double cross-strand stack. The Watson–Crick faces of G4319 and G4328 stabilize the SRL RNA through hydrogen bonds between the cross-strand base and the backbone. The flexible region and the G-bulged cross-strand A stack widen the major groove by 5 Å while maintaining the depth common to A-form RNA.
Figure 3.
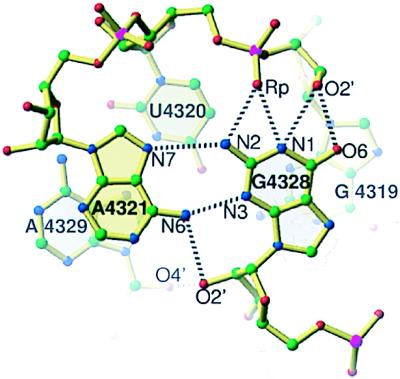
The bulged-G cross-strand A stack. The bulged-G4319 creates a second cross-strand stack: A4321 from the 5′-strand stacks on A4329 from the 3′-strand and G4328 from the 3′-strand stacks on G4319 from the 5′-strand.
The bulged G4319 modifies the conformation of the reversed Hoogsteen cross-strand A stack (Figs. 3 and 7) relative to the two nonbulged cross-strand A stacks found in E. coli 5S rRNA (12). In E. coli 5S rRNA, the 3′-side of a G from a sheared A⋅G pair is unstacked, whereas in the SRL RNA both sides of G4328 are stacked with a guanosine. Another distinguishing characteristic of the SRL motif is its direct cross-strand hydrogen bonds between the G and the backbone in contrast to a water-mediated hydrogen bond between the base and backbone that occurs in E. coli 5S rRNA.
Figure 7.
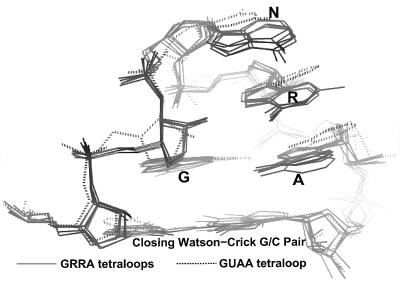
Superposition of GRRA (9, 10) and GUAA (29) tetraloop structures. The GRRA tetraloops are taken from SRL, three copies in the asymmetric unit of the GAAA tetraloop from a hammerhead ribozyme (11), a GAAA tetraloop from the P4–P6 domain (10), and a GUAA tetraloop from another hammerhead ribozyme (31).
In addition to the reversed Hoogsteen cross-strand A stack, there is a tandem sheared cross-strand A stack, which contains two tandem sheared purine pairs (12). While the reversed Hoogsteen type has only C3′-endo sugar puckers, the tandem sheared type usually contains one inverted C2′-endo sugar pucker (Fig. 4). Moreover, the backbone geometry is different for these two types (Fig. 4).
Figure 4.
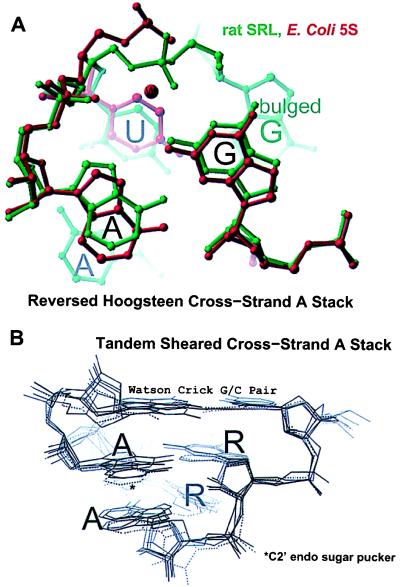
Comparison of cross-strand A stacks. (A) Superposition of the SRL (green) and the loop E reversed Hoogsteen cross-strand A stacks (12). (B) Tandem-sheared cross-strand A stacks: the P4-P6 domain (10) and the hammerhead ribozyme (11, 31). One of the two observed copies of the P4-P6 domain found in the asymmetric unit does not have an inverted sugar pucker (dashed lines) (10).
The base triple in the SRL RNA does not make a tertiary contact, instead the third base arises from an internal bulged G4319 within the helix (Fig. 5). In contrast, in base triples found in tRNA (24–27) and in the P4-P6 domain of the group I intron (10), the third base typically makes a tertiary contact in either groove to a helical base pair.
Figure 5.
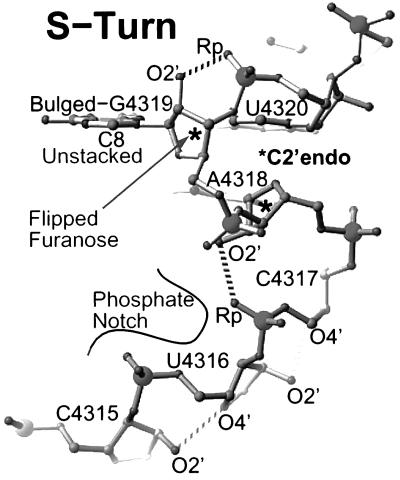
The S-turn structure. Both A4318 and G4319 have C2′-endo sugar puckers. Recognition opportunities abound in the S-turn: G4319 is unstacked on its 5′-side; the furanose ring of G4319 is flipped exposing its hydrophobic backside, which together with the accessible and adjacent C8 and C5 atoms creates a hydrophobic pocket; and the phosphates of U4316, C4317, and G4319 create a notch in the backbone ridge.
Tetraloop.
The GAGA (4323–4326) sequence forms a common RNA turn, a GNRA tetraloop (Fig. 6). The first and fourth nucleotides of the tetraloop adopt a modified sheared G4323·A4326 pair, in which G4323 makes a single bifurcated hydrogen bond that contacts both the base and the backbone of the A4326. The second, third, and fourth nucleotide of the tetraloop present their Watson–Crick faces to the minor groove, forming a triple purine stack that is perched on the 2′-hydroxyl of G4327 of the closing Watson–Crick C4322·G4327 pair. In addition to stabilization provided by stacking, there is a characteristic 2′-hydroxyl to purine N7 contact between the first and the third nucleotide.
Figure 6.
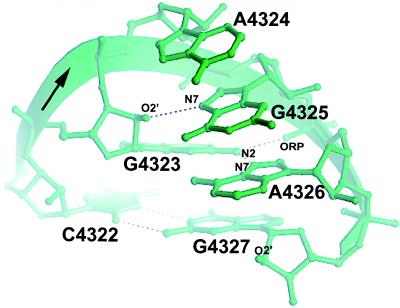
The structure of the GAGA tetraloop. A4324, G4325, and A4326 form a characteristic purine stack and present their Watson–Crick faces to the accessible minor groove.
GNRA turns can form tertiary contacts by docking with helices (28) or tetraloop receptors (10). An intermolecular lattice contact found in the rat SRL crystal demonstrates a third type of tertiary contact. The second and third base of one tetraloop form a ribose zipper (10) with a symmetry-related tetraloop. N3 of the purine of the second base forms a hydrogen bond with the 2′-hydroxyl of third base from a symmetry-related tetraloop. Because the O2 atom of pyrimidine bases and N3 atom of purines are isosteric and hydrogen bond acceptors, this tetraloop to tetraloop contact is possible between any two GNRA turns.
A comparison of different GRRA tetraloop structures determined by x-ray crystallography (10, 11, 29) as well as by NMR spectroscopy (30) demonstrates that these RNAs adopt a similar backbone structure whose pairwise rms structural differences range from 0.4 to 0.5 Å (Fig. 7). In contrast, the conformation of the one known GYRA tetraloop shows a 0.9 Å rms difference and thus lies outside this cluster.
The S-Turn.
The backbone adjacent to the bulged-G4319 is distorted by two sharp bends giving it an S-shaped appearance (Fig. 5). Each bend reverses the chain direction through backbone distortions and inverted sugar puckers. At the first bend, the A4318 base flips over, and the second bend restores the chain direction of the bulged G4319. The bulged G4319 and the 5′-A4318 present their 2′-hydroxyl groups in the major groove, in contrast to A-form RNA where the 2′-hydroxyls lie in the minor groove. This S-turn is stabilized in part by base stacking and 2′-hydroxyl to phosphate backbone hydrogen bonds.
Comparison of the Crystal Structure of the SRL with its NMR Conformation.
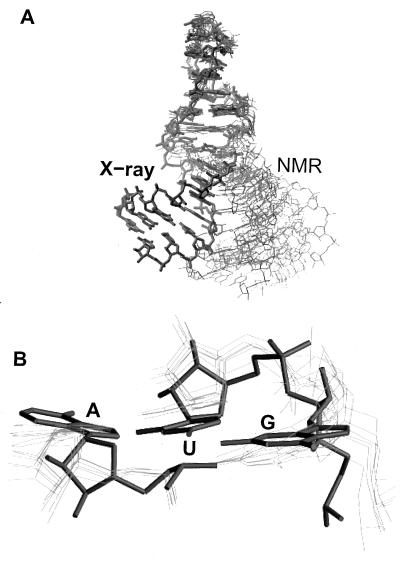
The overall 5.3-Å rms difference between the SRL conformation determined by NMR (8, 9) and that determined by crystallography is primarily a consequence of differences in the conformation of the flexible region, which results in different relative orientations of the flanking helical regions (Fig. 8). While the tetraloop at the top of the hairpin and the canonical terminal stem at the base show relatively low rms differences (0.9 and 1.1 Å) between the two structures, the rms structural difference is 2.3 Å for the flexible region. Superposition of all six NMR models (9) of the 12 nt-conserved sequence presents a large ensemble spread of terminal stems (Fig. 8A), due in part to lack of distance constraints in this region (P. B. Moore, personal communication). Surprisingly, when the crystal structure is included in this superposition, none of the NMR terminal stems superimpose on the crystal structure stem. Another difference is the width of the flexible region’s major groove, which in the NMR conformation is 8 Å larger at the narrowest point and 14 Å larger at the widest point than in the crystal structure.
Figure 8.
(A) Superposition of the SRL-conserved region structures determined by NMR (thin lines) (8, 9) on the x-ray structure (bold lines). (B) A close-up view of the base triple.
The two G-bulged cross-strand A stacks differ by 1.3 Å rms between the two structures. The most striking difference is the absence of the G4319·U4320·A4329 base triple in the NMR structure (Fig. 8B), which places G4319 and A4329 in roughly the same plane and staggers U4320 an average of 1.5 Å toward the tetraloop. This staggering retains the reversed Hoogsteen U4320·A4329 pair but prevents the base triple. The S-turn is more compact in the x-ray structure as demonstrated by the distance between phosphates at either end of the S-turn (C4317–A4320) that are on average 1.4 Å farther apart in the NMR structure than in the crystal structure.
The differences between the NMR and x-ray structures described here prompted a reevaluation of the NMR data by using new refinement techniques (31) that were not available when this pioneering NMR structure was determined (8, 9). As a consequence, the agreement between the NMR and x-ray determined structures has been dramatically improved (31).
Recognition Opportunities.
The structures of either EF or ribotoxin complexed with SRL is unknown; however, the purines identified biochemically provide recognition opportunities (Fig. 5). The non-Watson–Crick base pairs alter the surface features of RNA thereby providing direct base recognition opportunities in both grooves as well as providing opportunities for indirect recognition of unusual backbone features. The major groove is widened by a bulge (32) or non-Watson–Crick base pairs (12) to give proteins access. Moreover, non-Watson–Crick motifs increase the complexity of the hydrogen bond donors and acceptors presented by the bases into the nonregular minor groove (12). Finally, sequences can be recognized indirectly through contacts to the nonregular backbone conformations they specify.
Ricin Recognition.
The remarkable specificity of ricin, which depurinates one adenosine out of roughly 7,000 nts in eukaryotic ribosomes, might be due in part to direct base recognition. This specificity also may be partly due to steric hindrance since most of the potential ricin sites in the rRNA are sterically blocked by the folded rRNA structure complexed with proteins. Because GRRA tetraloops adopt a common fold (Fig. 9) and because only a GAGA tetraloop is both necessary and sufficient for ricin recognition and depurination of A4324 (33), direct recognition of the second and third base of the tetraloop is likely. Direct recognition is possible because all the hydrogen bond donors and acceptors of the second base are accessible and the Watson–Crick face of the third base is presented to the minor groove for inspection.
Figure 9.
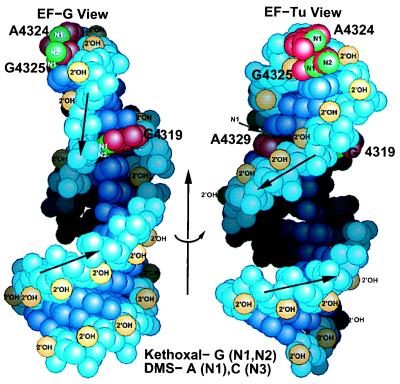
Mapping the chemical footprinting on the SRL structure. The left EF-G view is rotated 65° about the indicated arrow to generate the right EF-Tu view. The left drawing shows the unusual riboses of A4318 and G4319, which present their 2′-hydroxyls to the major instead of the minor groove. The structure is consistent with the chemical footprinting, since Watson–Crick faces of A4324, G4325, and A4329 are accessible. In the case of the bulged G4319, kethoxal is likely to approach perpendicular to the ring, since the 5′-side of G4319 is accessible and its Watson–Crick face is not.
Sarcin Recognition.
Restrictocin and its close relative sarcin are T1-like ribonucleases that contain insertions of lysine-rich loops thought to participate in recognition of the bulged G motif (34). Extensive mutational analysis (review in ref. 35) suggests this nuclease uses a molecular ruler because primary recognition occurs at the bulged G4319 and three base pairs (11.5 Å) away the toxin cleaves the backbone. Unlike a simple ruler, cleavage by this toxin depends on the type of base pairs between the primary recognition G4319 and the cut site. Thus in addition to the distance between G4319 and the site of cleavage, the fold may be also recognized. Direct recognition of the bulged G3219 is likely because this toxin is not active with a transition, transversion, or deletion mutation of this G.
Elongation Factor Recognition.
Elongation factors bind to overlapping sites in the conserved region of the SRL. Because biochemical and genetic data are available for the E. coli rRNA, these data are mapped on the rat SRL structure, which must be similar in the conserved region. EF-Tu and EF-G both protect the bulged-G as well as the second and third nucleotides of the tetraloop (Fig. 9) from chemical modification by kethoxal or dimethylsulfate (4). These two protected bases, A4324 and G4325, lie at the top of the tetraloop a quarter turn around the SRL helix axis and 4 bp away from G4319. In addition to these three nucleotides, EF-Tu also protects the A4329 of the base triple, which lies on the opposite side of the helix from the bulged G4319.
EF-G recognition of the SRL is thought to be indirect based on binding studies of mutant SRL oligomers (5). Only deletion and transversion of the bulged G have an affect on binding (5). A transition from G to A has virtually no growth phenotype (I.G.W., unpublished data) and a small affect on EF-G binding (5). This result is surprising because one would expect all three hydrogen bonds to be lost with a G4319A transition (Fig. 3). The absence of a strong phenotype for this transition is consistent with indirect recognition of the bulged G. Direct recognition by EF-G of A4324 and G4325 is unlikely, since transition and transversion mutations did not affect binding (5). Two possible points of indirect contact are the unstacked 5′-side of G4319 and A4324. A contact with the 5′-side of both purines, reminiscent of a c-clamp, would not be sensitive to transition or transversion mutations but would protect these purines from chemical modification. Additional indirect contacts are possible at the S-turn that provides an unusual cluster of phosphates and a hydrophobic patch around the flipped furanose ring of G4319 (Fig. 5).
Ribosome Translocation Switch.
During the elongation cycle of protein synthesis, ribosomes switch between the pre- and post-translocational state and have been hypothesized to adopt at least two conformations (36), one that favors binding of EF-1 (EF-Tu) and a second that favors binding of EF-2 (EF-G). Factors complexed with GTP bind to the ribosome. Hydrolysis of the GTP results in EF release and the switch between the pre- and post-translocational state. Because elongation factors catalyze the switch and the SRL is known to contact these factors (4), it is possible that the SRL participates in this switch. If the conformation change that is associated with the switch involves the SRL, then it seems most likely to occur in the flexible region.
Acknowledgments
We thank Peter B. Moore for helpful discussions. This work was supported by National Institutes of Health Grants GM-33702 (to I.G.W.) and GM-22778 (to T.A.S.).
ABBREVIATIONS
- SRL
sarcin/ricin loop
- EF
elongation factor
- GNRA
where N is any base and R is A or G
Footnotes
Data deposition: The atomic coordinates for the 2.1 Å resolution SRL structure have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 430D).
References
- 1.Hausner T P, Atmadja J, Nierhaus K H. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 2.Endo Y, Wool I G. J Biol Chem. 1982;257:9054–9060. [PubMed] [Google Scholar]
- 3.Endo Y, Tsurugi K. J Biol Chem. 1987;262:8128–8130. [PubMed] [Google Scholar]
- 4.Moazed D, Robertson J M, Noller H F. Nature (London) 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- 5.Munishkin A, Wool I G. Proc Natl Acad Sci USA. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo Y, Glück A, Wool I G. J Mol Biol. 1991;221:193–207. doi: 10.1016/0022-2836(91)80214-f. [DOI] [PubMed] [Google Scholar]
- 7.Glück A, Wool I G. J Mol Biol. 1996;256:838–848. doi: 10.1006/jmbi.1996.0130. [DOI] [PubMed] [Google Scholar]
- 8.Szewczak A A, Moore P B, Chan Y L, Wool I G. Proc Natl Acad Sci USA. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szewczak A A, Moore P B. J Mol Biol. 1995;247:81–98. doi: 10.1006/jmbi.1994.0124. [DOI] [PubMed] [Google Scholar]
- 10.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 11.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:68–74. doi: 10.1038/372068a0. [DOI] [PubMed] [Google Scholar]
- 12.Correll C C, Freeborn B, Moore P B, Steitz T A. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 13.Westman E, Strömberg R. Nucleic Acids Res. 1994;22:2430–2431. doi: 10.1093/nar/22.12.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 16.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 17.Brunger A T. x-plor, A System for X-Ray Crystallography and NMR. New Haven, CT: Yale University; 1992. , Version 3.1. [Google Scholar]
- 18.Pannu N S, Read R J. Acta Crystallogr A. 1996;52:659–668. [Google Scholar]
- 19.Carson M. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 20.Nicolls A, Bharadwaj R, Honig B. Biophys J. 1993;64:166. (abstr.). [Google Scholar]
- 21.Saenger W. Principles of Nucleic Acid Structure. New York: Springer; 1984. pp. 120–121. [Google Scholar]
- 22.Rould M A, Perona J J, Steitz T A. Nature. 1991;352:213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- 23.Baeyens K J, De Bondt H L, Holbrook S R. Nat Struct Biol. 1995;2:56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- 24.Kim S H, Quigley G J, Suddath F L, McPherson A, Sneden D, Kim J J, Weinzierl J, Rich A. Science. 1973;179:285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- 25.Robertus J D, Ladner J E, Finch J T, Rhodes D, Brown R S, Clark B F, Klug A. Nature (London) 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 26.Schevitz R W, Podjarny A D, Krishnamachari N, Hughes J J, Sigler P B, Sussman J L. Nature (London) 1979;278:188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- 27.Moras D, Comarmond M B, Fischer J, Weiss R, Thierry J C, Ebel J P, Giege R. Nature (London) 1980;288:669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- 28.Pley H W, Flaherty K M, McKay D B. Nature (London) 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 29.Scott W G, Finch J T, Klug A. Cell. 1995;81:991–1002. doi: 10.1016/s0092-8674(05)80004-2. [DOI] [PubMed] [Google Scholar]
- 30.Jucker F M, Heus H A, Yip P F, Moors E H, Pardi A. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- 31.Rife, J. P, Stallings, S. C., Correll, C. C., Dallas, A., Steitz, T. A. and Moore, P. M. (1998) Biophys. J., in press. [DOI] [PMC free article] [PubMed]
- 32.Battiste J L, Mao H, Rao N S, Tan R, Muhandiram D R, Kay L E, Frankel A D, Williamson J R. Science. 1996;273:1547–1551. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 33.Wool I G, Glück A, Endo Y. Trends Biochem Sci. 1992;17:266–269. doi: 10.1016/0968-0004(92)90407-z. [DOI] [PubMed] [Google Scholar]
- 34.Yang X, Moffat K. Structure (London) 1996;4:837–852. doi: 10.1016/s0969-2126(96)00090-1. [DOI] [PubMed] [Google Scholar]
- 35.Wool I G. In: Ribonuclease: Structure and Functions. D’Alessio G, Riordan J F, editors. New York: Academic; 1997. pp. 131–162. [Google Scholar]
- 36.Czworkowski J, Moore P B. Prog Nucleic Acid Res Mol Biol. 1996;54:293–332. doi: 10.1016/s0079-6603(08)60366-9. [DOI] [PubMed] [Google Scholar]