Abstract
The robust surface adherence property of the aquatic bacterium Caulobacter crescentus permits visualization of single cells in a linear microfluidic culture chamber over an extended number of generations. The division rate of Caulobacter in this continuous-flow culture environment is substantially faster than in other culture apparati and is independent of flow velocity. Analysis of the growth and division of single isogenic cells reveals that the cell cycle control network of this bacterium generates an oscillatory output with a coefficient of variation lower than that of all other bacterial species measured to date. DivJ, a regulator of polar cell development, is necessary for maintaining low variance in interdivision timing, as transposon disruption of divJ significantly increases the coefficient of variation of both interdivision time and the rate of cell elongation. Moreover, interdivision time and cell division arrest are significantly correlated between mother and daughter cells, providing evidence for epigenetic inheritance of cell division behavior in Caulobacter. The single-cell growth/division results reported here suggest that future predictive models of Caulobacter cell cycle regulation should include parameters describing the variance and inheritance properties of this system.
INTRODUCTION
Studies of single microbial cells have revealed remarkable variability in the level of individual gene expression, rate of cell growth, and timing of cell division (1–3). Microfluidic devices have recently emerged as tools for studying dynamic processes at the single-cell level (4–6), with a number of studies reporting the use of microfluidics in quantifying single-cell growth and division (7–9). Such studies of single-cell behavior have been extremely valuable, yielding insights into phenomena that are not revealed in population-wide measurements (10–14). Although experiments that image single cells over short timescales on either agarose/gelatin pads or in microfluidic devices have become relatively routine, long-term and multigenerational studies of single cells have been complicated by problems with perturbative cell immobilization protocols or by rapid accumulation of cells on the pad or inside the microfluidic device.
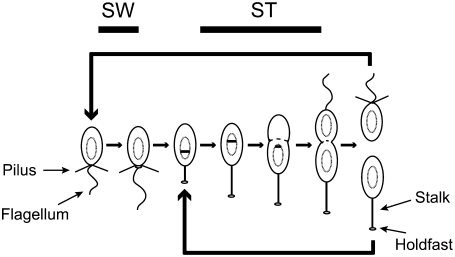
The freshwater α-proteobacterium Caulobacter crescentus (15) (henceforth referred to as Caulobacter) naturally allows for experiments that do not suffer from the aforementioned problems and is thus an ideal model to probe single-cell behavior across multiple generations (16). Caulobacter exists in two unique states during its cell cycle: a “swarmer” (SW) state, in which the cell possesses polar type IV pili and a single polar flagellum, and a nonmotile “stalked” (ST) state (Fig. 1). Differentiation from SW to ST occurs just before the initiation of DNA replication, at which time the flagellum is released, the pili are retracted, and a narrow cylindrical extension of the cell envelope known as the stalk is grown in their place. At the tip of the stalk is a structure known as the holdfast, which contains an exceptionally strong polysaccharide adhesive (17,18). Toward the end of the ST stage, a new flagellar assembly and pili are constructed at the pole opposite the stalk, and on division, a new motile, chemotactic SW cell is spawned. The SW cell then progresses through the full cell cycle, whereas the adhesive ST cell commences another round of DNA replication and division.
FIGURE 1.
Caulobacter cell cycle. A Caulobacter crescentus cell divides asymmetrically into two morphologically distinct cells: a stalked cell (ST) and a chemotactic swarmer cell (SW) with a single polar flagellum and type IV pili. The distal end of the stalk is capped with an adhesive holdfast. Differentiation from SW to ST occurs just before the initiation of DNA replication (replicating DNA is represented by the theta structure).
The natural adhesive properties of Caulobacter allowed us to conduct a multigenerational single-cell study of growth and division in a linear microfluidic culture chamber under temporally homogeneous and minimally perturbative conditions. We show that division of Caulobacter ST cells under constant medium flow is rapid and tightly regulated (i.e., exhibits low variance) relative to other bacterial species. Disruption of the gene encoding the DivJ histidine kinase, a core regulator of polar cell development (19,20), significantly increases variance in interdivision timing relative to the mean interdivision time. In addition, we show that factors controlling generational timing and division arrest are inherited from mother ST cells to daughter SW cells epigenetically, resulting in correlated cell division behavior between mother and daughter cells in the same generational window. This intragenerational correlation suggests that the network controlling Caulobacter cell division has determinsitic properties in which the current state of a cell influences future divisions.
METHODS
Microfluidic growth and division assays
Microfluidic channels measuring 200 μm wide by 50 μm deep by 2 cm long were made with polydimethylsiloxane (PDMS, Sylgard Brand 184 Silicone Elastomer Kit, Dow Corning, Midland, MI). The PDMS and glass coverslip were cleaned and sealed using a Plasma Prep II plasma cleaner (SPI Supplies, West Chester, PA). Sodium hydroxide (2 M solution), ethanol, and water were sequentially flowed into the channels to clean the interior before cell loading.
Individual colonies of wild-type Caulobacter strain CB15 (21) were taken from a peptone-yeast extract (PYE)-agar plate and grown overnight in 5 ml PYE medium at 30°C, diluted to 0.1 optical density at 660 nm (OD660), and regrown for 2 h. Cells were then loaded into the microfluidic chamber and incubated for an additional hour before imaging. A Harvard Apparatus (Holliston, MA) PHD2000 infuser was used to induce a constant flow of PYE medium at a rate of 12 μl/min for the duration of the experiment. This flow rate was found to be the minimum required for the cells to be maintained in a stable position at the glass surface (as shown in Fig. 2, B and C).
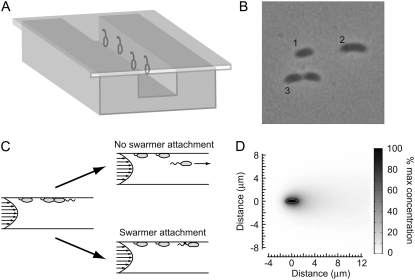
FIGURE 2.
Microfluidic device. (A) Microfluidic channels measuring 200 μm wide by 50 μm deep by 2 cm long were made with PDMS and sealed with a glass coverslip. Cells adhere to the glass surface via the adhesive holdfast. (B) A microscope image of cells in the chamber shows a young stalked cell (1), an early predivisional cell (2), and a late predivisional cell (3). Cells are aligned in the direction of medium flow. (C) The fluid flowing in the channel at a constant rate is laminar. The two-dimensional cross section of a laminar flow profile is parabolic, and the arrows represent the decreasing medium flow velocity from the center of the channel to the sides. On cell division, the majority of daughter cells are flushed out the far end of the channel. However, daughter cells occasionally attach to the glass coverslip before separation and remain attached downstream of their mothers when division is complete, rotated by ∼180° relative to their original orientation. (D) Simulations of diffusion under flow in the microfluidic culture channel show that small molecules emitted from the cell do not accumulate in the channel. The concentration of a simulated small molecule with diffusion coefficient of 10−9 m2/s was fixed at the bacterial surface. The concentration of the simulated small molecule is color graded from high (black) to low (white). For the flow rate used in our experiments (12 μl/min), the small molecule concentration quickly drops off outside the cell.
Cells were imaged with a Leica (Wetzlar, Germany) DM5000 at 630× magnification in phase-contrast mode. Images were collected at 2-min intervals on a Hamamatsu (Hamamatsu City, Japan) Orca-ER digital camera, and the light dosage was limited to 200 msec exposure and ∼5 s manual focus time per exposure. The temperature in the room was maintained at 30°C. Cell growth and division were monitored for 12–14 h during each of four independent experimental runs. This procedure was repeated with the CB15 divJ∷Tn5 (22) transposon insertion mutant strain on a shorter time scale because of accumulated errors in cell polarity and division in this genetic background.
Images were imported into ImageJ (National Institutes of Health, Bethesda, MD) for processing. The images were converted into binary stacks by subtracting the image backgrounds and adjusting the threshold pixel intensity. Cell areas were calculated in ImageJ, and data were further analyzed using Mathematica (Wolfram Research, Champaign, IL).
Simulations of diffusion under flow in the microfluidic channel
Simulations of flow around a bacterium in a microfluidic channel were created with a finite element analysis package (Comsol Multiphysics 3.2, Comsol, Stockholm, Sweden). A simulated channel was created with the same cross-sectional dimensions as the PDMS microfluidic: 200 μm wide by 50 μm deep. A single bacterium was modeled as an oval cylinder (i.e., a “pill-box”) with length 2 μm, width 0.5 μm, and height 0.5 μm, placed in the center of the channel width and on the top surface, with the major axis aligned in the direction of simulated flow. The flow profile was calculated using the Navier-Stokes equations for an incompressible fluid with a time-independent flow velocity:
 |
(1) |
where  is the fluid velocity, ρ is the density, p is the pressure, and η is the fluid viscosity.
is the fluid velocity, ρ is the density, p is the pressure, and η is the fluid viscosity.
To model the concentration of possible extracellular signaling molecules exported from the cell, the bacterial surface was held as a fixed source of a solute with diffusion coefficient D that was free to diffuse and undergo convective flow in the channel, according to the convective diffusion equation
 |
(2) |
The Navier-Stokes and convective diffusion equations were solved simultaneously until the simulator arrived at a stationary solution of the solute concentration c.
Batch cell growth measurements
To obtain the growth rate in a batch bioreactor culture, Caulobacter strain CB15 was cultured in PYE medium in a 10-liter bioreactor vessel (New Brunswick Scientific, Edison, NJ) maintained at 30°C. The pH of the growth medium was monitored and maintained at 7.1 by periodic titration of 0.5 M hydrochloric acid. The oxygen level was controlled at 95% of the oxygen level in air-saturated medium by automatically adjusting the mixing speed and the flow of air bubbled through the culture vessel. To obtain the growth rate of Caulobacter in a rolled test-tube culture, cells were also grown at 30°C in a standard test-tube roller in 5 ml PYE. Both the bioreactor and the rolled tubes were inoculated from a log-phase flask-grown culture to an OD660 of 0.03. Cells were monitored up to an OD660 of 0.2, and the growth rate was determined by fitting the data points to the exponential growth equation
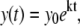 |
(3) |
where y0 is the initial optical density, and k is the rate of growth. Time points were collected at 20-min intervals. Optical density data were measured on a Thermo Genesys 20 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Calculations of flow-induced forces on bacteria in the microfluidic channel
The approximate shape of the velocity profile of a fluid flowing in a rectangular duct is
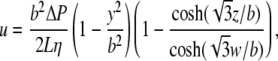 |
(4) |
where b is the half-height of the channel measured along coordinate y, w is the half-width measured along coordinate z, ΔP is the pressure drop over the channel length L, and η is the fluid viscosity (23). The total flow is the integral of the flow profile over the cross-sectional dimensions of the channel:
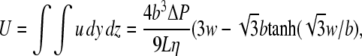 |
(5) |
and the profile can be rewritten as
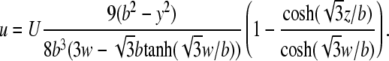 |
(6) |
The maximum flow rate occurs in the middle of the microfluidic channel, z = 0. For the cells to remain stationary, the drag forces induced by the flow must be counteracted by the adhesive force of the holdfast and the structural forces that maintain cellular integrity. In calculating the drag, we assume that each cell is a prolate ellipsoid of width 2a and length 2c, yielding a drag force in the direction of the flow (24):
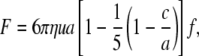 |
(7) |
where f ≈ 1.7 is a constant that takes into account the effect of the surface on the drag force (25).
There exists an additional lift force that tends to pull the cell away in a direction normal to the surface. However, it has been shown that for spherical particles with low Reynolds numbers (O(10−2)), the lift force is insignificant relative to the drag (26). We assume the same is true for the bacteria, as the lateral force on a prolate ellipsoid of total width 2a and length 2c (given above) is only 1.6 times the lateral force on a sphere of radius a, and the Reynolds number of a single bacterium (of dimension l ≈ 1 μm) in the channel is Re = luρ/η ≈ 5 × 10−3.
Nonparametric correlation analysis
We quantified the correlation between cells using Spearman's rank correlation coefficient ρ, a common measure of the strength of monotone association of two variables that is independent of the frequency distribution of the variables (i.e., it is nonparametric). The Spearman's rank correlation coefficient is defined as
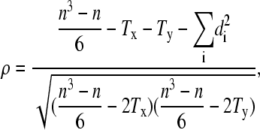 |
(8) |
where n is the number of elements and di is the rank difference between each of the corresponding values of the variables x and y (27). The Tx,y are the correction terms for ties in ranks on x and y,
 |
(9) |
where each tx,y is the number of objects in each tie set in x and y, and the sum is over the total number of tie sets.
The correlation of generation time between mother and daughter cells was determined by comparing cells' division times in the same generation window. For a mother cell m that produces a daughter cell d that attaches in generation g, the daughter's first generation is g + 1 with division time td,g+1, and the mother's division time in the same generation is tm,g+1. This labeling scheme can be seen in Fig. 3 B; for the sample mother and daughter “sawtooth” oscillations shown, generation g + 1 = 6. For each mother/daughter pair, two lists of division times {tm,g+1,tm,g+2, …} and {td,g+1, td,g+2, …} were created, with the list lengths determined by the number of generations in which both mother and daughter cells were dividing. Data taken on three separate days were combined, and the mother and daughter division times were correlated using Eq. 8. The effect of inheritance on the division time correlation was determined by randomly shuffling the daughter cells before their association and correlation with the mother cells.
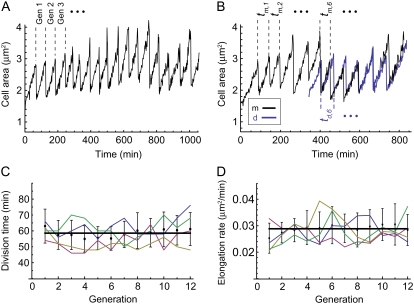
FIGURE 3.
Caulobacter division time and elongation rate. (A) The size of a single attached cell over time is a “sawtooth” oscillation with a period of the cell division time. (B) The majority of swarmer cells are flushed out of the microfluidic channel. However, there is a finite probability of attachment of the daughters downstream of their mothers. Growth of the mother cell m (black trajectory) and daughter cell d (blue trajectory) can thus be simultaneously measured. The division times are labeled as described in Methods: the daughter attaches in generation g, so that the daughter's first generation is g + 1 with division time td,g+1, and the mother's division time in the same generation is tm,g+1. (C) Division time in Caulobacter is tightly controlled: the average division time of 82 ST cells over 12 generations is 58.3 ± 9.5 min (total number of division events n = 727). The mean division time as a function of generation is shown in black, with error bars indicating the standard deviations. The trajectories of four single cells (green, blue, yellow, and red) are overlaid on the mean curve. (D) Cell elongation rates are also narrowly distributed and approximately constant over the course of the experiment, with an average elongation rate of 0.029 ± 0.006 μm2/min. Mean elongation rates are shown in black, and four individual cells trajectories are shown in color (green, blue, yellow, and red).
The significance of each correlation coefficient ρ′ was quantified by its p-value, i.e., the probability, under the null hypothesis, of obtaining a coefficient at least as extreme as the one calculated. Mathematically,
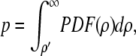 |
(10) |
with PDF(ρ) being the numerically determined probability density function.
RESULTS
Caulobacter growth in the microfluidic device is rapid and tightly controlled
The microfluidic channel shown in Fig. 2 A allows for the monitoring of growth and division of single Caulobacter cells over extended periods. ST cells are attached to the glass surface via the adhesive holdfast present at the stalk tip, and a constant flow of growth medium through the microfluidic device ensures that most cells born over the course of the experiment do not accumulate in the channel. After cell division, mother ST cells remain attached to the surface while the majority of daughter SW cells are flushed out of the channel (Fig. 2, B and C). However, daughter cells do occasionally attach to the glass coverslip before separation. This attachment appears random, likely mediated by the polar type IV adhesive pili and the flagellum (28,29). Daughter cells remain attached downstream of their mothers when division is complete, rotated by ∼180° relative to their original orientation (Fig. 2 C). The point around which the SW cells pivot is located an average of 0.3 μm from the flagellar pole, with no pivot located further than 2 μm from the pole, consistent with previously measured pili lengths between 1 and 4 μm (30,31). The cells then stay attached to the surface as they transition from the SW to the ST phase, and the stalk and holdfast develop.
The size of a single attached cell over time is a “sawtooth” oscillation with a period of the ST cell interdivision time (Fig. 3 A). We refer to this duration of time as a generation, the first generation being the first cycle for which a cell is present in the microfluidic chamber. The occasional attachment of daughter SW cells allows for simultaneous measurement of the growth and division of mothers and daughters (Fig. 3 B; Movie S1 in the Supplementary Material, Data S1).
The rate of Caulobacter cell division inside the microfluidic channel is the fastest reported to date, more rapid than Caulobacter cells grown in either shaken flasks or in a batch bioreactor culture (Table 1). The average interdivision time of 82 ST cells across 12 generations (total number of division events n = 727) is 58.3 ± 9.5 min. The complete cell cycle is longer, as the cells must progress through the SW phase and transition to a ST cell before chromosome replication and the process of cell division can begin (Fig. 1). We measure the complete cell cycle time for daughter SW cells that attach downstream of mother ST cells and find the average amount of time between attachment and first division to be 68.7 ± 8.6 min (n = 101). Thus, under these culture conditions, cells exist in the SW state for ∼10 min.
TABLE 1.
Comparison of mean division times 〈T〉 for Caulobacter cultured under three different conditions
| Growth conditions (cell type) | 〈T〉 (min) |
|---|---|
| Microfluidic (ST wild-type) | 58.3 ± 9.5* |
| Microfluidic (SW + ST wild-type) | 68.7 ± 8.6* |
| Microfluidic (ST divJ∷Tn5) | 76.6 ± 32.0* |
| Bioreactor (all wild-type) | 87.0 ± 1.3† |
| Rolled test-tube (all wild-type) | 126.5 ± 9.5† |
Errors reported are either *the population standard deviation or †the error in the exponential fit to the cell growth curve.
In comparison, a population of CB15 cells cultured under pH controlled and highly aerated conditions in a bioreactor doubles in size in 87.0 ± 1.3 min (from the measured growth rate k = 7.97 × 10−3 ± 1.24 × 10−4 min−1, coefficient of determination R2 = 0.99). The doubling time for cells grown in a rolled test tube is 126.5 ± 9.5 min (k = 5.48 × 10−3 ± 2.47 × 10−4 min−1, R2 = 0.99).
The mean interdivision time of wild-type ST cells as a function of generation number and the trajectories of four individual cells are shown in Fig. 3 C. Division timing over a full cell cycle in Caulobacter is tightly controlled, with a coefficient of variation (standard deviation divided by the mean) of 12.5%. As with division timing, the variance in the rate of cell elongation measured at the single cell level (determined by fitting a straight line to the cell size data between division events) is also narrowly distributed, with an average elongation rate of 0.029 ± 0.006 μm2/min (Fig. 3 D).
The microfluidic environment exerts minimal physical stress on cells
The rapid growth and regular division timing observed in microfluidic culture are not likely a result of physical forces applied to the bacteria by medium flow in the culture channel. Given a channel width of 200 μm, a height of 50 μm, and a flow of 12 μl/min, the maximum flow velocity experienced by cells attached at the channel surface is u ≈ 0.07 cm/s (cell midpoint at y = 24.75 μm). For a cell that is 2 μm long and 0.5 μm in diameter, and using the viscosity of water η = 1 × 10−3 N · s/m2, the maximum drag force on the cells in the channel is then <10 pN. For comparison, an air-liquid interface (i.e., air bubble) passing over an attached Caulobacter can exert up to 0.2 μN in surface tension forces (32), still smaller than the measured 0.59 ± 0.62 μN tensile force required to detach the cells from a substrate (18). The drag forces experienced by single Caulobacter cells inside the microfluidic device under laminar flow are sufficient to cause the cells to lie flat against the coverslip surface; however, they are small relative to what the cells may typically encounter in their natural freshwater environment (either at air-water interfaces or attached to stationary objects in moving waters). Indeed, changing the flow rate from 12 μl/min to a maximum of 50 μl/min had no discernable effect on cell behavior and did not change cell division statistics. The mean division time of ST cells subjected to a flow rate of 50 μl/min is 58.3 ± 8.1 min (n = 65).
In addition to exerting minimal physical stress on individual Caulobacter cells, our microfluidic culture channel has the added advantage of constant environmental conditions; a continuous flow of growth medium prevents the accumulation of cellular waste products. In simulated microfluidic conditions, we find that small molecules emitted from the cell do not accumulate in the channel. The concentration of a simulated diffusive molecule with diffusion coefficient of D = 10−9 m2/s (consistent with a ∼100-Da molecule (33)) was held fixed on the simulated bacterial surface but was free to evolve via diffusion and convective flow in the channel. For a flow rate of 12 μl/min, the concentration quickly drops off outside the cell (Fig. 2 D). Reducing the diffusion coefficient by a factor of 2 or increasing the flow rate by a factor of 5 had only negligible effects on the simulated concentration profile (data not shown).
Disruption of the divJ histidine kinase gene increases the coefficient of variation of cell interdivision time and cell elongation rate
The DNA-binding response-regulator protein CtrA is essential for Caulobacter viability and controls the transcription, either directly or indirectly, of 144 of 553 known cell cycle-regulated genes (34). Additionally, CtrA binds to five sites in the origin of chromosome replication and blocks the initiation of DNA replication in the SW cell (35).
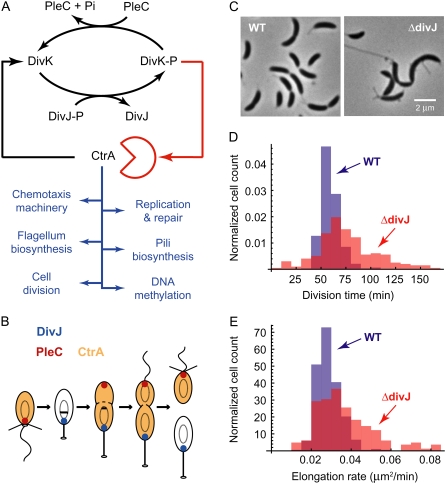
The histidine kinase DivJ is a component of a regulatory subnetwork in Caulobacter that also includes the PleC histidine kinase/phosphatase and the single-domain response regulator, DivK. Together, these proteins function to regulate the differential stability of CtrA in SW versus ST cells. DivJ acts as the primary kinase for DivK, which, in its phosphorylated state (DivK-P), cues CtrA proteolysis (Fig. 4 A) (36). DivJ is localized to the stalked pole of the cell, and PleC, a DivK-P phosphatase, is located at the flagellar pole (20,37,38) (Fig. 4 B). The asymmetric localization of DivJ and PleC ensures that on cell division, CtrA is cleared from the ST cell (allowing DNA replication to begin) but remains high in the SW, where replication is inhibited. It should be emphasized that there are multiple interlocking pathways involved in the transcriptional and posttranslational regulation of CtrA activity; Fig. 4 A presents just one module of the larger cell cycle regulatory network of Caulobacter.
FIGURE 4.
Caulobacter cell cycle control network and the DivJ sensor histidine kinase. (A) CtrA is essential for Caulobacter viability, controlling the transcription of 144 of 553 known cell cycle-regulated genes and acting as a negative regulator of DNA replication. The stability of CtrA is differentially regulated in swarmer versus stalked cells by the DivK response regulator, the histidine kinase DivJ, and the histidine phosphatase, PleC. (B) DivJ (blue) is localized to the stalked pole of the cell, and PleC (red) is localized to the flagellar pole. The asymmetric localization of DivJ and PleC ensures that on cell division, the level of CtrA (orange) drops in the ST cell while remaining high in the nonreplicative SW. (C) Phase contrast micrographs of wild-type (WT) Caulobacter and a Caulobacter strain carrying a Tn5 transposon insertion at the divJ locus (ΔdivJ). Mutations in the gene encoding DivJ result in cell morphology defects such as elongation, multiple constriction sites, and multiple or mislocalized stalks. (D) Transposon disruption of divJ results in a marked increase in the variance of the ST division time distribution (red). The mean ST division time in the divJ∷Tn5 strain is 76.6 ± 32.0 min (coefficient of variation = 0.42). The wild-type distribution (blue) has a mean of 58.3 min and standard deviation of 9.5 min (nWT = 727, nΔdivJ = 200). (E) The distribution of elongation rates is change in ΔdivJ (red). The mean elongation rate is increased from 0.029 μm2/min in the wild-type strain to 0.038 μm2/min in the mutant strain, whereas the standard deviation increases from 0.006 μm2/min to 0.016 μm2/min. The wild-type distribution is shown in blue.
Despite the role of DivJ as a regulator of DivK phosphorylation and, by extension, CtrA proteolysis, it is not essential for cell viability (20). However, mutation or disruption of the divJ gene does result in cell morphology defects, such as elongation, multiple constriction sites, and multiple or mislocalized stalks (19,20,39) (Fig. 4 C). We show that a Tn5 transposon insertion at the divJ locus of Caulobacter strain CB15 (22) results in a dramatic increase in the variance of single ST cell generation time and cell elongation rate. The interdivision time standard deviation increases from a wild-type value of 9.5 min to 32.0 min in the divJ∷Tn5 strain (Fig. 4 D), and the elongation rate standard deviation increases from 0.006 μm2/min in the wild-type strain to 0.016 μm2/min in the transposon mutant (Fig. 4 E). Despite these broadened distributions, the mean interdivision (i.e., generation) time and elongation rate exhibit relatively modest increases, from 58.3 min to 76.6 min (Table 1) and from 0.029 μm2/min to 0.038 μm2/min, respectively. This increase in mean generation time is consistent with the increase in population doubling time that has been previously reported for Caulobacter CB15N divJ∷Tn5 in batch culture (94 min and 103.5 min, respectively (39)). Disruption of the gene encoding the DivJ histidine kinase thus has a significantly greater effect on the variance (or “noise”) in interdivision timing than it does on the mean interdivision time of the population. This is reflected in the large increase in the coefficient of variation (COV = σ/μ) of Caulobacter CB15 divJ∷Tn5 interdivision timing and cell elongation rate relative to wild-type Caulobacter (Table 2 and Fig. 4, D and E). Our data therefore provide evidence for a correlation between maintaining low variance in division timing and proper polar development in Caulobacter.
TABLE 2.
Comparison of mean single-cell division times 〈T〉 and COV for different organisms
| Organism | 〈T〉 (min) | COV | References |
|---|---|---|---|
| Bacillus cereus | 49.0 | 0.490 | (54) |
| Escherichia coli | 52.0, 86.6 | 0.330, 0.441 | (40,55) |
| Proteus vulgaris | 28.2 | 0.309 | (41) |
| Enterobacter aerogenes | 30.0 | 0.300 | (54) |
| Enterococcus faecalis | 25.9 | 0.265 | (41) |
| Saccharomyces ellipsoideus | 107.0 | 0.200 | (54) |
| Saccharomyces cerevisiae | 99 | 0.18 | (3) |
| Methylobacterium extorquens | 187.2 | 0.176 | (2) |
| Caulobacter crescentus | 68.7 | 0.125 | This work |
| Schizosaccharomyces pombe | 108.3 | 0.047 | (47) |
It should be noted that the data presented were collected under a number of different growth conditions. Flow chambers of varying design were used in previous publications (2,40,55). All other data were collected from cells growing on agarose or gelatin pads. The value for S. cerevisiae is for haploid cells only.
Generation time and division arrest are significantly correlated between mother and daughter cells
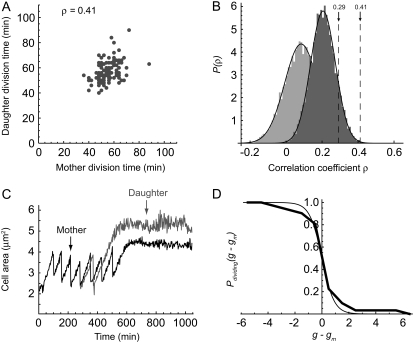
Attachment of daughter cells adjacent to their mothers allows one to study the relation between mother and daughter for many generations after the initial cell division. We find that for all generations, the mother and daughter cell size sawtooth functions remain in phase (Fig. 3 B). The degree to which the mothers and daughters stay in phase can be quantified by correlating the interdivision times for both mother and daughter across all generations (as described in Methods). A total of 20 mother/daughter cell pairs across 3 days were analyzed for each generation g + i in which both mother and daughter divided, yielding 122 pairs of division times {tm,g+i, td,g+i}, where i = 1, 2, …. To ensure that non-Gaussian outliers in the mother-daughter interdivision time distributions would not bias our correlation analysis, we chose to use the nonparametric Spearman rank correlation test, which is more conservative than Pearson's correlation (see Methods). Our data reveal a positive correlation between mother and daughter generation timing, with a Spearman rank correlation coefficient of ρ0 ≈ 0.41 (Fig. 5 A). Notably, the mother/daugher correlation is higher for early generations: the correlation coefficient for the first generation only is ρg=1 ≈ 0.65, that for two or fewer generations is ρg<2 ≈ 0.63, and three or fewer generations is ρg<3 ≈ 0.53. However, these correlation coefficients are determined with fewer data points than the number used to calculate ρ0 (ng=1 = 20, ng<2 = 39, and ng<3 = 57). In our numerical analysis to determine the significance of mother-daughter generation time correlation, we have considered all data across all generations to ensure the highest statistical confidence.
FIGURE 5.
Mother/daughter division control correlation. (A) Comparison of mother cell and daughter ST cell division times shows a significant positive correlation. The Spearman's rank correlation coefficient for 20 cell pairs across 12 generations is ρ0 ≈ 0.41 (n = 122). (B) The probability distribution P(ρ) generated from 10,000 random permutations of mothers with nondaughters across all days is Gaussian with a mean correlation coefficient 〈ρ〉 = 0.08 with standard deviation σρ = 0.09 (light gray). A more restricted probability distribution P(ρ) created from 10,000 random permutations of daughters with nonmothers, limited to within the same days, is also Gaussian with mean coefficient 〈ρ〉 = 0.21 and standard deviation σρ = 0.07 (dark gray), thus indicating day-to-day variation in mean population interdivision time. The observed mother/daughter correlation coefficient of 0.41 is significant relative to these two probability distributions at the level of 3.6σ (p ≈ 0.0002) and 3σ (p ≈ 0.001), respectively. The division time correlation coefficient of 18 proximal, unrelated cell pairs is 0.29 (n = 169; p > 0.1), indicating that the effect of cell proximity on division behavior is not statistically significant. (C) Mother cells that stop dividing in the microfluidic channel are more likely to produce daughter cells that also exhibit early division arrest. The growth trajectories of a selected mother cell (black) and daughter cell (gray) that stop dividing over the course of the experiment are shown. In this figure, the daughter stops dividing two generations before its mother. (D) Probability that a daughter cell still divides in generation g, given that the mother arrests in generation gm. At the generation of arrest of the mother, ∼50% of daughter cells are no longer dividing. The data fit the Fermi-Dirac distribution function P(g − gm) = 1/(1 + exp[(g − gm)/g0]), where g0 ≈ 0.45. Data were taken from 31 offspring of 26 terminal mother cells.
The significance of the specific mother/daughter generation time correlation was determined using a permutation test in which daughter cells were randomly paired with nonmothers and correlation coefficients were calculated for each randomized realization. With the null hypothesis that the cell division time distributions across the 3 days are equivalent, there are a total of 20 mother cells and 20 daughter cells, and thus 20! > 2.4 × 1018 ways to associate nonmother/daughter pairs. We performed 10,000 random permutations of daughters with nonmothers and calculated the correlation coefficients for the resultant mixed populations. The distribution of correlation coefficients is Gaussian with a mean of 〈ρ〉 = 0.08 with standard deviation σρ = 0.09 (Fig. 5 B). The correlation coefficient for the “correct” mother/daughter pairing of ρ0 ≈ 0.41 is therefore 3.6σ above our numerically simulated mean (p ≈ 0.0002).
However, this initial analysis ignores the possibility that the cell division statistics are different on each day of data collection. To determine the effect of day-to-day variation, we used a more restricted randomization process, permuting cells within each day before combining the data sets and calculating correlation coefficients. The total number of mother/daughter pairing realizations is then reduced to ∼1.8 × 1010. We performed 10,000 random permutations of daughters with nonmothers, limiting the permutations to within the same days, and calculated the correlation coefficients for the randomized cell population. As before, the distribution of correlation coefficients is Gaussian; however, the mean coefficient is increased to 〈ρ〉 = 0.21 and standard deviation σρ = 0.07 (Fig. 5 B), indicating that there is indeed measurable variability from day to day. However, day-to-day variability is not sufficient to explain the significant mother/daughter correlation of ρ0 ≈ 0.41, which is still >3σ above the mean (p ≈ 0.001).
From the simulation of diffusion under flow detailed in the previous section, we find that the concentration of a small molecule at the average mother/daughter distance of ∼1 μm is ∼30% of that at the surface of the mother cell (Fig. 2 C). Thus, we considered the possibility that the correlation in generation time between mothers and daughters is the result of cell proximity and some unknown diffusive molecule that affects the timing of cell growth and division. We applied the correlation analysis described above to nonmother/daughter cell pairs that are within the typical mother/daughter distance of each other. The division time correlation coefficient of 18 of these cell pairs (average intercell distance of d = 1.4 ± 1.2 μm) over 12 generations is 0.29 (n = 169) (Fig. 5 B), which is not significantly greater than the numerically simulated nonmother/daughter mean correlation coefficient of 0.21 (p > 0.1). Thus, we conclude that the division correlation is not caused by mother/daughter proximity but is instead caused by an unknown heritable term.
Although the division times of mother and daughter cells are significantly correlated within the same generation, the division times of individual cells are not correlated with themselves across generations. For example, the correlation of division times of all generations i with those of generations i + 1 yields no significant value (p > 0.1). The coefficients associated with larger generational separations are similarly insignificant. A similar lack of cross-generational self-correlation has been reported for Escherichia coli (40). However, the fact that mother and daughters are correlated within generations suggests that there is some degree of deterministic behavior in cell division timing in Caulobacter, in which the state of the cell in generation i affects i + n. Indeed, deterministic behavior in E. coli cell division was first reported over 40 years ago in a number of pioneering studies by Powell, Kubitschek, and the group of Arthur Koch (41–44).
In addition to the correlation in division timing between mother and daughter, we also show that there is a high probability that factors resulting in cell division arrest are similarly inherited. Compared with the majority of cells in the microfluidic channel that grow for the duration of the experiment (∼95%), mother cells that cease growing and dividing during the experiment are more likely to have offspring that also stop growing and dividing. An example of this inherited, premature division arrest can be seen in Fig. 5 C. From a population of 31 daughter cells born to 26 terminal mother cells, we determined the probability that the daughter cell continues to divide normally in a generation relative to the generation of its mother's death. Given a mother cell that stops growing in generation gm, the probability that a daughter cell still divides in generation g follows the form
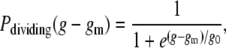 |
(11) |
where g0 is a constant that was determined to be ∼0.45 (Fig. 5 D). Not included in Fig. 5 D were five mother cells that ceased dividing, but whose daughter cells were born within four generations of the end of the experiment and did not arrest in that time.
DISCUSSION
Chemically inert microfluidics such as the device reported here have a number of advantages over other experimental methods for the study of single Caulobacter cells, most importantly a homogeneous and minimally perturbative environment. This system has the added advantage that experiment duration is not limited by cell population growth. Because cells do not quickly accumulate in the microfluidic device over the course of the experiment, there is nothing physically constraining the individual cells' growth. Thus, one can study single Caulobacter cells for even more generations than reported here. A striking result of the use of microfluidics is the drastic reduction in Caulobacter generation time relative to other culture methods (Table 1). Because changing the medium flow rate by a factor of ∼4 had no effect on division rate, we propose that this fast generation time results from the continuous flow of fresh medium to and removal of waste products from the cell. The Caulobacter chromosome replicates once and only once during a cell division cycle (45) (in contrast with other model prokaryotes such as E. coli), giving an upper bound on the rate of division. With a chromosome containing 4.01 million base pairs undergoing bidirectional replication from a single origin, and assuming a maximal rate of DNA replication by DNA polymerase of ∼1000 nucleotides/s (46), the theoretical minimum Caulobacter cell interdivision time is 33 min. A number of ST cell division events in our microfluidic culture channel approached this theoretical minimum time (Fig. 4 D). It should be noted that in the case of the divJ∷Tn5 strain, we see a number of divisions occurring faster than the theoretical minimum time. However, these times occur only in elongated cells that form multiple constriction sites, partitioning into several attached compartments, which then detach in quick succession.
The “tightness” of division timing control can be assessed by considering the coefficient of variation, a metric that describes the dispersion of a probability distribution (COV = σ/μ). A comparison of the COV for Caulobacter single-cell division times with those of other single-cell organisms (Table 2) shows that the Caulobacter COV is lower than those of all but one other microbial model organism. Data for a number of additional microbial species grown under various conditions can be found in Powell (41) and Schaechter et al. (42); however, none has a lower COV than Caulobacter. Only the fission yeast Schizosaccharomyces pombe exhibits a lower COV in division timing at the single-cell level (47).
Clearly, the cell cycle control network of Caulobacter generates an oscillatory output with low levels of noise. However, Caulobacter cells must be able to adapt to changing or heterogeneous environments, and it has been argued that populations exhibiting phenotypic variability have an adaptive advantage over those that do not (48). Disruption of the developmental regulator divJ dramatically increases the coefficient of variation in interdivision timing to levels observed in bacteria that undergo binary fission during vegetative growth (Table 2). This increased noise in division timing is correlated with the defects in polar morphogenesis that are evident in ΔdivJ strains. Our results suggest that if adaptive noise exists in Caulobacter regulatory systems, the magnitude of this noise must not exceed what is permitted to ensure proper development. Although none of the Caulobacter cell cycle control network models currently published (49,50) addresses noise in the regulation of cell division and development, it provides an excellent test for future models. More generally, it may be that the level of regulatory noise tolerated by a species is related to its developmental complexity.
We also show an unpredicted division time correlation between mother and daughter Caulobacter cells within the same generation (postdivision), indicating a degree of determinism in division time control in this bacterium. The correlation across 12 generations is small in that variation in mother cell division time accounts for only ∼16% of the variation in daughter cell division time (coefficient of determination ρ2 ≈ 0.16). However, the low probability of achieving a correlation of ρ ≈ 0.41 between two random cells (p ≈ 0.001) means that we can confidently reject the hypothesis that mother and daughter cell division times are independent. Furthermore, we find additional evidence for inherited division control factors in the observed inheritance of cell division arrest.
These results, although striking, are not difficult to explain conceptually given that mother and daughter cells share the same membrane and cytoplasm just before cell separation. Because the population of cells being monitored in the microfluidic device is isogenic, and there is an extremely small probability of mutations in genes affecting division time occurring over the course of a single experiment, we conclude that the observed inheritance in interdivision timing and division arrest is epigenetic. The mother/daughter division time and division arrest correlations can be explained by the inheritance of a quasistable molecule or molecules that regulate cell division, or perhaps by other factors that are known to be transmitted epigenetically, such as the state of DNA methylation (51–53). The fact that our calculated mother/daugher correlation coefficients are higher at early generational gaps and exhibit a per-generation decrease (see results) adds to the evidence for a quasistable inherited factor that regulates division timing.
Although the molecular mechanisms responsible for inheritance of growth and division behavior are currently unknown, it is clear that future characterization of these mechanisms will require innovative single-cell experiments. Furthermore, our results indicate that Caulobacter growth, development, division, and senescence can be more completely described using mathematical models that take noise and epigenetic inheritance into account.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Acknowledgments
We thank Cara Boutte, Erin Purcell, Kevin Dalton, Rachel Applefield, and Aretha Fiebig for assistance with data collection, and Aaron Dinner, Norbert Scherer, Tao Pan, John McDonald, and Harley McAdams for helpful discussions. Kinneret Keren and Patrick McGrath provided insightful criticism of drafts of this manuscript. We also thank Rustem Ismagilov and the Chicago Materials Research Science and Engineering Facility for help with microfluidic construction.
S.C. is funded by a Beckman Young Investigator Award and a grant from the Mallinckrodt Foundation.
Editor: Alexander Mogilner.
References
- 1.Elowitz, M. B., A. J. Levine, E. D. Siggia, and P. S. Swain. 2002. Stochastic gene expression in a single cell. Science. 297:1183–1186. [DOI] [PubMed] [Google Scholar]
- 2.Strovas, T. J., L. M. Sauter, X. Guo, and M. E. Lidstrom. 2007. Cell-to-cell heterogeneity in growth rate and gene expression in Methylobacterium extorquens AM1. J. Bacteriol. 189:7127–7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiTalia, S., J. M. Skotheim, J. M. Bean, E. D. Siggia, and F. R. Cross. 2007. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 448:947–951. [DOI] [PubMed] [Google Scholar]
- 4.Groisman, A., C. Lobo, H. J. Cho, J. K. Campbell, Y. S. Dufour, A. M. Stevens, and A. Levchenko. 2005. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods. 2:685–689. [DOI] [PubMed] [Google Scholar]
- 5.Hansen, C., and S. R. Quake. 2003. Microfluidics in structural biology: smaller, faster, better. Curr. Opin. Struct. Biol. 13:538–544. [DOI] [PubMed] [Google Scholar]
- 6.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science. 305:1622–1625. [DOI] [PubMed] [Google Scholar]
- 7.Cookson, S., N. Ostroff, W. L. Pang, D. Volfson, and J. Hasty. 2005. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol. Syst. Biol. epub 2005.0024. [DOI] [PMC free article] [PubMed]
- 8.Eriksson, E., J. Enger, B. Nordlander, N. Erjavec, K. Ramser, M. Goksor, S. Hohmann, T. Nystrom, and D. Hanstorp. 2007. A microfluidic system in combination with optical tweezers for analyzing rapid and reversible cytological alterations in single cells upon environmental changes. Lab Chip. 7:71–76. [DOI] [PubMed] [Google Scholar]
- 9.Umehara, S., I. Inoue, Y. Wakamoto, and K. Yasuda. 2007. Origin of individuality of two daughter cells during the division process examined by the simultaneous measurement of growth and swimming property using an on-chip single-cell cultivation system. Biophys. J. 93:1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colman-Lerner, A., A. Gordon, E. Serra, T. Chin, O. Resnekov, D. Endy, C. G. Pesce, and R. Brent. 2005. Regulated cell-to-cell variation in a cell-fate decision system. Nature. 437:699–706. [DOI] [PubMed] [Google Scholar]
- 11.Pedraza, J. M., and A. van Oudenaarden. 2005. Noise propagation in gene networks. Science. 307:1965–1969. [DOI] [PubMed] [Google Scholar]
- 12.Rosenfeld, N., J. W. Young, U. Alon, P. S. Swain, and M. B. Elowitz. 2005. Gene regulation at the single-cell level. Science. 307:1962–1965. [DOI] [PubMed] [Google Scholar]
- 13.Korobkova, E., T. Emonet, J. M. G. Vilar, T. S. Shimizu, and P. Cluzel. 2004. From molecular noise to behavioural variability in a single bacterium. Nature. 428:574–578. [DOI] [PubMed] [Google Scholar]
- 14.Longo, D., and J. Hasty. 2006. Dynamics of single-cell gene expression. Mol. Syst. Biol. 4:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackermann, M., S. C. Stearns, and U. Jenal. 2003. Senescence in a bacterium with asymmetric division. Science. 300:1920. [DOI] [PubMed] [Google Scholar]
- 17.Merker, R. I., and J. Smit. 1988. Characterization of the adhesive holdfast of marine and freshwater Caulobacters. Appl. Environ. Microbiol. 54:2078–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsang, P. H., G. L. Li, Y. V. Brun, L. B. Freund, and J. X. Tang. 2006. Adhesion of single bacterial cells in the micronewton range. Proc. Natl. Acad. Sci. USA. 103:5764–5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohta, N., T. Lane, E. G. Ninfa, J. M. Sommer, and A. Newton. 1992. A histidine protein-kinase homolog required for regulation of bacterial-cell division and differentiation. Proc. Natl. Acad. Sci. USA. 89:10297–10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell. 4:683–694. [DOI] [PubMed] [Google Scholar]
- 21.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. Eisen, J. F. Heidelberg, M. R. K. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. Deboy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA. 98:4136–4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, J. C., A. K. Hottes, H. H. McAdams, P. T. McGrath, P. H. Viollier, and L. Shapiro. 2006. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 25:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi, T., and W. N. Gill. 1980. Hydrodynamic chromatography—3-dimensional laminar dispersion in rectangular conduits with transverse flow. Chem. Eng. Comm. 5:367–385. [Google Scholar]
- 24.Li, G., and J. X. Tang. 2006. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys. J. 91:2726–2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oneill, M. E. 1968. A sphere in contact with a plane wall in a slow linear shear flow. Chem. Eng. Sci. 23:1293–1298. [Google Scholar]
- 26.Leighton, D., and A. Acrivos. 1985. The lift on a small sphere touching a plane in the presence of a simple shear-flow. Z. Angew. Math. Phys. 36:174–178. [Google Scholar]
- 27.Kendall, M., and J. Gibbons. 1990. Rank Correlation Methods, 5th ed. Oxford University Press, London.
- 28.Janakiraman, R. S., and Y. V. Brun. 1999. Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J. Bacteriol. 181:1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodenmiller, D., E. Toh, and Y. V. Brun. 2004. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186:1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagenaur, C., and N. Agabian. 1977. Caulobacter crescentus pili: structure and stage-specific expression. J. Bacteriol. 131:340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sommer, J. M., and A. Newton. 1988. Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J. Bacteriol. 170:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez-Suarez, C., H. J. Busscher, and H. C. van der Mei. 2001. Analysis of bacterial detachment from substratum surfaces by the passage of air-liquid interfaces. Appl. Environ. Microbiol. 67:2531–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, G., S. J. Yao, Y. X. Guan, and D. Q. Lin. 2005. Preparation and characterization of NaCS-CMC/PDMDAAC capsules. Colloid. Surf. B. Biointerfaces. 45:136–143. [DOI] [PubMed] [Google Scholar]
- 34.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 290:2144–2148. [DOI] [PubMed] [Google Scholar]
- 35.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. USA. 95:120–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hung, D. Y., and L. Shapiro. 2002. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc. Natl. Acad. Sci. USA. 99:13160–13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hecht, G. B., T. Lane, N. Ohta, J. M. Sommer, and A. Newton. 1995. An essential single-domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J. G., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. USA. 95:1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierce, D. L., D. S. O'Donnol, R. C. Allen, J. W. Javens, E. M. Quardokus, and Y. V. Brun. 2006. Mutations in DivL and CckA rescue a divJ null mutant of Caulobacter crescentus by reducing the activity of CtrA. J. Bacteriol. 188:2473–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wakamoto, Y., J. Ramsden, and K. Yasuda. 2005. Single-cell growth and division dynamics showing epigenetic correlations. Analyst. 130:311–317. [DOI] [PubMed] [Google Scholar]
- 41.Powell, E. O. 1955. Some features of the generation times of individual bacteria. Biometrika. 42:16–44. [Google Scholar]
- 42.Schaechter, M., J. P. Williams, J. R. Hood, and A. L. Koch. 1962. Growth, cell and nuclear divisions in some bacteria. J. Gen. Microbiol. 29:421–434. [DOI] [PubMed] [Google Scholar]
- 43.Kubitschek, H. E. 1962. Normal distribution of cell generation rate. Exp. Cell Res. 26:439–450. [DOI] [PubMed] [Google Scholar]
- 44.Kubitschek, H. E. 1966. Generation times - ancestral dependence and dependence upon cell size. Exp. Cell Res. 43:30–38. [DOI] [PubMed] [Google Scholar]
- 45.Marczynski, G. T. 1999. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 181:1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marians, K. J. 1992. Prokaryotic DNA replication. Annu. Rev. Biochem. 61:673–719. [DOI] [PubMed] [Google Scholar]
- 47.Kovar, D. R., J.-Q. Wu, and T. D. Pollard. 2005. Profilin-mediated competition between capping protein and formin Cdc12p during cytokinesis in fission yeast. Mol. Biol. Cell. 16:2313–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smits, W. K., O. P. Kuipers, and J. W. Veening. 2006. Phenotypic variation in bacteria: the role of feedback regulation. Nat. Rev. Microbiol. 4:259–271. [DOI] [PubMed] [Google Scholar]
- 49.Brazhnik, P., and J. J. Tyson. 2006. Cell cycle control in bacteria and yeast - a case of convergent evolution? Cell Cycle. 5:522–529. [DOI] [PubMed] [Google Scholar]
- 50.Li, S., P. Brazhnik, B. Sobral, and J. J. Tyson. 2008. A quantitative study of the division cycle of Caulobacter crescentus stalked cells. PLoS Comput. Biol. 4:111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chernoff, Y. O. 2001. Mutation processes at the protein level: is Lamarck back? Mutat. Res. 488:39–64. [DOI] [PubMed] [Google Scholar]
- 52.Jablonka, E., and M. J. Lamb. 1998. Epigenetic inheritance in evolution. J. Evol. Biol. 11:159–183. [Google Scholar]
- 53.Jablonka, E., M. Lachmann, and M. J. Lamb. 1992. Evidence, mechanisms and models for the inheritance of acquired characters. J. Theor. Biol. 158:245–268. [Google Scholar]
- 54.Kelly, C. D., and O. Rahn. 1932. The growth rate of individual bacterial cells. J. Bacteriol. 23:147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Inoue, I., Y. Wakamoto, and K. Yasuda. 2001. Non-genetic variability of division cycle and growth of isolated individual cells in on-chip culture system. Proc. Jpn. Acad. 77:145–150. [Google Scholar]