Abstract
Conduction of ions through the NaK channel, with M0 helix removed, was studied using both Brownian dynamics and molecular dynamics. Brownian dynamics simulations predict that the truncated NaK has approximately a third of the conductance of the related KcsA K+ channel, is outwardly rectifying, and has a Michaelis-Menten current-concentration relationship. Current magnitude increases when the glutamine residue located near the intracellular gate is replaced with a glutamate residue. The channel is blocked by extracellular Ca2+. Molecular dynamics simulations show that, under the influence of a strong applied potential, both Na+ and K+ move across the selectivity filter, although conduction rates for Na+ ions are somewhat lower. The mechanism of conduction of Na+ differs significantly from that of K+ in that Na+ is preferentially coordinated by single planes of pore-lining carbonyl oxygens, instead of two planes as in the usual K+ binding sites. The water-containing filter pocket resulting from a single change in the selectivity filter sequence (compared to potassium channels) disrupts several of the planes of carbonyl oxygens, and thus reduces the filter's ability to discriminate against sodium.
INTRODUCTION
The crystal structures of several types of potassium channels, including KcsA (1), MthK (2), and KvAP (3), have been presented in recent years. All of these highly selective tetrameric channels contain a central pore and a selectivity filter that is strongly conserved in terms of both sequence and structure (1,4–6). However, other channels with closely related sequences, such as cyclic-nucleotide-gated channels, are permeable to both K+ and Na+ (7). Recently, Shi et al. (8) determined the crystal structure of the NaK channel from Bacillus cereus, a channel which has the selectivity filter sequence of a cyclic-nucleotide-gated channel but which is otherwise quite similar to K+ channels. Using a radioactive tracer technique, they show that NaK is permeable to both K+ and Na+. Also, the structure is the same when the channel is crystallized with either K+ or Na+ in the pore, unlike the KcsA K+ channel which assumes different structures in the presence of different ions (9). Comparison of structures and permeation mechanisms between NaK and K+ channels should help us understand more about the physical basis of ion selectivity.
For potassium channels, the signature sequence forming the selectivity filter is threonine-valine-glycine-tyrosine-glycine (i.e., TVGYG). The NaK selectivity filter differs by only one residue: the tyrosine is replaced by aspartate, and the sequence becomes TVGDG. The substitution of tyrosine with the acidic aspartate residue causes pronounced changes in the structure of the filter and possibly alters the energy landscape encountered by permeant ions (8). The KcsA crystal structure (9) and theoretical calculations (10–16) reveal the presence of multiple K+ ions within the selectivity filter of the KcsA potassium channel. Usually, two K+ ions occupy binding sites S1 and S3 or S2 and S4. Sites S1–S3 in the KcsA filter are formed between pairs of planes of carbonyl oxygens, respectively, from Tyr78/Gly77, Gly77/Val76, and Val76/Thr75, while S4 is formed by the Thr75 carbonyl oxygens and the Thr75 side-chain hydroxyl groups. In NaK, the structure of the selectivity filter is essentially identical to KcsA in the region of sites S3 and S4, but sites S1 and S2 are replaced by an enlarged, water-filled filter pocket.
The basis of selectivity and the mechanisms of K+ conduction in potassium channels have been explored using a range of simulation techniques including molecular dynamics (MD), Brownian dynamics (BD), and quantum-mechanical simulations. Each technique has its strengths and limitations involving tradeoffs between the detail in which the protein, cell membrane, and solutions are modeled, and the system size and time spanned by the simulations. Only BD or other semimicroscopic simulation methods can calculate a continuous flow of ions through a channel and membrane system (17–19). However, BD mainly considers electrostatic effects and cannot model selectivity between ions of the same valency. Quantum-mechanical simulations have the potential to model selectivity at a fundamental level (20,21), but they are limited to small numbers of atoms and short time-spans. MD can model a large enough system, but is still too slow for continuous conduction of ions under normal conditions, and it is not certain that its empirical force fields are capable of fully modeling the atomic-level interactions essential for selectivity. Nevertheless, several important features in the mechanisms of permeation and the basis for selectivity in the potassium channels have been uncovered using MD. We now have a reasonable estimate of the landscape encountered by two K+ ions in the KcsA filter (22–25). The involvement of KcsA site S2 in discriminating between K+ and Na+, and some effects of its absence in NaK, have been detailed using classical MD and quantum mechanical simulations (20,25,26). Also, several steps in the conduction of K+ through the Kv1.2 channel have been simulated with classical MD (27).
Here we study the conduction of ions through NaK using Brownian dynamics, showing current-voltage and current-concentration relationships. Compared to the KcsA potassium channel, the magnitude of currents flowing through NaK is small. The permeant ions need to surmount the large energy barrier between the selectivity filter and the intracellular gate. We demonstrate that the height of this barrier can be reduced by substituting the glutamine residue near the intracellular entrance of the pore with a glutamate residue, which increases current significantly. We then analyze conduction of ions through the selectivity filter in greater detail using molecular dynamics, noting the differences between Na+ and K+ behavior. Under the influence of a strong applied potential, both K+ and Na+ ions traverse the pore. The sites in the selectivity filter where K+ and Na+ ions dwell preferentially differ, as do their coordination patterns.
METHODS
Channel models
We create computational models from the crystal coordinates (8) of the NaK channel, PDB accession code 2AHZ. For our study, we use a truncated form of the channel, where the M0 helix is removed from the structure. Shi et al. (8) report that, with the M0 helices removed, there is a significantly higher flux through the channel. The M0 helix contains many positively charged residues, creating a large energy barrier encountered by cations near the intracellular gate. Our BD simulations show that the inward current across the channel with the M0 helix attached is small, 0.8 pS at −100 mV, compared to 6.8 pS when the M0 helix is removed. The truncated form of NaK has been used for all results in this article.
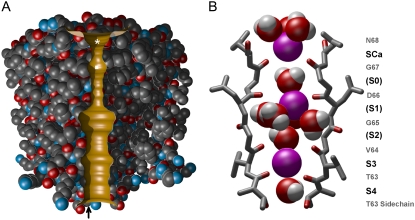
The published crystal structure contains two subunits, which we duplicate using the MD program CHARMM (28) to create a functional tetrameric channel. For the BD model, we increase the radii of the intracellular gate and the selectivity filter to produce a conducting state channel. Using the MD procedures detailed previously (16), we expand the radii of the filter and intracellular gate regions to 1.5 Å and 3.0 Å, respectively. To insert this model into our BD program, we use van der Waals radii to trace the boundary separating the water-filled ion conduction pore from the protein. The minimum radius of the pore for each z value is determined, and the resulting outline is rotated by 360° to create a three-dimensional, cylindrically symmetric pore. Fig. 1 A shows a slice through the channel pore, with two adjacent protein subunits surrounding it. In Fig. 1 B, we show the NaK filter containing several water molecules and potassium ions, and define some ion binding sites (S0–S4). Sites S3 and S4, formed by planes of carbonyl oxygen atoms from Thr63 and Val64 and the Thr63 side-chain oxygens, are the same as in KcsA (23,24). In NaK, the oxygens from Gly65 and Asp66 are more spread out than their equivalents from KcsA. The center of their carbonyl oxygens are at radius 4.64 and 3.84 Å from the pore axis, whereas the corresponding positions of Gly77 and Tyr78 are 2.33 and 2.56 Å, respectively. Thus, the sites S0–S2 from KcsA merge and form a filter pocket in NaK that contains several water molecules and usually an ion that occupies no definite position within the pocket. Outside Gly67, aided by Asn68, is a further site not found in KcsA, which can attract divalent cations such as Ca2+ as well as monovalent cations (8). We refer to this binding site as SCa. It is usual to find more than one ion in the KcsA filter (10,13,18,16), but we expect to find at least three in the longer NaK filter with SCa included.
FIGURE 1.
Model of the NaK channel used in Brownian dynamics and molecular dynamics. (A) Two adjacent subunits of the full protein are shown, with the water-protein interface in gold. The asterisk marks the Ca2+ binding site, and the square marks the filter pocket. The upward arrow indicates the approximate location of the residue 103. (B) Structure of the NaK selectivity filter during MD equilibration. Residues Thr63–Asn68 from two of the four monomers are shown in stick format with pore-lining oxygens colored red. K+ ions are represented as purple spheres and water molecules as red/white. K+ binding sites S3 and S4 are virtually identical to the same sites in potassium channels, whereas the equivalents of other potassium channel sites S0–S2 have become wider, and merge to form a filter pocket large enough to contain an ion surrounded by several water molecules as shown. Site SCa at the extracellular end is the NaK Ca2+ blocking site.
Brownian dynamics
To the conducting state model of the NaK channel, we attach two large cylindrical reservoirs, 30 Å in radius, on each side, to mimic the intracellular and extracellular solutions. These reservoirs are then filled with a fixed concentration of K+ and Cl− ions. We employ, when needed to maintain low concentrations of Ca2+ in the external reservoir, the grand canonical Monte Carlo method (29). The implementation of this method is detailed in Corry et al. (30). The protein and water are assigned dielectric constants of ɛp = 2 and ɛw = 60, respectively, and the Langevin equation is solved to calculate the trajectories of the ions. For the justification of using these values for the dielectric constant of the pore and the protein, see Ng et al. (31). In BD, the channel is assumed to be rigid in that all protein atoms remain in their fixed position. The validity of this simplifying assumption has been tested recently by Chung and Corry (32). They compared the conductance properties of KcsA obtained by allowing the carbonyl oxygen atoms lining the selectivity filter to remain flexible with those obtained with the rigid selectivity filter. The slope conductance of the channel, the number of resident ions in the channel and the dynamics of ion permeation in the selectivity filter derived under these conditions do not differ appreciably.
The motion of the individual ions is simulated using the Langevin equation,
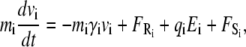 |
(1) |
where mi, vi, γi, and qi are the mass, velocity, friction coefficient, and charge on an ion with index i, while 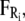 Ei, and
Ei, and 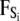 are the random stochastic force, systematic electric field, and short-range forces experienced by the ion, respectively. In the above equation, the first term −miγivi represents the frictional retardation experienced by an ion in a solution. The second term
are the random stochastic force, systematic electric field, and short-range forces experienced by the ion, respectively. In the above equation, the first term −miγivi represents the frictional retardation experienced by an ion in a solution. The second term 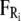 models forces that arise from random collisions between the ion and water molecules (note that water is otherwise treated as a continuum in BD). The third term qiEi is the electrostatic force term, described below in more detail. The final term
models forces that arise from random collisions between the ion and water molecules (note that water is otherwise treated as a continuum in BD). The third term qiEi is the electrostatic force term, described below in more detail. The final term 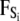 is the short-range force which arises from close interactions of ion i with other ions and the protein (equivalent to van der Waals repulsive forces).
is the short-range force which arises from close interactions of ion i with other ions and the protein (equivalent to van der Waals repulsive forces).
The electrostatic force term encountered in the Langevin equation arises from Coulomb interactions between the ions and from the fixed charges, and image forces from the dielectric boundaries. It is computed by solving Poisson's equation,
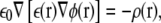 |
(2) |
where ε(r), φ(r), and ρ(r) are the space-dependent dielectric constant, electric potential, and charge density. The equation is solved using the boundary element method, as implemented by Hoyles et al. (33,34). To avoid repetitive calculations, the values are computed for 0, 1, and 2 ion combinations at a number of grid positions, and saved in lookup tables. During the simulations, these are used to interpolate between grid points.
All partial charges on the protein atoms are assigned using the CHARMM parameter set (Ver. 19), and treated as point charges surrounded by the protein dielectric. As an ion moves from the reservoir to the channel, the dielectric constant ɛw changes from 80 to 60. Since the implementation of a space-dependent water dielectric is complicated for channel-like geometries, we assign a Born energy barrier at the entrance and exit of the channel. This is incorporated as a ramp function extending for 3 Å into the pore along the channel axis (18). The K+ and Cl− ions are assigned random positions and velocities within the reservoirs, and their motion is calculated using the Langevin equation and an algorithm devised by van Gunsteren et al. (35,36). A more complete description of our implementation of Brownian dynamics can be found in the literature (17,18,37).
Simulations under various conditions, each lasting 2,000,000 time-steps (0.2 μs), are repeated numerous times. Initially, a fixed number of ions are assigned random positions in the reservoirs, with velocities also assigned randomly according to the Maxwellian distribution. The current is determined from the number of ions traversing the channel during the simulation period. For construction of the current-voltage curve, 150 mM KCl was used with 16 pairs of ions in each reservoir. Their trajectories are calculated every 100 fs, with a shorter time step of 2 fs used inside the pore where the force encountered by an ion changes rapidly. Also, a short time-step ensures that the average distance a particle traverses in one time interval is small compared to the dimensions of the system. If an ion is inside the short time-step region at the beginning of a 100-fs period, then that ion is simulated by 50 short steps while the other ions in the long-time regions are frozen to maintain the synchronicity. A temperature of 298 K is assumed throughout, and the list of the other parameters used in the BD simulations is given elsewhere (37).
Molecular dynamics
For MD simulations, the tetrameric channel including all hydrogens is first embedded in a POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine) membrane, and then bathed in either a KCl or NaCl solution. The TIP3 model is used for water molecules. The simulation system we use is similar to the arrangement used by Noskov and Roux (26), except that, for the reasons described previously, the M0 helices, composed of residues 1–19, are deleted. Simulations are performed using the CHARMM v32b1 program (28) and the PARAM27 all-hydrogen force field (38), with periodic boundary conditions and particle mesh Ewald electrostatics.
The salt solutions contain 28 pairs of ions, plus four additional K+ or Na+ so that the total charge of the simulation assembly is neutral. Apart from early stages of equilibration, no constraints are used on the pore loops and pore helices of the channel, while harmonic constraints of 10 kcal/mol/Å2 are applied to the M1 and M2 helices so that the channel is kept centered. We note here that the nearest constrained atoms are at radius 10 Å from the pore, and most are 15–20 Å away. Thus, the motion of the atoms lining the pore is unlikely to be affected by the constraints placed on the M1 and M2 helices. Equilibration at neutral or low membrane voltages is carried out separately for 12 ns for the Na+ and K+ systems, with ions restrained initially in SCa, the filter pocket, and S3 during the first 5 ns. We then remove constraints on the ions in the later stages of equilibration, so that they could move to energetically favorable positions.
Since ion conduction under normal conditions is too slow for obtaining useful results in a feasible simulation time, we adopt two procedures for accelerating conduction: we use a very high voltage gradient, and we preposition ions ready for conduction at the intra- or extracellular pore mouth. In the previous MD studies by Crozier et al. (39) and Khalili-Araghi et al. (27), a membrane potential of 5 to 6 × 108 V/m was used to simulate conduction of ions through channels. For the NaK channel, a field strength of ±1.2 × 109 V/m or ± 1.4 × 109 V/m gives adequate conduction rates. The electric field is applied only to the 3 or 4 ions that are inside or approaching the filter, while a physically realistic ±5.0 × 107 V/m is applied to the filter protein. No gradient is applied to the remainder of the system.
In BD, the rate-limiting step for a conducting channel is usually the time taken for ions to enter the intra- or extracellular pore mouths or to climb out of internal energy wells, but our main interest for MD is the conduction process through the selectivity filter. Therefore, we make the MD conduction simulations more efficient by prepositioning ions close to site SCa for inward conduction or the intracellular gate for outward conduction. To allow as many ions to move across the filter as possible during a simulation period, we position an ion near the internal or external entrance of the filter after a conduction event occurs. We apply a voltage gradient to the nearest ion in solution and also apply a harmonic potential to bring it close to the pore axis. The resulting force encountered by the selected ion is proportional to the distance between its position and the central axis. Thus, the initial movement of the chosen ion is rapid, but it slows down as the ion approaches the channel mouth. The positions of the ions in the selectivity filter are checked at least twice per nanosecond during the simulation or four times per nanosecond at the higher gradient of ±1.4 × 109 V/m, and a new ion is prepositioned if one or more ions leave the filter and all other ions move forward significantly. By using short simulation periods of either 0.25 or 0.5 ns, we ensure that not too much simulation time passes before a new ion is prepositioned. Prepositioning obviously saves computation time by eliminating the rate-limiting step, but it also means that conduction rates are now determined solely by the filter and cannot be extrapolated to the whole channel system. For each combination of ion type and voltage polarity, we simulate for a total period of either 108 ns (applied field ±1.2 × 109 V/m) or 40 ns (±1.4 × 109 V/m). A conduction event is counted when an ion in the filter reaches the intra- or extracellular solution.
RESULTS
Brownian dynamics
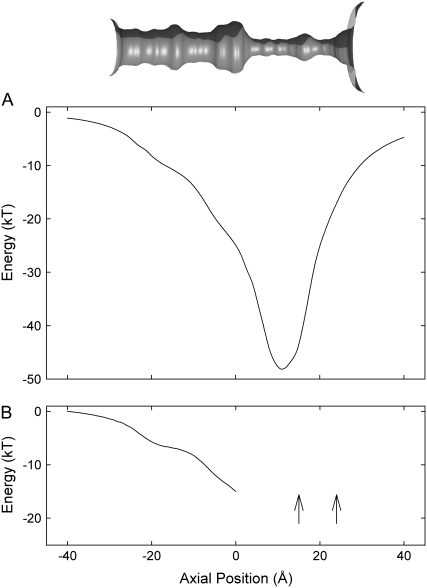
To study the potential energy landscape seen by ions as they traverse the channel, we move a single K+ along the central axis and solve Poisson's equation at 1 Å steps. The profile does not take account of the frictional and random forces acting on ions, and attractive and repulsive ion-ion interactions taking place in the reservoir. Also, the probability of an ion entering the intracellular gate cannot be ascertained from such profiles. As shown in Fig. 2 A, a permeating ion encounters a deep energy well created in part by the carbonyl oxygen atoms lining the selectivity filter as it moves across the conduit. The depth of the well at z = 12 Å, in the vicinity of the filter pocket, is −48 kT (1 kT = 4.11 × 10−21 J or 0.59 kcal/mol), which is somewhat shallower than the well of −67 kT in the selectivity filter of the KcsA potassium channel (40). The depth of this well ensures that at least three ions are attracted into the filter region. Thus, we can predict that a conduction event will be a multiion process. The multiion energy profile reveals that, at a given potential, outward current will be larger than the inward current. The energy landscape encountered by a third ion, with two ions in the selectivity filter, one at z = 15 Å (near the binding sites S0-S2) and the other at z = 22 Å (near SCa), is illustrated in Fig. 2 B. Starting at z = 0, the third ion is moved toward the intracellular gate 1 Å at time, while the two resident ions are allowed to adjust their positions. The profile is constructed with no applied potential. To reach the intracellular gate (z = – 20 Å), the innermost ion encounters a steep energy barrier of ∼8 kT. In contrast, an ion from the intracellular reservoir, once it is attracted to the inner gate of the channel, can readily move toward the selectivity filter, as it will encounter an energy well. This steep barrier presented to the ion attempting to move inward will be reduced if several negatively charged residues are placed near the intracellular gate. It is, however, not possible to deduce channel conductances accurately under various conditions or predict the degree of asymmetry in the current-voltage curve by examining one-dimensional potential or free energy profiles.
FIGURE 2.
Single and multiion energy profiles. The inset at the top shows water-protein interface, reproduced from Fig. 1 A and scaled to match the energy profiles shown below. (A) Potential energy encountered by a single K+ ion traversing the channel is constructed in the absence of an applied potential. (B) A potential energy seen by a K+ when moved between 0 and −40 Å (solid line) is constructed with two other K+ located in the selectivity filter (arrows). The positions of the two resident ions are allowed to adjust as the test ion is moved toward the intracellular entrance.
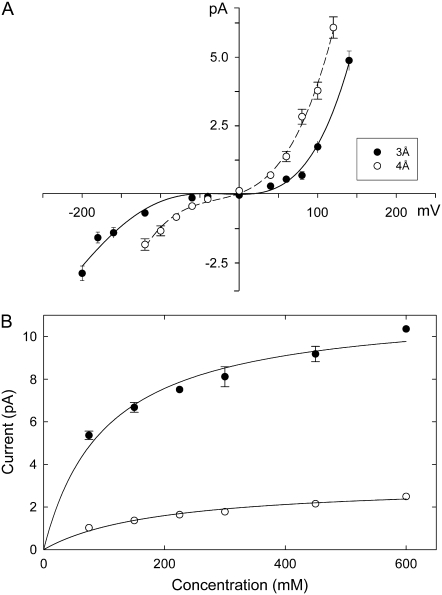
To characterize the conductance properties of the NaK channel, we carry out Brownian dynamics simulations. Fig. 3 A shows the current-voltage curve determined from BD simulations. The channel is outwardly rectifying, with an outward conductance of 17.3 ± 2.0 pS at 100 mV, and a much smaller inward conductance value of 3.5 ± 0.9 pS at −100 mV. The relationship is pronouncedly nonlinear, deviating systematically from Ohm's law with an increase in the applied potential. This nonlinearity results from the presence of an energy barrier in the channel. A barrier becomes less of an impediment to a permeating ion when the driving force is large (17). The current across the NaK channel is only ∼30% of that across the KcsA potassium channel. No physiological data on the conductance properties of the NaK channel are currently available. Since the magnitude of currents flowing across the channel is shown to be critically dependent on the radius of the intracellular gate (16,40), we also constructed a current-voltage curve with the radius of the intracellular gate widened from 3 Å to 4 Å. The relationship obtained with this model is shown in Fig. 3 A (open circles). At the applied potential of 100 mV and −100 mV, the conductance is 35 ± 3 pS and 13 ± 2 pS, respectively.
FIGURE 3.
Current-voltage-concentration profiles of NaK. (A) The current-voltage relationships are obtained with solutions of 150 mM KCl in each reservoir. The radii of the intracellular gate are set at 3 Å (solid circles) and 4 Å (open circles). Error bars in this and the following figures have a length of mean ±2 SE and are not shown when they are smaller than the data points. (B) Current-concentration curves are obtained with applied potentials of ±160 mV. Inward and outward currents are shown as open and solid circles, respectively. Lines are fitted from the Michaelis-Menten equation.
We also construct current-concentration curves by varying the concentration in the two reservoirs at an applied potential of ±160 mV. Results are shown in Fig. 3 B. Outward currents (solid circles) are much larger than inward currents (open circles). The solid lines fitted through the data points are calculated from the Michaelis-Menten equation and have the same shape as those observed experimentally from potassium channels. Theoretically, the current-concentration curve is expected to saturate if the transport of ions across the channel is determined by two independent processes, one of which depends on ionic concentrations in the extra- and intracellular regions and one that does not. In potassium channels, as in many other ion channels, the time it takes for an ion to enter the pore depends on ionic concentrations as well as the applied potential, whereas the time it takes for an ion in the pore to traverse it depends solely on the applied potential. If the energy barrier encountered by the permeant ion is large, the half-saturation concentration [K]s is expected to be lower (16). The values of [K]s for both curves are ∼75 mM. The experimental values for various types of potassium channels range from 40 mM for inward rectifiers (41) to 300 mM for Shaker K+ channels (42).
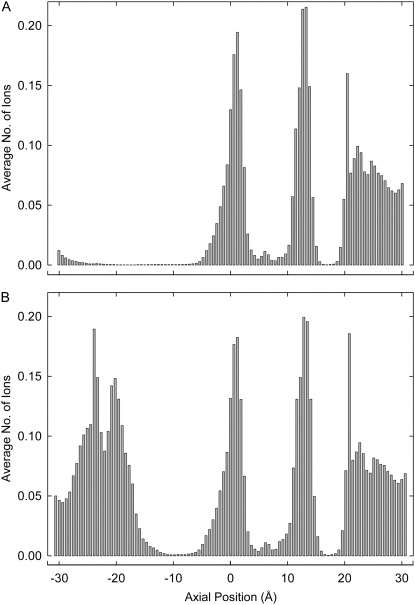
To determine the pore regions preferentially occupied by ions, we divide the channel into 100 thin sections and count the average number of ions in each section during a simulation period of 1.6 μs. Fig. 4 A shows histograms of the positions of ions inside the channel with an applied potential of −100 mV. There are on average 3.5 ions inside the channel, mainly concentrated within and close to the filter region. One ion is at the entrance to the selectivity filter from the intracellular chamber (near site S4), one is in the filter pocket region formed by residues GDG, and the remaining ions are on the extracellular side of the filter (site SCa and vicinity). The shape of the dwell histogram obtained during outward conduction, not shown here, is virtually identical to that shown in Fig. 4 A.
FIGURE 4.
Dwell histograms for potassium ions in the BD system at an applied potential of −100 mV. The number of ions in each of 100 thin sections during a conduction period of 1.6 μs is tabulated and plotted in the form of histograms. (A) The histogram displayed is obtained from the wild-type NaK channel. (B) The four glutamine residues at position 103 are replaced with fully charged glutamate residues and the dwell histogram is obtained under the same condition as in panel A. Note the prominent peak near the intracellular gate.
There is one pronounced difference between the single-ion energy profile (Fig. 2 B) obtained from the NaK channel and that obtained from KcsA (16). In the KcsA profile, there is an energy well near the intracellular gate, created by the glutamate residues Glu118. The depth of this well ranges from 8 kT to 1 kT, depending on the radius of the intracellular entrance (40). This intracellular well created by the charged residues guarding the gate attracts K+ ions from the inner reservoir and also lowers the energy barrier presented to permeating ions crossing the wider hydrophobic region. Dwell histograms for KcsA show a peak at this point that is not present for NaK. The residue corresponding to Glu118 from KcsA is Gln103 in the NaK channel. To check if the conductance properties of NaK can be altered by introducing four charged residues near the intracellular gate, we make a Q103E mutant channel and measure the current flowing across it under various conditions. The dwell histogram, shown in Fig. 4 B, is obtained with the Q103E mutant during inward conduction. The regions in the selectivity filter where ions dwell preferentially are essentially the same in wild-type and mutant channels. On average, two ions are in the selectivity filter, and 1.4 ions are near the external entrance (z > 20 Å) of the selectivity filter. There is now, however, a prominent peak at the intracellular gate, centered near z = −20 Å. This peak contains, on average, 2.3 ions. The presence of two or more ions in this region facilitates the conduction event.
The single-ion energy profile for the mutant channel, shown in Fig. 5 A, reveals the presence of an energy well at z = −20 Å. The depth of the profile at its minimum (z = 12 Å) is now −56 kT, 8 kT deeper than that obtained from the wild-type channel, which is reproduced from Fig. 2 B (broken line). The current-voltage curves, illustrated in Fig. 5 B, derived from the Q103E mutant channel are obtained under two different conditions. The four glutamate residues are either fully charged or their charges reduced to −0.2e, corresponding to partial protonation. The relationship obtained from the wild-type channel is reproduced from Fig. 3 A (broken line). Currents are systematically reduced when the charges on the glutamates are lowered. Outward and inward conductances at ±100 mV, when each mutated residue is fully charged, are 54.5 ± 0.2 pS and 30.5 ± 2.0 Ps, respectively. When the charges on the glutamate residues are reduced to −0.2e, the corresponding conductances are 33.0 ± 2.8 pS and 10.5 ± 0.3 pS, respectively. Compared to the wild-type channel, we see a threefold increase in the outward current and an even greater increase in the inward current by replacing the polar residues with fully charged acidic residues. The pronounced outward rectification seen in Fig. 3 A persists in the mutant channel.
FIGURE 5.
Single-ion potential energy profile and current-voltage relationships obtained from the Q103E mutant channel. (A) The potential profile obtained from the Q103E mutant, with each of the four glutamate residues carrying the full electronic charge of 1.6 × 10−19 C, shows a deep energy well near the intracellular gate (solid line). For comparison, the profile obtained from the wild-type channel is reproduced from Fig. 2 (broken line). (B) Current-voltage relationships from the Q103E mutant channel are obtained under two different conditions. The data shown in solid circles are obtained with the four glutamate residues fully charged, while those shown in open circles are obtained with each of the four glutamate carrying 0.2e. The relationship obtained from the wild-type NaK channel is reproduced from Fig. 3 (broken line). All current-voltage relations are obtained with symmetric solutions of 150 mM in the reservoirs.
Next, we study the effects of external Ca2+ ions on the conductance properties of NaK channel, using the Q103E mutant. Conduction of monovalent ions through cyclic-nucleotide-gated channels is blocked by low concentrations of Ca2+ ions, and similar behavior is expected for NaK based on crystallographic evidence (8). As shown in Fig. 6 A, Ca2+ ions are attracted to the binding site SCa located just outside the selectivity filter. The dwell histograms of Ca2+ (solid bars) and K+ (shaded bars) are obtained with Ca2+ and K+ concentrations in the extracellular reservoir of 3.2 and 150 mM, respectively, and the applied potential of −180 mV. Under these conditions, there are on average 0.3 Ca2+ ions in the binding site. The presence of a divalent ion there occludes the pore and prevents the passage of K+ across the channel. The current is attenuated to 60% of the control current. In Fig. 6 B, the inward current flowing across the Q103E mutant channel is plotted against the concentration of Ca2+ ions in the external reservoir. The current declines exponentially as a function of calcium concentration. Externally applied Ca2+ ions bind to the binding site and become unbound according to a first-order Markovian process. Conduction events occur only when no Ca2+ ion is in the binding site. We tabulate the time the channel is not blocked by a Ca2+ ion at different Ca2+ concentrations. The results are plotted in the inset of Fig. 6 B. With 3.2 mM of Ca2+ in the reservoir, the channel stays open on average for 140 ns before it becomes blocked by a divalent ion. Once a Ca2+ enters the binding site, it remains there on average 100 ns. Attempts to measure currents of monovalent ions through single NaK channels have not yet succeeded (43), and therefore Ca2+ blocking has not been studied experimentally. It has been established by a radioactive isotope technique that NaK conducts Ca2+ at a very low rate (43). Again, this is similar to cyclic-nucleotide-gated channels, and hence Ca2+ blocking at low concentrations is expected.
FIGURE 6.
Mechanisms of calcium blockade of the mutant NaK channel. (A) Dwell histograms for potassium (shaded bars) and calcium (solid bars) ions in the channel are constructed during 0.8 μs BD simulation with an applied potential of −180 mV. The concentration of CaCl2 in the external reservoir is controlled by the grand canonical Monte Carlo method and is maintained at 3.2 mM. The reservoirs contain 150 mM KCl. (B) Inward potassium currents obtained with an applied potential of −180 mV are plotted against calcium concentrations in the external reservoir. In the inset, the mean time the channel stays open is plotted against five different concentrations of Ca2+ ions in the external reservoir.
Molecular dynamics
The NaK channel, unlike the KcsA channel, conducts both K+ and Na+, although the selectivity ratio between these two ionic species is unknown. Also not known are the binding sites of each ionic species in the selectivity filter. As BD cannot properly discriminate between Na+ and K+, we use MD to detail some of the salient differences in the conduction processes of the monovalent species.
Both K+ and Na+ ions move across the selectivity filter of the NaK channel under the influence of a strong applied potential. Inward and outward conduction rates for both ionic species, determined at two different levels of applied potential, are summarized in Table 1. Conduction rates for K+ are appreciably higher than those for Na+, and inward rates are higher than the corresponding outward rates. We note here that the conduction rate shown in Table 1 does not change linearly with the driving force. For example, a 14% decrease in applied electric field causes a 25% reduction in outward K+ conduction, and 80% reduction in Na+ conduction. This nonlinear current-voltage relationship, also found in the curves obtained with BD simulations, stems from a large energy barrier each permeating ionic species encounters in traversing the selectivity filter. The larger the barrier height is, the greater the nonlinearity becomes. Thus, we can infer that the barrier seen by a Na+ ion is higher than that seen by a K+ ion. It is not possible, however, to deduce what the K+/Na+ selectivity ratio will be at a physiological range of applied potentials. As a control, we compare the conduction rates of K+ and Na+ ions in the KcsA potassium channel, using the same protocol. In KcsA, the voltage gradient of 0.5 × 109 V/m results in an outward K+ conduction rate of 0.9 ions per ns. During a 28-ns simulation period, no outward conduction of Na+ is observed. With an applied membrane potential of −1.2 × 109 V/m, the ratio of the inward conduction rate for K+ and Na+ is 17 for KcsA, compared to 7 for NaK.
TABLE 1.
Conduction rates through the selectivity filter determined from MD simulations
| Ion type | Voltage gradient (V/m) | Inward conduction rate | Outward conduction rate |
|---|---|---|---|
| K+ | ±1.2 × 109 | 1.4 | 0.3 |
| ±1.4 × 109 | 2.4 | 0.4 | |
| Na+ | ±1.2 × 109 | 0.2 | 0.06 |
| ±1.4 × 109 | 0.9 | 0.3 |
The conduction rate is expressed as the number of ions crossing the filter per ns.
All of these rates obtained from NaK are far higher than would be expected for simulations of conduction at normal membrane potentials and without prepositioning of the ions, but in the absence of the modified conditions, only one or at most two ions would have moved across the filter during the entire set of simulations. We note that, on a few occasions, ions began to enter the intracellular gate by themselves, or moved into site SCa without any prepositioning. These observations suggest that prepositioning is simply a mechanism used to speed up a natural process. The results of our MD simulations show that rates for K+ and Na+ are within the same order of magnitude, but the channel conducts K+ ions several times faster than Na+. Increasing the applied potential from 1.2 × 109 V/m to 1.4 × 109 V/m causes ∼50% increase in conduction rate for K+ ions. For Na+ ions, however, the increase in conduction rate is nearly fivefold. The steep dependency of conduction rates of Na+ ions on the applied potential implies that ions need to surmount large energy barriers to move from one binding site to the next. The current-voltage relationship obtained with BD, as we have seen in Fig. 3, shows outward rectification. In contrast, the results of MD simulations show that the rate of transport outside to inside is greater than that of the opposite direction. This seemingly contradictory rectification behavior is the result of sidestepping certain stages of conduction in the MD simulations to accelerate the process.
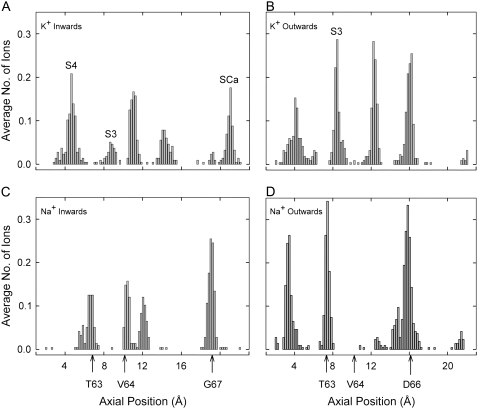
There are several positions in the selectivity filter at which permeant ions dwell preferentially. Histograms of the axial positions (z coordinate) occupied by ions passing through the filter are shown in Fig. 7. The preferred binding sites vary depending on ion type and direction of conduction. The dwell histograms obtained with MD reveal details that are not shown in the histograms obtained BD (see Fig. 4). The histograms obtained with molecular dynamics reveal five K+ peaks in the selectivity filter during inward conduction (Fig. 7 A) and four during outward conduction (Fig. 7 B). There are three Na+ peaks during both inward and outward conduction (Fig. 7, C and D). Not all peaks represent distinct ion binding sites, because the filter pocket (12 < z < 17) is wide enough to accommodate ions in a range of off-axis positions, especially when two Na+ ions are present in the pocket. Defined K+ sites SCa, S3, and S4 are indicated. Although Fig. 7 A contains five peaks, a close examination of the raw data shows that sites S3 and S4 are almost never occupied simultaneously. The average number of ions in the selectivity filter region, derived by integration of the four histograms in Fig. 7, is 3.5–4 ions. We note here that although the total numbers of ions observed in the filter for BD and MD are nearly the same, the binding sites are smeared out in the BD simulations by the uniform dielectric protein and the cylindrically symmetric pore outline.
FIGURE 7.
Dwell histograms of axial positions occupied by ions passing through the filter during MD simulations. Axial position is measured from the average center of the membrane. Arrows show approximate positions of carbonyl oxygens from the indicated residues. All histograms are obtained with an applied electric field of ±1.2 × 109 V/m. Potassium sites SCa, S3, and S4 are labeled where relevant. The four histograms are obtained during: inward conduction of K+ (A), outward conduction of K+ (B), inward conduction of Na+ (C), and outward conduction of Na+ (D).
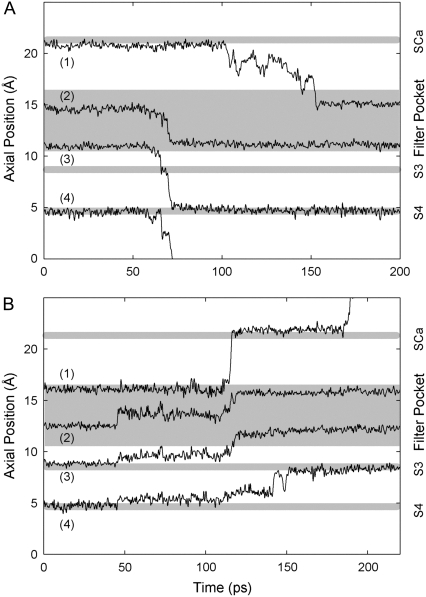
Fig. 8 shows typical trajectories for K+ ions during inward and outward conduction. The ions come to rest at positions corresponding to the peaks in Fig. 7. During inward conduction, shown in Fig. 8 A, ions 2 and 3 in the filter pocket begin to move downwards almost simultaneously, and also appear to trigger the exit of ion 4 from site S4. Ion 3 jumps to site S3 for ∼10 ps and then moves on the newly-vacant S4, while ion 2 takes up the former position of ion 3 at the bottom of the filter pocket. Site S3, located between Thr63 and Val64, is not usually occupied by K+ during inward conduction; once a K+ reaches S3 it rapidly moves on to S4. Within the next 80 ps, ion 1 from site SCa gradually moves down to the upper part of the filter pocket. Thus, the motion of one ion triggers the motion of another through a change in the ion-ion repulsive force, and a conduction event results from several ions moving in concert. During outward conduction (see Fig. 8 B), the key triggering event seems to be the sudden movement of ion 2 at 50 ps. This ion moves around sideways (not shown) within the filter pocket as well as upwards, until it eventually dislodges ion 1. Ion 3 jumps into the filter pocket behind 2, allowing ion 4 to advance outwards soon afterwards. We often find two ions present simultaneously in the filter pocket. Ion 1 stays in site SCa for a short time before moving into solution.
FIGURE 8.
Axial positions of K+ ions in the filter region during typical MD conduction events. Shaded strips indicate binding sites or the filter pocket. The trajectories of the resident K+ ions are obtained during inward (A) and outward conduction (B) under the influence of an applied electric field of magnitude 1.2 × 109 V/m.
Other conduction events differ in detail but display the same general trends. Sodium ions also move more-or-less simultaneously in sudden jumps (results not shown), but they generally sit at different positions from potassium ions when not moving, as can be inferred from the histograms of Fig. 7. The key steps during conduction of both K+ and Na+ seem to be taken by ions that are moving into or out of the filter pocket, which involves partial rehydration or dehydration of the ions.
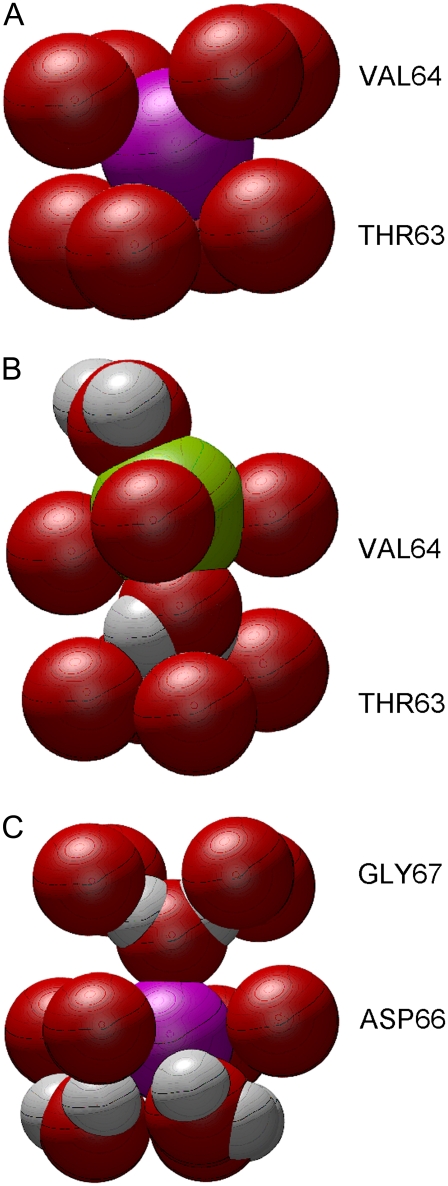
As can be gleaned from Fig. 7, Na+ ions, unlike K+ ions, do not dwell within either of the defined sites S3 and S4. Instead, they are usually found either inside the filter pocket or at the level of one of the planes of carbonyl oxygens from Thr63, Val64, or Gly67. Typical binding situations for K+ and Na+ are shown in Fig. 9. In Fig. 9 A, a K+ ion is sitting in S3, coordinated by all eight surrounding carbonyl oxygens, and not in touch with any water molecules. However, it is very unusual to find Na+ in site S3, as shown by the dwell histograms in Fig. 7. Na+ is normally coordinated by only four of the carbonyl oxygens forming S3, plus two water molecules (see Fig. 9 B). Shrivastava et al. (15) also found such differences between K+ and Na+ binding in KcsA. The same contrast between K+ and Na+ occurs at the outer end of the filter, where a K+ ion typically sits in site SCa, whereas Na+ sits lower, within the plane of the Gly67 oxygens.
FIGURE 9.
Typical positions of ions in the filter relative to pore-lining carbonyl oxygens (red spheres) from the indicated residues. Potassium ion is shown as a purple sphere, sodium ion as a green sphere. Illustrated in panels A–C are, respectively, a K+ ion in site S3 during inwards conduction, a Na+ ion and two water molecules during inward conduction, and a K+ ion and water molecules at the top of the filter pocket during outward conduction.
Most likely the close coordination of a Na+ ion by water molecules as in Fig. 9 B makes it more difficult for a Na+ ion to jump to adjacent positions, especially when a water molecule forms a kind of bridge propped against the next layer of oxygens (from Thr63 in this case). However, a K+ ion faces a similar hydration problem during outward conduction, as shown in Fig. 9 C. Here it is surrounded by the four Asp66 carbonyl oxygens, three water molecules from the filter pocket, and one water molecule bridging the Gly67 oxygens above. The situation arises from the wider spacing of the carbonyl oxygens from Asp66 that help to form the filter pocket. This type of coordination pattern occurs often, and it appears to create a significant barrier at this position during outward conduction of K+. Na+ ions are detained around the same point, but they rarely form the neat structure shown in Fig. 9 C. Often, two Na+ ions end up at nearly the same height, each touching one or two different carbonyl oxygens and several water molecules. Note that all of the differences between K+ and Na+ binding are also found at low or zero membrane potential, and they occur irrespective of the direction of conduction.
DISCUSSION
In carrying out both BD and MD simulations of conduction through the NaK channel, our main aim is to draw from the complementary strengths of the two methods. With BD, we are able to simulate sustained conduction through the entire channel under various combinations of membrane potential and ion concentration. With this simulation method, however, it is not possible to study how the channel discriminates between Na+ and K+. Utilizing MD, we show that the channel allows the passage of both ionic species across the selectivity filter, albeit at somewhat different conduction rates. We observe some differences between the mechanisms of conduction of Na+ and K+ ions.
The results of our BD simulations on the NaK channel, with the M0 helices removed, predict that the channel has a small conductance, is outwardly rectifying and obeys the Michaelis-Menten current-concentration relationship (Fig. 3). The permeation dynamics of the NaK channel we uncovered here broadly mirror those of the KcsA channel. In both channels, the deep energy well created by the carbonyl oxygens lining the selectivity filter attracts 2–3 permeant ions, which dwell preferentially at well-defined binding sites and establish a stable equilibrium with each other (Figs. 2 and 4). This equilibrium is disrupted when an additional ion under the influence of an applied potential enters the pore, and a conduction event occurs as a multiion shuttling process. However, the NaK channel, unlike the KcsA channel, exhibits no pronounced energy well near the intracellular gate. This well, in KcsA, is generated by four acidic residues (Glu118) located near the intracellular gate. The absence of such a well in the vicinity of the internal entrance of the channel severely reduces both inward and outward currents. An ion attempting to move inward from the selectivity filter encounters a large energy barrier in the wider, hydrophobic segment of the channel. The absence of a well there also reduces the probability of an ion entering the intracellular entrance, thus reducing the frequency of outward conduction. By replacing the polar glutamine near the intracellular gate with the acidic glutamate, we show that the magnitudes of both inward and outward current can be appreciably enhanced (Fig. 5). Using the mutant channel, we also show that the channel is blocked by the presence of millimolar concentrations of extracellular calcium ions (Fig. 6).
We demonstrate with MD simulations that the NaK channel indeed conducts both K+ and Na+ ions, with preference for the former. However, there are a number of caveats to the application of MD to assess the conductance properties of the channel. A large, physiologically unrealistic electric field needs to be applied to observe the passage of ions across the channel on a manageable timescale. Moreover, ions have to be prepositioned to enhance conduction, an approach that has also been used, among others, by Khalili-Araghi et al. (27). Despite the use of these procedures to facilitate ion conduction, we can nevertheless glean some important details of the behavior of ions as they move through the selectivity filter.
Our simulations reveal that Na+ preferentially avoids the K+ binding sites and binds instead to just one plane of carbonyl oxygens (Fig. 9). This observation appears not to be in accord with the structures presented by Shi et al. (8), which show identical binding sites for K+ and Na+. On the other hand, Noskov and Roux (26) report that a Na+ ion confined to the vicinity of site S3 moves up or down toward the adjacent planes of carbonyl oxygens, whereas a K+ ion placed in the same position tends to remain centered in the site. Possibly the conditions under which the channels are crystallized and then illuminated in the synchrotron are sufficiently different from cell conditions to alter the preferred positions of Na+ in the protein crystal. If the two water molecules next to a Na+ in the binding site are absent during crystal structure determination, the effect on Na+ binding could be quite significant. The issue of crystallization conditions and their effects on protein structure has been discussed by several authors recently. For example, Halle (44) suggests that hydration is significantly affected during flash cooling of protein, and Boda et al. (45) find large differences in ion behavior during channel simulations at 100 K or 300 K.
Recently, Varma and Rempe (20) analyzed the coordination of K+ and Na+ by oxygen atoms from various molecules, especially sets of glycine dipeptides, which form a cavity similar to site S3 in the NaK channel. Their quantum chemical calculations show that, whereas sites formed by eight oxygens always favor K+ over Na+, sites formed by five or six oxygens favor Na+. Their phase diagram suggests that the free energy for K+ and oxygens does not vary much over the range of four to eight oxygens, but for Na+ the free energy rises significantly when it is surrounded by more than six oxygens. Their results support our findings that K+ sits in the traditional eight-oxygen binding sites but Na+ resides in sites with fewer oxygens. Thomas et al. (21), having obtained similar results from ab initio calculations, point out that any channel (including KcsA) that forces permeating ions to coordinate with eight oxygens at some point will discriminate against Na+.
How does the NaK filter structure allow both Na+ and K+ to pass, in contrast to the standard potassium channels? In the NaK channel, there is only one site (S3) where Na+ faces a discriminatory energy barrier, and that barrier is not very large. The greatest Na+ barrier in the KcsA potassium channel appears to be S2, in the middle of the filter (25), but S2 does not exist in NaK, having merged into the filter pocket along with S0 and S1. Both types of ion are required to rehydrate partially and then dehydrate again as they pass through the filter pocket. S4, formed partly by Thr63 side chains and adjacent to the intracellular solution in the channel's central chamber, is not able to control coordination of ions as strictly as the other sites and is not a barrier for Na+. Thus, both types of ion are conducted through NaK, but the conductivity for Na+ is lower as a result of the residual discrimination from site S3.
In conclusion, the study of the NaK channel using the recently unveiled crystal structure with the combined methods of molecular and Brownian dynamics reveals many of its salient conductance properties. The channel is indeed permeable to both sodium and potassium ions, with a slightly higher permeability for the latter. The current across the wild-type channel is small but it can be appreciably enhanced by substituting one polar residue near the intracellular entrance with an acidic residue. The simulations also demonstrate dynamically that the conduction mechanisms used by Na+ and K+ as they cross the selectivity filter are different, confirming the coordination processes predicted by ab initio calculations (20,21).
Acknowledgments
We thank Dan Gordon for his helpful comments on the manuscript.
This work was supported by grants from the National Health & Medical Research Council of Australia. The calculations upon which this work is based were carried out using the SGI Altix cluster of the Australian National University Supercomputing Facility.
Editor: Peter C. Jordan.
References
- 1.Doyle, D. A., J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- 2.Jiang, Y., A. Lee, J. Chen, M. Cadene, B. T. Chait, and R. MacKinnon. 2002. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 417:515–522. [DOI] [PubMed] [Google Scholar]
- 3.Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. [DOI] [PubMed] [Google Scholar]
- 4.Heginbotham, L., T. Abramson, and R. MacKinnon. 1992. A functional connection between the ores of distantly related ion channels as revealed by mutant K+ channels. Science. 258: 1115–1152. [DOI] [PubMed] [Google Scholar]
- 5.Schrempf, H., O. Schmidt, R. Kümmerlen, S. Hinnah, D. Müller, M. Betzler, T. Steinkamp, and R. Wagner. 1995. A prokaryotic potassium ion channel with two predicted transmembrane segment from Streptomyces lividans. EMBO J. 14:5170–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacKinnon, R., S. L. Cohen, A. Kuo, A. Lee, and B. T. Chait. 1998. Structural conservation in prokaryotic and eukaryotic potassium channels. Nature. 280:106–109. [DOI] [PubMed] [Google Scholar]
- 7.Gamel, K., and V. Torre. 2000. The interaction of Na+ and K+ in the pore of cyclic nucleotide-gated channels. Biophys. J. 79:2475–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi, N., S. Ye, A. Alam, L. Chen, and Y. Jiang. 2006. Atomic structure of a Na+- and K+-conducting channel. Nature. 440:570–574. [DOI] [PubMed] [Google Scholar]
- 9.Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. [DOI] [PubMed] [Google Scholar]
- 10.Allen, T. W., A. Bliznyuk, A. P. Rendell, S. Kuyucak, and S. H. Chung. 2000. The potassium channel: structure, selectivity and diffusion. J. Chem. Phys. 112:8191–8204. [Google Scholar]
- 11.Shrivastava, I. H., and M. S. P. Sansom. 2000. Simulations of ion permeation through a potassium channel: molecular dynamics of KcsA in a phospholipid bilayer. Biophys. J. 78:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guidoni, L., V. Torre, and P. Carloni. 2000. Water and potassium dynamics inside the KcsA (K+) channel. FEBS Lett. 447:37–42. [DOI] [PubMed] [Google Scholar]
- 13.Bernéche, S., and B. Roux. 2001. Energetics of ion conduction through the K+ channel. Nature. 414:73–77. [DOI] [PubMed] [Google Scholar]
- 14.Guidoni, L., and P. Carloni. 2002. Potassium permeation through the KcsA channel: a density functional study. Biochim. Biophys. Acta Biomembr. 1563:1–6. [DOI] [PubMed] [Google Scholar]
- 15.Shrivastava, I. H., D. P. Tieleman, P. C. Biggin, and M. S. P. Sansom. 2002. K+ versus Na+ ions in a K channel selectivity filter: a simulation study. Biophys. J. 83:633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung, S. H., T. W. Allen, and S. Kuyucak. 2002. Conducting-state properties of the KcsA potassium channel from molecular and Brownian dynamics simulations. Biophys. J. 82:628–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung, S. H., M. Hoyles, T. W. Allen, and S. Kuyucak. 1998. Study of ionic currents across a model membrane channel using Brownian dynamics. Biophys. J. 75:793–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung, S. H., T. W. Allen, M. Hoyles, and S. Kuyucak. 1998. Permeation of ions across the potassium channel: Brownian dynamics studies. Biophys. J. 77:2517–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burykin, A., C. N. Schutz, J. Villa, and A. Warshel. 2002. Simulations of ion current in realistic models of ion channels: the KcsA potassium channel. Proteins Struct. Funct. Genet. 47:265–280. [DOI] [PubMed] [Google Scholar]
- 20.Varma, S., and S. B. Rempe. 2007. Tuning ion coordination architectures to enable selective partitioning. Biophys. J. 93:1093–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas, M., D. Jayatilaka, and B. Corry. 2007. The predominant role of coordination number in potassium channel selectivity. Biophys. J. 93:2635–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen, T. W., S. Kuyucak, and S. H. Chung. 1999. Molecular dynamics study of the KcsA potassium channel. Biophys. J. 77:2502–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Åqvist, J., and V. Luzhkov. 2000. Ion permeation mechanism of the potassium channel. Nature. 404:881–884. [DOI] [PubMed] [Google Scholar]
- 24.Bernèche, S., and B. Roux. 2000. Molecular dynamics of the KcsA K+ channel in a bilayer membrane. Biophys. J. 78:2900–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noskov, S. Y., S. Bernéche, and B. Roux. 2004. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 431:830–834. [DOI] [PubMed] [Google Scholar]
- 26.Noskov, S. Y., and B. Roux. 2007. Importance of hydration and dynamics on the selectivity filter of the KcsA and NaK channels. J. Gen. Physiol. 129:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khalili-Araghi, F., E. Tajkhorshid, and K. Schulten. 2006. Dynamics of K+ ion conduction through Kv1.2. Biophys. J. 91:72–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- 29.Im, W., S. Seefeld, and B. Roux. 2000. A grand canonical Monte Carlo-Brownian dynamics algorithm for simulating ion channels. Biophys. J. 79:788–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Corry, B., M. Hoyles, T. W. Allen, M. Walker, S. Kuyucak, and S. H. Chung. 2002. Reservoir boundaries in Brownian dynamics simulations of ion channels. Biophys. J. 82:1975–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng, J. A., T. Vora, V. Krishnamurthy, and S. H. Chung. 2008. Estimating the dielectric constant of the channel protein and pore. Eur. Biophys. J. 37:213–222. [DOI] [PubMed] [Google Scholar]
- 32.Chung, S. H., and B. Corry. 2007. Conduction properties of KcsA measured using Brownian dynamics with flexible carbonyl groups in the selectivity filter. Biophys. J. 93:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyles, M., S. Kuyucak, and S. H. Chung. 1996. Energy barrier presented to ions by the vestibule of the biological membrane channel. Biophys. J. 70:1628–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoyles, M., S. Kuyucak, and S. H. Chung. 1998. Computer simulation of ion conductance in membrane channels. Phys. Rev. E. 58:3654–3661. [Google Scholar]
- 35.van Gunsteren, W. F., and H. J. C. Berendsen. 1982. Algorithms for Brownian dynamics. Mol. Phys. 45:637–647. [Google Scholar]
- 36.van Gunsteren, W. F., H. J. C. Berendsen, and J. A. Rullmann. 1982. Stochastic dynamics for molecules with constraints: Brownian dynamics of n-alkanes. Mol. Phys. 4:69–95. [Google Scholar]
- 37.Corry, B., T. W. Allen, S. Kuyucak, and S. H. Chung. 2001. Mechanisms of permeation and selectivity in calcium channels. Biophys. J. 80:195–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacKerell, A. D., Jr., D. Bashford, M. Bellot, R. L. Dunbrack, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, D. J.-M. S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, R. E. Reiher, III, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin, and M. Karplus. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. [DOI] [PubMed] [Google Scholar]
- 39.Crozier, P. S., R. L. Rowley, N. B. Holladay, D. Henderson, and D. D. Busath. 2001. Molecular dynamics simulation of continuous current flow through a model biological membrane channel. Phys. Rev. Lett. 86:2467–2470. [DOI] [PubMed] [Google Scholar]
- 40.Chung, S. H., T. W. Allen, and S. Kuyucak. 2002. Modeling diverse range of potassium channels with Brownian dynamics. Biophys. J. 83:263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stampe, P., J. Arreola, P. Perez-Cornejo, and T. Begenisich. 1998. Non-independent K+ movement through the pore in IRKI potassium channels. J. Gen. Physiol. 112:475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heginbotham, L., and R. MacKinnon. 1993. Conduction properties of the cloned Shaker K+ channel. Biophys. J. 65:2089–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alam, A., N. Shi, and Y. Jiang. 2007. Structural insight into Ca2+ specificity in tetrameric cation channels. Proc. Natl. Acad. Sci. USA. 104:15334–15339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halle, B. 2004. Biomolecular cryocrystallography: structural changes during flash-cooling. Proc. Natl. Acad. Sci. USA. 101:4793–4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boda, D., W. Nonner, M. Valiskó, D. Henderson, R. Eisenberg, and D. Gillespie. 2007. Steric selectivity in Na+ channels arising from protein polarization and mobile side chains. Biophys. J. 93:1960–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]