Abstract
Living systems rely on fluid dynamics from embryonic development to adulthood. To visualize biological fluid flow, devising the proper labeling method compatible with both normal biology and in vivo imaging remains a major experimental challenge. Here, we describe a simple strategy for probing microscopic fluid flows in vivo that meets this challenge. An all-optical procedure combining femtosecond laser ablation, fast confocal microscopy and 3D-particle tracking was devised to label, image and quantify the flow. This approach is illustrated by studying the flow generated within a micrometer scale ciliated vesicle located deep inside the zebrafish embryo and involved in breaking left-right embryonic symmetry. By mapping the velocity field within the vesicle and surrounding a single beating cilium, we show this method can address the dynamics of cilia-driven flows at multiple length scales, and can validate the flow features as predicted from previous simulations. This approach provides new experimental access to questions of microscopic fluid dynamics in vivo.
The analysis of biological fluid flows is becoming critical since it is increasingly recognized that cell responses to mechanical flows are involved in major events of embryonic development and adult homeostasis (1). A classical case is the specification of the left-right axis in vertebrates, where beating cilia generate a directional flow necessary for breaking the embryonic symmetry in the so-called left-right organizer (2). Yet, investigating flow dynamics in vivo requires the use of the appropriate procedure for flow labeling. Common methods involve the injection of tracer particles with a needle. This procedure is challenging when targeting micrometer scale structures located deep inside a living organism because it can seriously damage the targeted tissue.
To circumvent this limitation, we devised an all-optical strategy that relies on subcellular femtosecond laser ablation (3) to generate fluorescent microdebris that seeds the flow. The nonlinear effect involved in this technique provides a high spatial confinement and a low invasiveness, thus allowing the targeting of a single cell. Subsequent fast confocal imaging and three-dimensional (3D) particle tracking were used to image and quantify the seeded flow. We illustrate this approach by investigating the cilia-driven flow generated within the Kupffer's vesicle (KV), the left-right organizer of zebrafish embryos (2).
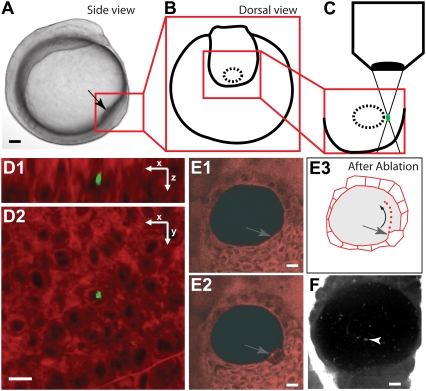
To minimize the possible artifacts due to experimental manipulations, we took advantage of the properties of femtosecond laser ablation (3), which can disrupt cell integrity with tight spatial confinement and low collateral damage. Nanometer-scale dissections have been applied in cell biology (4,5). Thanks to the relatively low scattering of near infrared light in tissue, this technique can be used to perform 3D-confined dissections in thicker specimens, permitting useful applications in neuroscience (6) and developmental biology (7). The spatial extent of the ablation depends on the tissue's optical properties; thus, we first investigated the depth where a subcellular 3D-confinement was feasible. We focused femtosecond laser pulses (300 fs, 820 nm, 80 MHz) on mesodermal cells, 70 μm beneath the epidermal cell layer inside a living zebrafish embryo at bud stage. The extent of each ablation was determined by attenuating the laser power and recording the two-photon excited fluorescence (2PEF) images of the endogenous fluorescence around the targeted area (Fig. 1 D; Movie S1 in the Supplementary Material). Successful ablations generate intense fluorescence in the targeted region (as described in Supatto et al. (7)), with an emission spectrum larger than the spectrum of the fluorescent dye (collected in the 500–550 nm wavelength range). A subcellular spot (∼20 fL) was observed, with no visible damage between the focal volume and the surface of the embryo (Fig. 1 D; Movie S1), demonstrating the 3D confinement of the ablation at this depth. We characterized the optical properties of the embryonic tissue by measuring a phenomenological 2PEF attenuation length (l) of the excitation light at 820 nm (see the Supplementary Material, Data S1) and found that l ∼ 70 μm (N = 4) in the zebrafish tail bud. Thus, the subcellular micrometer-scale ablation was achievable down to a depth comparable to l.
FIGURE 1.
Femtosecond laser ablation procedure for flow fluorescent seeding. (A) Side view of a zebrafish embryo at the stage of ablation. The box outlines the location of the KV. (B) Schematic view of the KV before the ablation (dorsal view). (C) Magnified region of the KV corresponding to the boxed region in B during the ablation step, showing the laser excitation in the tissue. (D) Two views, in x-z (D1) and in x-y (D2), showing the 3D-confined ablation (green, <5 μm in each direction) generated 70 μm deep inside a dye-loaded embryo. The green fluorescence is generated as a result of the laser ablation. (E) Results of the ablation when performed on the cells lining the KV (gray arrows), raw images (E1 and E2), depicting the cells before (E1) and after ablation (E2 and E3). The drawing (E3) illustrates the flow seeding after laser ablation. (F) Fast confocal microscopy allows imaging of the fluorescent microdebris (white arrowhead) seeding the flow. Black and white scale bars 50 μm and 10 μm, respectively.
In fish, the KV is a cavity (∼250 pL) located beneath several layers of mesodermal cells (70–120 μm deep) in the embryonic tail bud (Fig. 1, A–C) (8,9). The 3D confinement of femtosecond-induced ablations was used to lesion the plasma membrane of a single cell lining the cavity (N = 20, Fig. 1 and data not shown). This optical disruption induced the release of fluorescently labeled cell debris inside the vesicle, seeding the flow with fluorescent particles (Fig. 1, E and F). We evaluated the invasiveness of this approach by looking at the vesicle morphology after ablation and by checking left-right axis specification in the manipulated embryos. We found that the vesicle morphology remained the same after ablation (Fig. 1, E and F) and that no left-right defects resulted (N = 8).
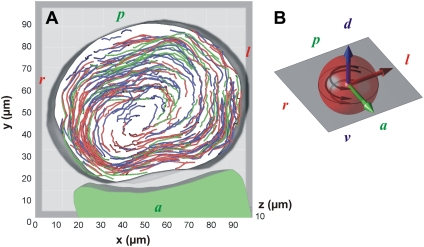
After seeding the fluid with fluorescent particles, we investigated the 3D motion of the flow within the cavity. Particles were imaged in 3D after ablation using a fast confocal microscope at 44 frames per second (four z-stacks per second) and tracked in 3D using image processing (see the Supplementary Material, Data S1). The particles located 10 μm away from the vesicle surface exhibited a circular motion following a counterclockwise rotation (dorsal view) around the dorso-ventral axis (Fig. 2; Movie S2 and Movie S3), confirming previous observations (8,9). The time-independent feature of this flow is demonstrated in Fig. 2 A by merging particle tracks recorded during different time windows. The maximum speeds ranged up to 10–50 μm/s, leading the particles to perform a full vesicle revolution every 15 s. Assuming that the viscosity within the vesicle is close to the viscosity of pure water, the corresponding Reynolds number is on the order of 10−3.
FIGURE 2.
Kupffer's vesicle flow extracted from fluorescence data. (A) Particle tracks (N = 266) indicating the fluid streamlines of the steady-state flow within the KV. The three different track colors correspond to the superposition of tracks obtained at different time windows (3 × 15 s over a 45-s period) showing that tracks align over time, indicating a steady state. The particles exhibit a circular motion around the dorso-ventral direction. (B) Schematic display of the spatial orientation (antero-posterior, dorso-ventral, and left-right axis are represented in green, blue, and red, respectively) of the vesicle (red sphere). The black arrows indicate the counterclockwise rotation of the flow (when viewed from the dorsal side). d, dorsal; v, ventral; a, anterior; p, posterior, l, left; and r, right.
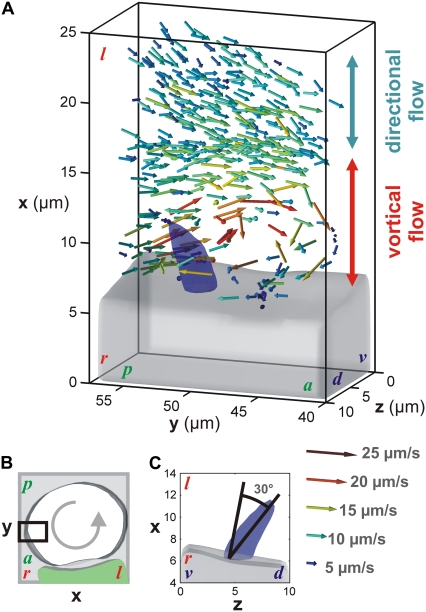
Understanding how the directional flow is generated within the KV requires knowledge of the spatial position, the direction of rotation, and the tilt of the beating cilia. Yet, the direct fluorescence imaging in 3D of these cilia is challenging because they beat fast (∼25 Hz (9)) and are localized deep below the tissue surface. Fortunately, the particles traveling near the envelope of a beating cilium can be trapped in a vortical flow with the same rotation direction, but with a significantly slower speed, as predicted by simulations (10). Fast confocal microscopy (11) was sufficient to image the trajectories of these particles and indirectly probe cilia characteristics. Indeed, particles trapped in vortical flows were observed close to the cell surface and localized to a potential beating cilium (Movie S4). Particle tracking revealed a transition between the directional flow (>10 μm) and the vortical flow closer to the surface (<10 μm, Fig. 3 A, Movie S5). The presence of chaotic advection generated around the envelope of the cilium was observed by following the quick divergence of nearby particle tracks (Fig. 1 in the Supplementary Material (Data S1), see legend for details; Movie S6). In the context of Stokes flow at low Reynolds number, this chaotic behavior is an original characteristic predicted by previous simulations (10).
FIGURE 3.
Velocity vector map around a single cilium extracted from 3D particle tracking (100 tracks). (A) Dorsal view of the velocity field surrounding a beating cilium on the right side of the vesicle (black box in B, same vesicle as in Fig. 2 A) and showing a transition between directional and vortical flow close to the cell surface. The gray volume represents the cell surface and the blue volume the cilium position. (C) Posterior view showing the 30° dorsal tilt of the beating axis. The vector map corresponds to the accumulation of instantaneous particle velocities recorded at different time points. The color of the arrows encodes for the norm of the velocity vectors. d, dorsal; v, ventral; a, anterior; p, posterior; l, left; and r, right. The axis scales and position are kept as in Fig. 2 A.
Directional flow can be driven by cilia with an axis of rotation tilted in a plane perpendicular to the flow (12–14). To define the rotation axis of the observed cilium, we identified the main direction of the volume around which the particles spin. Its surface was manually reconstructed by drawing the boundary of the space wherein no particles were detected (blue surface in Fig. 3; Supplementary Material Fig. 1 (Data S1); and Movie S5 and Movie S6). The axis exhibits a 30° angle tilt toward the dorsal direction (Fig. 3 C). This angle is close to the 35° value anticipated by simulations to generate the optimal directional flow (15). Considering the direction of the directional flow measured inside the vesicle (Fig. 2), the dorsal tilt is in accordance with the theoretical model (12). Furthermore, the counterclockwise rotation (view from the tip of the cilium) of the trapped particles corroborates previous observations on the rotation direction of the cilium (13,14).
In conclusion, the good agreement between our experimental observations and the features recently predicted by numerical simulations confirms that this approach can reveal characteristics of ciliary hydrodynamics, and argues favorably for the noninvasiveness of this approach. Such detailed descriptions of cilia-driven fluid movement will be valuable in unraveling the relationships between flow and signal transduction crucial for maintaining the asymmetry of the embryo. Finally, this approach opens new opportunities for investigating microscopic flow in living tissues.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
We are grateful to D. Wu and T. Truong for critical comments, the Caltech Biological Imaging Center, and P. Björkman for sharing equipments.
J.V. was supported by the Human Frontier Science Program Fellowship. This work was supported by the Biological Imaging Center of the Caltech Beckman Institute and a Centers of Excellence in Genomic Sciences grant from the National Institutes of Health (P50 HG004071).
Editor: Michael Edidin.
References
- 1.Bartman, T., and J. Hove. 2005. Mechanics and function in heart morphogenesis. Dev. Dyn. 233:373–381. [DOI] [PubMed] [Google Scholar]
- 2.Hirokawa, N., Y. Tanaka, Y. Okada, and S. Takeda. 2006. Nodal flow and the generation of left-right asymmetry. Cell. 125:33–45. [DOI] [PubMed] [Google Scholar]
- 3.Vogel, A., J. Noack, G. Huttman, and G. Paltauf. 2005. Mechanisms of femtosecond laser nanosurgery of cells and tissues. Appl. Phys. B. 81:1015–1047. [Google Scholar]
- 4.Tirlapur, U. K., and K. König. 2002. Cell biology: targeted transfection by femtosecond laser. Nature. 418:290–291. [DOI] [PubMed] [Google Scholar]
- 5.Kumar, S., I. Z. Maxwell, A. Heisterkamp, T. R. Polte, T. P. Lele, M. Salanga, E. Mazur, and D. E. Ingber. 2006. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys. J. 90:3762–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanik, M. F., H. Cinar, H. N. Cinar, A. D. Chisholm, Y. Jin, and A. Ben-Yakar. 2004. Neurosurgery: functional regeneration after laser axotomy. Nature. 432:822. [DOI] [PubMed] [Google Scholar]
- 7.Supatto, W., D. Débarre, B. Moulia, E. Brouzes, J. L. Martin, E. Farge, and E. Beaurepaire. 2005. In vivo modulation of morphogenetic movements in Drosophila embryos with femtosecond laser pulses. Proc. Natl. Acad. Sci. USA. 102:1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Essner, J. J., J. D. Amack, M. K. Nyholm, E. B. Harris, and J. Yost. 2005. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 132:1247–1260. [DOI] [PubMed] [Google Scholar]
- 9.Kramer-Zucker, A. G., F. Olale, C. J. Haycraft, B. K. Yoder, A. F. Schier, and I. A. Drummond. 2005. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer's vesicle is required for normal organogenesis. Development. 132:1907–1921. [DOI] [PubMed] [Google Scholar]
- 10.Smith, D. J., E. A. Gaffney, and J. R. Blake. 2007. Discrete cilia modeling with singularity distributions: application to the embryonic node and the airway surface liquid. Bull. Math. Biol. 69:1477–1510. [DOI] [PubMed] [Google Scholar]
- 11.Vermot, J., S. E. Fraser, and M. Liebling. 2008. Fast fluorescence microscopy for imaging the dynamics of embryonic development. HFSP J. 2:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cartwright, J. H. E., O. Piro, and I. Tuval. 2004. Fluid-dynamical basis of the embryonic development of left-right asymmetry in vertebrates. Proc. Natl. Acad. Sci. USA. 101:7234–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nonaka, S., S. Yoshiba, D. Watanabe, S. Ikeuchi, T. Goto, W. F. Marshall, and H. Hamada. 2005. De novo formation of left-right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 3:1467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okada, Y., S. Takeda, Y. Tanaka, J. C. I. Belmonte, and N. Hirokawa. 2005. Mechanism of nodal flow: a conserved symmetry breaking event in left-right axis determination. Cell. 121:633–644. [DOI] [PubMed] [Google Scholar]
- 15.Smith, D. J., J. R. Blake, and E. A. Gaffney. 2008. Fluid mechanics of nodal flow due to embryonic primary cilia. J. R. Soc. Interface. 5:567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.