Abstract
Cells can sense mechanical force in regulating focal adhesion assembly. One vivid example is the force-induced recruitment of vinculin to reinforce initial contacts between a cell and the extracellular matrix. Crystal structures of the unbound proteins and bound complex between the vinculin head subdomain (Vh1) and the talin vinculin binding site 1 (VBS1) indicate that vinculin undergoes a conformational change upon binding to talin. However, the molecular basis for this event and the precise nature of the binding pathway remain elusive. In this article, molecular dynamics is used to investigate the binding mechanism of Vh1 and VBS1 under minimal constraints to facilitate binding. One simulation demonstrates binding of the two molecules in the complete absence of external force. VBS1 makes early hydrophobic contact with Vh1 by positioning the critical hydrophobic residues (L608, L615, and L622) in the groove formed by helices 1 and 2 of Vh1. The solvent-exposed hydrophobic residues (V619 and L623) then gradually penetrate the hydrophobic core of Vh1, thus further separating helix 1 from helix 2. These critical residues are highly conserved as large hydrophobic side groups in other vinculin binding sites; studies also have demonstrated that these residues are essential in Vh1-VBS1 binding. Similar binding mechanisms are also demonstrated in separate molecular dynamics simulations of Vh1 binding to other vinculin binding sites both in talin and α-actinin.
INTRODUCTION
The ability of a cell to sense mechanical force is central to a wide variety of biophysical processes and is implicated in disease progression. Although the process by which a cell senses mechanical force and converts it into a biochemical signal, or mechanotransduction, has been extensively studied, its basic molecular mechanism remains elusive. One hypothesis is that applied mechanical force induces a conformational change in a force-sensitive protein, which then alters its interactions with other intracellular molecules. Focal adhesion regulation in response to mechanical force is one example that is being intensely investigated (1,2). However, the precise mechanism by which a cell regulates its focal adhesions by mechanical force is still a subject of considerable debate.
Talin and vinculin are essential to the formation of stable focal adhesions. Talin is a cytoplasmic protein with binding sites to other focal adhesion proteins including β-integrin (3) and F-actin (4); talin also contains 11 possible vinculin binding sites (VBSs), many of which are cryptic (5,6). Vinculin likely provides structural reinforcement because it can simultaneously bind to talin and F-actin. It consists of a globular head, a proline-rich neck region, and a rod-like tail domain that contains binding sites for many other cytoplasmic proteins (7,8). The vinculin head is known to bind to α-actinin (9) and talin (10), whereas the vinculin tail is known to bind to paxillin (11), F-actin (12), and phosphatidylinositol 4,5 bisphosphate (PIP2) (13). The neck region binds to the vasodilator-stimulated phosphoprotein, or VASP (14), vinexin (15), and ponsin (16). Vinculin forms an autoinhibitory head-tail interaction within cytosol, which masks many of its binding sites for other proteins (17–20). Recent findings show that the high-affinity autoinhibition interaction in a full-length vinculin is due to the cooperative effect of two low-affinity binding interfaces (21). Therefore, complete vinculin activation requires a combinatory signaling pathway of vinculin interacting with one or more of its binding partners (17). Cells with disrupted talin function fail to form focal adhesions and exhibit spreading defects (22), whereas cells with vinculin disruption can form focal adhesions but display reduced ability to spread and increased cell motility (23).
Because it has been demonstrated that mechanical force is needed for vinculin recruitment to focal adhesions (1), force-induced activation of cryptic VBSs on talin through conformational change may be the mechanosensing pathway leading to recruitment (24). Such recruitment could also lead to reinforcement of the focal adhesion. Indeed, talin1 is critical in force-dependent vinculin recruitment to adhesion sites independent of Src family kinase and focal adhesion kinase activities (2). Initial contacts are particularly attractive for study because they consist of a small number of proteins in which forces are transmitted via extracellular matrix-integrin-talin-F-actin linkages (25).
Some of the talin VBSs are inactive and unable to bind to the vinculin subdomain (Vh1; residues 1–258) (6). Vh1 is a subdomain of the vinculin head that contains the binding site for talin and is used in various talin-binding experiments (6,26). The first vinculin binding site (VBS1; residues 606–636) is the fourth helix (H4) of a stable N-terminal five-helix bundle (TAL5) of talin rod (27). VBS1 has hydrophobic residues that, upon binding to Vh1, become deeply embedded in the hydrophobic core of Vh1 (26). Izard et al. (26) have also demonstrated that VBS1 can bind to Vh1 in the Vh1-vinculin tail domain (Vt; residues 883–1066) complex and effectively sever the Vh1-Vt interaction. Purified talin, however, binds to full-length vinculin at a low affinity, suggesting that talin is only one of a number of binding partners needed for full vinculin activation (17). The same vinculin binding residues of VBS1 that become embedded within Vh1 form a tight hydrophobic core within TAL5 (27). Experiments have shown that isolated TAL5 has a low binding affinity for Vh1, whereas a four-helix bundle with helix 5 removed from TAL5 (6), a mutated TAL5 with an unstable hydrophobic core (27), or the wild-type TAL5 in elevated temperature solvent (6) can each disrupt TAL5 stability and strongly bind to Vh1.
Recent molecular dynamics simulations have demonstrated a mechanism by which transmitted mechanical force disrupts TAL5 stability and activates it to bind to Vh1 by a process in which a torque is applied to helix 4, causing it to rotate and thereby making the binding site accessible (28). The hydrophobic residues exposed under applied force are those that are known to be important in binding to Vh1 (5). For VBS1 to bind to Vh1, however, VBS1 must separate two helices of Vh1 and embed itself in between. How this occurs, has been a matter of considerable speculation, especially with the VBS1 cryptic within talin (6,27).
In this article, molecular dynamics is used to investigate the binding mechanism of Vh1 to VBS1, which is the immediate step following force-induced talin activation (28). Together, these two steps comprise the early mechanotransduction events in the force-induced recruitment of vinculin (1).
METHODS
Vh1-VBS1 binding simulation with effective energy function 1
Unbound and bound models of the Vh1-VBS1 complex (Fig. 1) were obtained from crystal structures of Vh1-Vt (Protein Data Bank (PDB) ID: 1RKE) and the Vh1-VBS1 complexes (PDB ID: 1T01), respectively. When Vh1 binds to VBS1, the N-terminal four-helix bundle of Vh1 bends over and undergoes considerable conformational change, whereas the C-terminal four-helix bundle of Vh1 remains unchanged (26). Therefore, the Vh1 structures from the bound and unbound models were aligned by the backbone atoms of the C-terminal four-helix bundle of Vh1 to highlight the conformational difference between the two models. The vinculin tail domain (Vt) was removed from the Vh1-Vt complex. VBS1 was aligned with its binding site but translated 12 Å away from Vh1 to obtain the unbound Vh1-VBS1 model (Fig. 1 A), whereas the known Vh1-VBS1 crystal structure was used as the bound Vh1-VBS1 model (Fig. 1 B). Views from the top show the separation of the molecules (Fig. 1 C) and the associated conformational change upon VBS1 binding toVh1 (Fig. 1 D). All of the visualizations presented in this article were performed with visual molecular dynamics (29).
FIGURE 1.
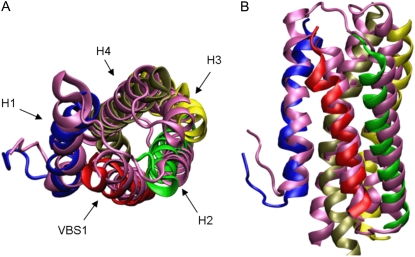
(A) Vh1 (PDB ID: 1RKE) and VBS1 (PDB ID: 1T01) unbound structures viewed from the front. VBS1 is translated by 12 Å from its corresponding position within the Vh1-VBS1 complex. (B) Vh1 and VBS1 bound complex (PDB ID: 1T01) viewed from the front. (C) Vh1 and VBS1 unbound structures viewed from the top. Only the first four helices of Vh1 (a seven-helix bundle) are shown for clarity. (D) Vh1 and VBS1 bound complex viewed from the top.
Commercial molecular dynamics software, CHARMM (Harvard University, Cambridge, MA) (30), was used with the effective energy function 1 (EEF1) (31) solvent model and the CHARMM19 force field (32). The default nonbonded cutoff parameters CTONNB = 7.0, CTOFNB = 9.0, and CUTNB = 10.0 were used, where the correction for the long-range effect beyond the 9 Å cutoff had been hardcoded in the EEF1 method. Energies of these models were minimized by alternating the steepest decent and adopted basis Newton Raphson methods with 3000 steps. Bond lengths between hydrogen and heavy atoms were fixed using the SHAKE constraint (33), and a 2 fs timestep was used. Molecules were heated to 300 K over 40 ps, followed by a 560 ps equilibration period at 300 K. During the heating and equilibrium process, weak harmonic constraints (0.1 kcal/mol/Å2) were applied to the Cα atoms to minimize deviations from the original position. After equilibration, Cα constraints were removed, and the production simulations were run using the Nosé-Hoover (34,35) thermostat for constant temperature control at 300 K.
Vh1-VBS1 binding simulations were performed beginning with the unbound Vh1 and VBS1 model (Fig. 1, A and C). In some simulations, VBS1 was initially rotated around the helix axis ±2° to determine the effect on binding of VBS1 orientation relative to Vh1. Two types of Vh1-VBS1 binding simulations were carried out. In one, all constraints were removed after equilibration to determine how the two molecules, initially separated, might interact in the complete absence of external forces. In the other, distance constraints were imposed. The parts of the atom pair were pulled toward each other when they were separated by the prespecified reference distance, which was between residues on VBS1 (L608, L615, and L622) and Vh1 (V16, L23, V44, L116, and F126), for 800 ps after equilibration to enhance the probability of Vh1-VBS1 binding. In a distance constraint, force of F = kmax × (dmax − d) was applied to the atom pair only if d > dmax, where kmax is the force constant, dmax is the maximum reference distance, and d is the current distance between the atom pair. Low-level force constraints were imposed (kmax = 0.5 kcal/mol/Å2 and dmax = 3.0Å) between Vh1 and VBS1 pair selections for a short period (800 ps) at the beginning of the simulation. Distance constraints with kmax = 0.5 kcal/mol/Å2 and dmax of the pair distance obtained from the crystal structure were imposed on the VBS1 backbone hydrogen bonding pairs, which were intended to retain its helicity. All of these constraints were completely removed after 800 ps, and the simulations were continued for 32 ns free of constraints. A 30 ns simulation was conducted on the Vh1-VBS1 bound complex (Fig. 1, B and D) with no constraints to characterize the binding interaction between the two molecules.
Mutational studies on Vh1-VBS1 binding and Vh1 binding to other VBSs
A number of Vh1-VBS1 unbound models were obtained with various VBS1 mutations: 1), L622 mutated to alanine (L622A); 2), L623 mutated to alanine (L623A); and 3), K613 mutated to proline (K613P). Unbound models of Vh1 with other VBSs were constructed by the same method used for obtaining the Vh1-VBS1 unbound structures but with the following crystal structures: 1), Vh1-Vt; 2), Vh1-VBS2 (PDB ID: 1U6H); 3),Vh1-VBS3 (PDB ID: 1RKC); and 4), Vh1-αVBS (PDB ID: 1YDI). The αVBS is the binding segment within α-actinin, with its sequence running in the opposite direction. Identical constraints like the ones applied between Vh1 and VBS1 were applied for 800 ps to Vh1 and other or modified VBSs. Only in the case of the Vh1-VBS3 binding simulation, for which the C-terminal end of VBS3 tended to unfold and did not bind to Vh1, were additional distance constraints on the hydrogen bonding pairs of the VBS3 backbone helix imposed to retain helicity. Specifically, a distance constraint with force constant of k = 0.5 kcal/mol/Å2 was applied between helix hydrogen bonding O and N atom pairs on the VBS3 backbone. As before, these constraints were removed after 800 ps, and simulations continued for 32 ns.
Simulation on Vh1-TAL5 binding
The structure of activated TAL5 (i.e., hydrophobic residues exposed to solvent) was obtained from the end state of force-induced activated TAL5 (28). VBS1 of TAL5 was aligned to VBS1 of the Vh1-VBS1 bound complex model (Fig. 1, B and D). After the heating and equilibration, stronger distance constraints (kmax = 1.0 kcal/mol/Å2 instead of kmax = 0.5 kcal/mol/Å2 as in the Vh1-VBS1 binding simulation) were applied between Vh1 and VBS1 of TAL5 throughout the simulation to enforce the binding of Vh1 to TAL5 through VBS1. Additional helicity constraints were also applied to Vh1 and TAL5 helices to force them to retain the secondary structures. Helicity constraints were specified in the identical manner as in the Vh1-VBS3 binding model.
RESULTS
Unconstrained binding
In the first series of Vh1-VBS1 simulations with no external constraints and the initial condition of Fig. 1 A, one simulation proceeded to complete binding, for which the end configuration is very similar to the bound Vh1-VBS1 crystal structure (Fig. 1, B and D). For this binding simulation and equilibration simulation starting from the bound complex (Fig. 1 B), the average root mean-square deviation (RMSD) of the Cα atoms of Vh1's N-terminal four-helix bundle and VBS1 were 2.33 Å and 1.97 Å, respectively. The superimposed configurations from near the end of the binding simulation and equilibration simulation show that the two are very close in their relative orientations (Fig. 2, A and B).
FIGURE 2.
(A) One conformation from Vh1-VBS1 binding simulation (same coloring scheme as Fig. 1) is superimposed with a conformation from Vh1-VBS1 equilibrium simulation (purple) viewed from the top and (B) same viewed from the front.
In the constraint-free Vh1-VBS1 binding simulation, VBS1 is displaced by 12 Å from its initial bound position to ensure no atomic clashes, but the closest distance between the two molecules (side-chain atoms of the two molecules) is much shorter than 12 Å. Within the first few picoseconds of simulation, Vh1 and VBS1 were brought together and formed an initial interaction. A large hydrophobic patch is exposed to solvent on the H1 and H2 interface, and the hydrophobic residues of VBS1 become inserted between helices H1 and H2. Through this hydrophobic insertion, L608, L615, and L622 of VBS1 form contacts with V16, L23, V44, A50, and L54 of Vh1, and this contact stabilizes the interaction of the two molecules (Fig. 3, A and D). Hydrogen bonds are formed between Q627 of VBS1 to H22 of Vh1 and between Q610 and K613 of VBS1 to N53 and R56 of Vh1. VBS1 moves farther between H1 and H2 with time as it separates H1 and H2 (Fig. 3, B and E). During this stage, VBS1 also moves closer to H4 of Vh1. Binding of Vh1 and VBS1 is complete when VBS1 rotates and effectively locks the exposed hydrophobic residues (L619 and L623) into the hydrophobic core of Vh1 (Fig. 3, C and F), which happened 34 ns into the production run. When L623 moves into the hydrophobic core, R624 of VBS1 swings over to form hydrogen bonds with Q19 and H22 of Vh1's H1. This Vh1-VBS1 binding mechanism viewed from the front and on a cross-sectional plane is shown in Fig. 3.
FIGURE 3.
Snapshots from one Vh1-VBS1 binding simulation with ribbon representations for the helical backbone, stick representations for polar and charged residues, and spherical representations for hydrophobic residues at (A) VBS1 hydrophobic insertion (t = 0.8 ns) between the hydrophobic patch of H1 and H2, (B) Vh1's H1 and H2 separation (t = 23.6 ns), and (C) VBS1 rotation (t = 39.2 ns) to snap in the exposed hydrophobic residues, i.e., L623, into the hydrophobic core. Some of the critical residues in Vh1-VBS1 binding are labeled: residues on VBS1 (red), residues on H1 (blue) and residues on H2 (green). (D–F) Corresponding cross-sectional views to (A–C) at the plane represented by the dashed line in Fig. 3 C. Corresponding animations are available (Movie S1, Movie S2, Movie S3, and Data S1).
Constrained binding
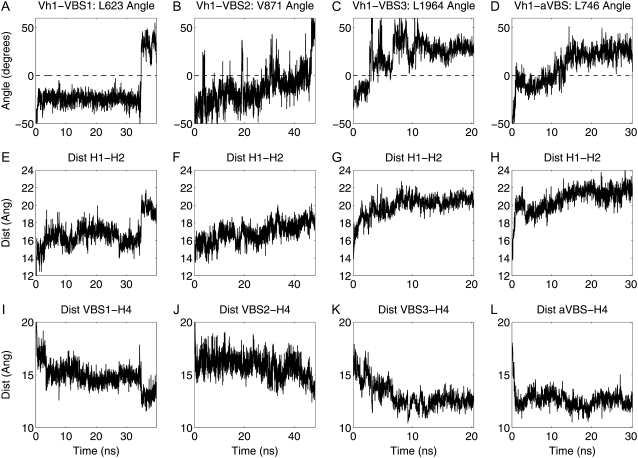
Vh1-VBS1 binding is enhanced when VBS1 is constrained so that its hydrophobic residues are inserted between H1 and H2 of Vh1 in the beginning of the binding simulations. Even though the constraints between Vh1 and VBS1 are completely removed after 800 ps, this proved sufficient to induce Vh1-VBS1 binding that occurred many nanoseconds later. The 800 ps of applying constraints increased the chances of VBS1 forming the initial and necessary hydrophobic insert into Vh1 (Fig. 3, A and D); the separation of H1 and H2 of Vh1, however, did not occur during this 800 ps but happened much later in the simulations. The 30 ns equilibration simulation of the Vh1-VBS1 complex (Fig. 1 B) was analyzed to determine the characteristics of the Vh1-VBS1 complex. Three measures were chosen to be the indicators of Vh1-VBS1 binding status: 1), the angle formed by L623 with VBS1-H1 vector (A(623)); 2), the distance between H1 and H2 of Vh1 (D(1-2)); and 3), the distance between VBS1 and H4 (D(V-4)) (Fig. 3 E). Average values for A(623), D(1-2), and D(V-4) from the equilibration simulation were evaluated to be 30.1°, 20.9 Å, and 12.2 Å, respectively. In a given Vh1-VBS1 binding simulation, these three indicators were monitored to decide whether the molecule underwent binding. For example, the first instant when A(623) > 30.1°, D(1-2) > 20.9 Å, and D(V-4) < 12.2 Å simultaneously is defined as the time when the Vh1-VBS1 binding is complete. Note that this is only used to define Vh1-VBS1 binding, and the three values actually fluctuate about the threshold values described above. Of 12 simulations with slightly different initial conditions, 6 simulations (50%) underwent binding with the averages of these values to be A(623) = 36.9° ± 6.9°, D(1-2) = 20.2 ± 0.8 Å, and D(V-4) = 12.6 ± 0.8 Å; all values are very close to those obtained from the Vh1-VBS1 complex equilibration simulation. The average time for binding to occur was 13.9 ± 8.0 ns.
Binding of vinculin with other VBSs
Simulations between Vh1 with VBS2, VBS3, and αVBS all underwent complete binding similar to that observed with Vh1-VBS1 (see Figs. 3–5). The plots of the three indicators defined above provide evidence to support that all VBSs bind to Vh1 through a combination of hydrophobic insertion, H1-H2 displacement, and VBS rotation (Fig. 6). Simulations between Vh1 and VBS1 with various mutations on VBS1 (K613P, L622A, and L623A) and Vh1 (A50I), however, did not bind, as expected (Fig. 7). To investigate VBS1 secondary structure stability, the extent of helicity, as measured by the number of hydrogen bonds, was evaluated for 1), Vh1-VBS1 complex equilibration; 2), Vh1-VBS1 constraint-free binding simulation; and 3), Vh1-VBS1 with K613P mutation binding simulation (Fig. 8).
FIGURE 5.
Crystal structures of Vh1 bound to various VBSs. The backbones of helical sequences are shown in ribbon representation; polar and charged residues are shown in stick representation; and the hydrophobic residues are shown as spheres. (A) Vh1-VBS1 complex (PDB ID: 1T01) with critical residues important in binding of the two molecules. VBS1 is shown as red ribbon. (B) Vh1-VBS2 complex (PDB ID: 1U6H) and corresponding critical residues are labeled. VBS2 is shown as a gray ribbon. (C) Vh1-VBS3 complex (PDB ID: 1RKC) and the corresponding critical residues are labeled. VBS3 is shown as a purple ribbon. (D) Vh1-αVBS complex (PDB ID: 1YDI) and the corresponding critical residues are labeled. αVBS is shown as a silver ribbon.
FIGURE 4.
Snapshots from Vh1-VBS1 dissociation simulation with same color representation as in Fig. 3. Five Cα atoms on VBS1 (cyan stars) are pulled out of the page at constant velocity. (A) At t = 0 ns, (B) at t = 5.12 ns, and (C) at t = 8.70 ns.
FIGURE 6.
(A) Angle formed by L623 with a vector connecting H1-VBS1 defined as A(623) in Fig. 3 E. (B) Angle formed by V871 with a vector connecting H1-VBS2. (C) Angle formed by L1964 with a vector connecting H1-VBS3. (D) Angle formed by L746 with a vector connecting H1-αVBS. (E–H) Distance between H1 and H2 in Vh1-VBS1, Vh1-VBS2, Vh1-VBS3, and Vh1-αVBS binding simulations. Distance defined as D(1-2) in Fig. 3 E. (I–L) Distance of VBS and H4 in Vh1-VBS1, Vh1-VBS2, Vh1-VBS3, and Vh1-αVBS binding simulations. Distance of VBS1-H4 defined as D(V-4) in Fig. 3 E.
FIGURE 7.
Results from the binding simulation of Vh1 with mutated VBS1 similar to Fig. 3. (A) Angle formed by L623 of wild-type VBS1 with vector connecting H1-VBS1. (B) Angle formed by L623 with vector connecting H1-VBS1, where VBS1 has K613P mutation. (C) Angle formed by L623 with vector connecting H1-VBS1, where VBS1 has L622A mutation. (D) Angle formed by A623 with vector connecting H1-VBS1, where VBS1 has L623A mutation. (E) Angle formed by A623 with vector connecting H1-VBS1, where Vh1 has I50A mutation. (F–J) Distance of H1 and H2 in Vh1-VBS1 binding simulations with wild-type, K613P, L622A, and L623A mutations on VBS1 and I50A mutation on Vh1, respectively. (K–O) Distance of VBS1 and H4 in Vh1-VBS1 binding simulations with wild-type, K613P, L622A, and L623A mutations on VBS1 and I50A mutation on Vh1, respectively.
FIGURE 8.
Helicity of VBS1 at Vh1-VBS1 bound complex equilibrium simulation; Vh1-VBS1 binding simulation; and Vh1-VBS1 with K613P mutation binding simulation.
DISCUSSION AND CONCLUSIONS
The simulations demonstrate that the critical early interaction between Vh1 and VBS1 is the insertion of the hydrophobic residues of VBS1 between H1 and H2 of Vh1 (Fig. 3 A). After the hydrophobic interaction of Vh1 with VBS1, H1 and H2 in the N-terminal four-helix bundle of Vh1 are displaced to make room for VBS1 in between them. By packing their hydrophobic residues in the core, the bound N-terminal four-helix bundle and VBS1 form a new five-helix bundle structure, as previously suggested by Izard et al. (26). This binding mechanism occurred during a simulation time of 34 ns in only one of the constraint-free simulations with Vh1 and VBS1 initially displayed by 12 Å from the original bound position (Supplementary Material, Movie S1, Movie S2, and Data S1). However, the low success rate from constraint-free simulations alone is insufficient to confirm that this is indeed the Vh1-VBS1 binding mechanism. To examine whether the unbinding of VBS1 from the Vh1-VBS1 bound complex follows the same pathway of the proposed binding mechanism but in reverse, a constant velocity unbinding simulation of VBS1 was performed. Interestingly, the unbinding mechanism did closely follow the reverse path of binding, i.e., VBS1 rotation, H1 and H2 of Vh1 closing due to VBS1 being pulled away, and complete unbinding (Fig. 4; Movie S3 and Data S1). After observing the constraint-free simulation results, we theorized that the initial hydrophobic insert of VBS1 is the critical step in the Vh1-VBS1 binding mechanism. To test this significant hypothesis, we performed constrained binding simulations to enhance this initial interaction. With the aid of an initial 800 ps of distance constraints to position VBS1 between H1 and H2 (without yet displacing H1 and H2) and to retain VBS1 helicity, 50% of the simulations (6 of 12) underwent binding by the same mechanism. This took an average of 13.9 ± 8.0 ns after the constraints were completely removed. Although these simulations do not represent the full binding of Vh1-VBS1, they do strongly support our suggested hypothesis. Therefore, the critical step in the binding mechanism is the hydrophobic insertion of VBS1 into Vh1; once this occurs, VBS1 continues to push its way into the hydrophobic core and finally snaps in by rotating the remaining exposed hydrophobic residues (L619 and L623) into the core. One should note that the measured time (13.9 ± 8.0 ns) only represents the time it took for VBS1 to be inserted into Vh1 in the constrained simulations with the EEF1 implicit model; it would not be representative of the binding timescale of the two molecules in solution. It is also important to note that all of the secondary structures of both proteins remain largely intact during binding. This observation leads us to believe that the forced activation of talin is a subtle change in conformation and that complete unfolding is not necessary.
One constraint-free simulation actually underwent the Vh1-VBS1 binding mechanism out of 20 attempted constraint-free simulations. In a majority of the unsuccessful binding simulations, the hydrophobic residues of VBS1 (L608, L615, and L622) were displaced away from the H1-H2 groove of Vh1. For example, simulations in which VBS1 hydrophobic residues interacted only with the opposite side of H2 failed to displace H1 from H2 and did not bind. It is important to note that most of the key hydrophobic residues remained exposed to solvent during the simulation, because they are not situated in between H1 and H2 of Vh1. In no simulations did the VBS1 diffuse away, even though the molecules did not undergo complete binding. Although there have been numerical studies of binding by induced fit between two molecules (36), this article is the first, to our knowledge, to demonstrate constraint-free binding of focal adhesion proteins with a particular emphasis on mechanotransduction. This study suggests that the key to simulating molecular binding is to ensure the correct orientation and initial contact between the two molecules. Although this could be achieved without knowledge of the structure of the bound complex, it would require a large number of calculations to identify the correct approach, which would be computationally prohibitive in most cases. In addition, the fact that 50% of the constrained simulations did not undergo binding is not surprising, given that the binding process, even in these implicit water simulations, will have a wide distribution of timescales. In addition, real binding in the presence of solvent could take orders of magnitude longer to occur. It is quite likely that protein-protein binding may not be observable in many systems unless some constraints are imposed to enforce the initial contact between the two molecules.
Our proposed binding mechanism is further supported by successful binding simulations of Vh1 with four different VBS peptides through the identical binding mechanism: hydrophobic insertion, displacement of H1 and H2, and rotation of VBS (Figs. 3 and 6). All the critical hydrophobic residues involved in the hydrophobic insertion are also found in nearby positions for VBS1, VBS2, VBS3, and αVBS (Fig. 5). Interestingly, the residue sequence in αVBS is reversed to that of talin VBS, but the critical hydrophobic residues are still found in the corresponding positions needed to undergo the proposed binding mechanism to Vh1. Similar to talin, α-actinin contains a cryptic VBS and is possibly subjected to mechanical force within cell-cell junction (37). Therefore, vinculin and α-actinin binding may proceed in the similar mechanism within cell-cell junctions as well as cell-matrix junctions. Also, this generality provides a critical insight into how talin, which contains 11 potential VBSs (5), might modify its conformation when subjected to tensile force to recruit multiple vinculins with a concomitant increase in adhesion strength, as has been observed experimentally (38).
When Vh1 and VBS1 form the initial hydrophobic contact, a number of hydrogen bonds are formed surrounding the hydrophobic interface. In particular, the hydrogen bond formed between Q627 and H22 was persistent throughout the binding simulations and was initially suspected to be a critical interaction that stabilized the initial hydrophobic insert in place. However, this hypothesis was disproved by mutating either Q627 on Vh1 or H22 on VBS1 to alanine and demonstrating through additional simulations that binding still occurs in these mutation simulations. The hydrogen bond persisted because hydrophobic insertion stabilized the interaction of the two molecules and placed Q627 and H22 in close proximity, allowing the bond to remain intact rather than the reverse. This is yet another example that supports the importance of the initial hydrophobic insertion of VBS1 and demonstrates the power of using molecular dynamics to quickly test hypotheses.
Gingras et al. (5) identified the critical residues on VBS1 for binding to Vh1 through a comprehensive mutational study. The identified critical residues were mostly hydrophobic, a finding that is consistent with this numerical study. The mutational study by Gingras et al. (5) showed that L608, L615, L622, and L623 are each individually critical for the stable binding of VBS1 to Vh1, the mechanism for which can be derived from our numerical results. L622 is apparently important in the hydrophobic insertion of VBS1 and, when it is mutated to alanine (L622A), Vh1-VBS1 binding does not proceed because the hydrophobic insertion is inhibited (Fig. 7, C, H, and M). In contrast, the simulation with L623A underwent the initial hydrophobic insertion and nearly completed the entire binding process. The smaller alanine residue, however, was insufficient to snap into the core and remain bound, hence destabilizing the Vh1-VBS1 bound complex (Fig. 7, D, I, and N). An interesting finding in the mutational study (5) was that binding was abolished when any one of VBS1 residues was mutated to a proline. A binding simulation with K613P mutation on VBS1 shows that the proline induces a break in the α-helix, which significantly reduces VBS1 helicity and prevents VBS1 hydrophobic insertion into Vh1 (Figs. 7, B, G, and L, and 8). One mutation on vinculin, A50I, also has been observed to inhibit vinculin-talin binding by stabilizing the interaction between H1 and H2 of vinculin (17). Indeed, the A50I mutation simulation did not undergo Vh1-VBS1 binding as the bulk of Ile prevented L615 of VBS1 from inserting itself between H1 and H2 (Fig. 7, E, J, and O).
Interestingly, the hydrophobic residues (L608, L615, and L622) of VBS1 involved in the hydrophobic insertion between H1 and H2 of Vh1 are the exact same residues that are exposed to solvent in force-induced activation of TAL5 (28). In the activated TAL5 structure, however, the hydrophobic residues (V619 and L623) that snap into the hydrophobic core in the later stages of binding are still embedded in the hydrophobic core of TAL5 (Fig. 9). Therefore, there must be a secondary conformational change in TAL5, which was not captured in the previous simulations, that exposes these two residues. Based on these observations, one hypothesis is that the transmitted tensile force altering TAL5 structure to expose hydrophobic residues may be one of the critical factors in the initial interaction with vinculin for binding. However, further numerical and experimental studies are needed to verify the proposed mechanism. Our previous study of force-induced activation of TAL5 (28) also relied on EEF1 as the implicit water model for the calculations, although some explicit water simulations also were performed. Comparison between the different implicit and explicit water models showed consistency in terms of the identified critical interactions and provided the motivation to use EEF1 in the current simulations. To approximate the bound configuration of Vh1 and TAL5, we performed binding simulations on Vh1 and force-activated TAL5 with excessive distance constraints between Vh1 and VBS1 of TAL5 in this study to force binding. The resulting configuration of the Vh1-TAL5 complex is shown in Fig. 9 C, which forms a nine-helical bundle together with VBS1 donated to the N-terminal four-helix bundle of Vh1. The binding mechanism viewed from the side is shown in Fig. 9, D–F.
FIGURE 9.
Proposed binding model for Vh1 and TAL5. (A) The N-terminal four-helix bundle of Vh1 is shown with the same coloring scheme as in Fig. 1. Inactivated TAL5 containing VBS1 is shown in red as viewed from the top. The hydrophobic residues necessary for Vh1 binding are shown as orange spheres. (B) With application of mechanical force on TAL5, VBS1 undergoes rigid body rotation to expose the hydrophobic residues (28). Applied force is represented by the red arrows, and VBS1 rotation is indicated by the black arrow. (C) Final configuration of Vh1 with TAL5 in a binding simulation. (D–F) The corresponding configurations to (A–C) viewed from the side.
All the simulations presented here are conducted without Vt; therefore, we are proposing a binding mechanism of Vh1 and VBSs. Further studies are needed to determine the vinculin activation mechanism in the presence of talin VBS (26,37). Recent evidence shows that vinculin autoinhibition and vinculin activation are achieved by cooperative efforts (17,21), that is, talin binding must be accompanied by other molecular binding (e.g., PIP2 (18)) to vinculin for full vinculin activation.
In conclusion, a Vh1-VBS binding mechanism has been proposed that involves hydrophobic insertion of VBS1 into Vh1, separation of H1 and H2 of Vh1, and VBS1 rotation to snap in exposed hydrophobic residues into the hydrophobic core. Results from mutational simulations and binding simulations with other VBSs suggest that the proposed mechanism may be more generally valid. This work constitutes the potential early stages of force-induced focal adhesion strengthening by vinculin recruitment immediately after the force-induced talin activation mechanism (28).
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health, grants HL064858 and GM076689 (R.D.K and M.R.K.M.), the Gates Millennium Scholarship, and a Department of Energy Computational Science Graduate Fellowship.
Editor: Gregory A. Voth.
References
- 1.Galbraith, C. G., K. M. Yamada, and M. P. Sheetz. 2002. The relationship between force and focal complex development. J. Cell Biol. 159:695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giannone, G., G. Jiang, D. H. Sutton, D. R. Critchley, and M. P. Sheetz. 2003. Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J. Cell Biol. 163:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xing, B. D., A. Jedsadayanmata, and S. C. T. Lam. 2001. Localization of an integrin binding site to the C terminus of talin. J. Biol. Chem. 276:44373–44378. [DOI] [PubMed] [Google Scholar]
- 4.Hemmings, L., D. J. G. Rees, V. Ohanian, S. J. Bolton, A. P. Gilmore, B. Patel, H. Priddle, J. E. Trevithick, R. O. Hynes, and D. R. Critchley. 1996. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 109:2715–2726. [DOI] [PubMed] [Google Scholar]
- 5.Gingras, A. R., W. H. Ziegler, R. Frank, I. L. Barsukov, G. C. Roberts, D. R. Critchley, and J. Emsley. 2005. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J. Biol. Chem. 280:37217–37224. [DOI] [PubMed] [Google Scholar]
- 6.Patel, B. C., A. R. Gingras, A. A. Bobkov, L. M. Fujimoto, M. Zhang, R. C. Liddington, D. Mazzeo, J. Emsley, G. C. Roberts, I. L. Barsukov, and D. R. Critchley. 2006. The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 281:7458–7467. [DOI] [PubMed] [Google Scholar]
- 7.Bakolitsa, C., J. M. de Pereda, C. R. Bagshaw, D. R. Critchley, and R. C. Liddington. 1999. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 99:603–613. [DOI] [PubMed] [Google Scholar]
- 8.Winkler, J., H. Lunsdorf, and B. M. Jockusch. 1996. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol. 116:270–277. [DOI] [PubMed] [Google Scholar]
- 9.Kroemker, M., A. H. Rudiger, B. M. Jockusch, and M. Rudiger. 1994. Intramolecular interactions in vinculin control alpha-actinin binding to the vinculin head. FEBS Lett. 355:259–262. [DOI] [PubMed] [Google Scholar]
- 10.Gilmore, A. P., P. Jackson, G. T. Waites, and D. R. Critchley. 1992. Further characterization of the talin-binding site in the cytoskeletal protein vinculin. J. Cell Sci. 103:719–731. [DOI] [PubMed] [Google Scholar]
- 11.Wood, C. K., C. E. Turner, P. Jackson, and D. R. Critchley. 1994. Characterization of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J. Cell Sci. 107:709–717. [PubMed] [Google Scholar]
- 12.Menkel, A. R., M. Kroemker, P. Bubeck, M. Ronsiek, G. Nikolai, and B. M. Jockusch. 1994. Characterization of an F-actin binding domain in the cytoskeletal protein vinculin. J. Cell Biol. 126:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, R. P., V. Niggli, P. Durrer, and S. W. Craig. 1998. A conserved motif in the tail domain of vinculin mediates association with and insertion into acidic phospholipid bilayers. Biochemistry. 37:10211–10222. [DOI] [PubMed] [Google Scholar]
- 14.Brindle, N. P. J., M. R. Holt, J. E. Davies, C. J. Price, and D. R. Critchley. 1996. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 318:753–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kioka, N., S. Sakata, T. Kawauchi, T. Amachi, S. K. Akiyama, K. Okazaki, C. Yaen, K. M. Yamada, and S. Aota. 1999. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J. Cell Biol. 144:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mandai, K., H. Nakanishi, A. Satoh, K. Takahashi, K. Satoh, H. Nishioka, A. Mizoguchi, and Y. Takai. 1999. Ponsin/SH3P12: an 1-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J. Cell Biol. 144:1001–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakolitsa, C., D. M. Cohen, L. A. Bankston, A. A. Bobkov, G. W. Cadwell, L. Jennings, D. R. Critchley, S. W. Craig, and R. C. Liddington. 2004. Structural basis for vinculin activation at sites of cell adhesion. Nature. 430:583–586. [DOI] [PubMed] [Google Scholar]
- 18.Gilmore, A. P., and K. Burridge. 1996. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature. 381:531–535. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, R. P., and S. W. Craig. 1994. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J. Biol. Chem. 269:12611–12619. [PubMed] [Google Scholar]
- 20.Johnson, R. P., and S. W. Craig. 1995. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 373:261–264. [DOI] [PubMed] [Google Scholar]
- 21.Cohen, D. M., H. Chen, R. P. Johnson, B. Choudhury, and S. W. Craig. 2005. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280:17109–17117. [DOI] [PubMed] [Google Scholar]
- 22.Priddle, H., L. Hemmings, S. Monkley, A. Woods, B. Patel, D. Sutton, G. A. Dunn, D. Zicha, and D. R. Critchley. 1998. Disruption of the talin gene compromises focal adhesion assembly in undifferentiated but not differentiated embryonic stem cells. J. Cell Biol. 142:1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, W. M., J. L. Coll, and E. D. Adamson. 1998. Rescue of the mutant phenotype by reexpression of full-length vinculin in null F9 cells; effects on cell locomotion by domain deleted vinculin. J. Cell Sci. 111:1535–1544. [DOI] [PubMed] [Google Scholar]
- 24.Vogel, V. 2006. Mechanotransduction involving multimodular proteins: converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 35:459–488. [DOI] [PubMed] [Google Scholar]
- 25.Jiang, G. Y., G. Giannone, D. R. Critchley, E. Fukumoto, and M. P. Sheetz. 2003. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 424:334–337. [DOI] [PubMed] [Google Scholar]
- 26.Izard, T., G. Evans, R. A. Borgon, C. L. Rush, G. Bricogne, and P. R. J. Bois. 2004. Vinculin activation by talin through helical bundle conversion. Nature. 427:171–175. [DOI] [PubMed] [Google Scholar]
- 27.Papagrigoriou, E., A. R. Gingras, I. L. Barsukov, N. Bate, I. J. Fillingham, B. Patel, R. Frank, W. H. Ziegler, G. C. K. Roberts, D. R. Critchley, and J. Emsley. 2004. Activation of a vinculin-binding site in the talin rod involves rearrangement of a five-helix bundle. EMBO J. 23:2942–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, S. E., R. D. Kamm, and M. R. Mofrad. 2007. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 40:2096–2106. [DOI] [PubMed] [Google Scholar]
- 29.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27–38. [DOI] [PubMed] [Google Scholar]
- 30.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4:187–217. [Google Scholar]
- 31.Lazaridis, T., and M. Karplus. 1999. Effective energy function for proteins in solution. Proteins. 35:133–152. [DOI] [PubMed] [Google Scholar]
- 32.Neria, E., S. Fischer, and M. Karplus. 1996. Simulation of activation free energies in molecular dynamics system. J. Chem. Phys. 105:1902–1921. [Google Scholar]
- 33.Krautler, V., W. F. Van Gunsteren, and P. H. Hunenberger. 2001. A fast SHAKE: algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J. Comput. Chem. 22:501–508. [Google Scholar]
- 34.Hoover, W. G. 1985. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 31:1695–1697. [DOI] [PubMed] [Google Scholar]
- 35.Nosé, S. 1984. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52:255–268. [Google Scholar]
- 36.Latzer, J., G. A. Papoian, M. C. Prentiss, E. A. Komives, and P. G. Wolynes. 2007. Induced fit, folding, and recognition of the NF-kappaB-nuclear localization signals by IkappaBalpha and IkappaBbeta. J. Mol. Biol. 367:262–274. [DOI] [PubMed] [Google Scholar]
- 37.Bois, P. R., R. A. Borgon, C. Vonrhein, and T. Izard. 2005. Structural dynamics of alpha-actinin-vinculin interactions. Mol. Cell. Biol. 25:6112–6122. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 38.Ziegler, W. H., R. C. Liddington, and D. R. Critchley. 2006. The structure and regulation of vinculin. Trends Cell Biol. 16:453–460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.