Abstract
Nuclear hormone receptors comprise a characteristic family of transcription factors found in vertebrates, insects and nematodes. Here we show by cDNA and gene cloning that a Cnidarian, Tripedalia cystophora, possesses a retinoid receptor (jRXR) with remarkable homology to vertebrate retinoic acid X receptors (RXRs). Like vertebrate RXRs, jRXR binds 9-cis retinoic acid (Kd = 4 × 10−10 M) and binds to the DNA sequence, PuGGTCA as a monomer in vitro. jRXR also heterodimerizes with Xenopus TR beta on a thyroid responsive element of a direct repeat separated by 4 bp. A jRXR binding half-site capable of interacting with (His6)jRXR fusion protein was identified in the promoters of three T. cystophora crystallin genes that are expressed highly in the eye lens of this jellyfish. Because crystallin gene expression is regulated by retionoid signaling in vertebrates, the jellyfish crystallin genes are candidate in vivo targets for jRXR. Finally, an antibody prepared against (His6)jRXR showed that full-length jRXR is expressed at all developmental stages of T. cystophora except the ephydra, where a smaller form replaces is. These data show that Cnidaria, a diploblastic phylum ancestral to the triploblastic invertebrate and subsequent vertebrate lineages, already have an RXR suggesting that RXR is an early component of the regulatory mechanisms of metazoa.
Nuclear hormone receptors (NHRs)comprise a large family of transactivating genes found in vertebrates, an echinoderm, arthropods and nematodes. NHRs include receptors for steroid and thyroid hormones, Ecdysone, retinoic acid, vitamin D, prostaglandin J2 (1), and other small lipophillic molecules recently identified (2–5). Homologies in their DNA binding domain (DBD) in particular but also in their ligand binding domain suggest that the family evolved from a single ancestor gene (6, 7). More than 300 distinct members of the NHR family have been identified and deposited in public databases. Only a small minority of NHR have known ligands, with others being referred to as orphan nuclear receptors. Unlike receptors that bind to their response elements as homodimers or heterodimers, some orphan receptors receptors bind the response elements as monomers (1). Retinoic acid X receptor (RXR), a NHR member, heterodimerizes with multiple NHRs and binds 9-cis-retinoic acid (8–10). The closest homologue of vertebrate RXRs among the invertebrates, Ultraspiracle, is present in Drosophila and binds insect juvenile hormone III and juvenile hormone III acid rather than retinoids (11, 12). While similarities in DBDs of NHRs indicate a clear evolutionary tree branching from a single ancestor gene to six subfamilies, the specificity for chemically similar ligands seems to be a function gained later rather than concurrently with the evolution of DBD. Indeed, it has been suggested that the NHR family evolved from an orphan receptor capable of binding DNA as a monomer or homodimer and that the ability to bind a ligand was achieved later during evolution (7, 13). Here we report a close homologue of vertebrate RXRs that binds 9-cis-retinoic acid as a ligand and also binds a vertebrate consensus DNA response element in one of the most primitive Cnidarian jellyfish Tripedalia cystophora. We also show that this jellyfish RXR (jRXR) binds to potential regulatory element of crystallin gene expressed highly in the eye lens of T. cystophora (14), raising the possibility that retinoid signaling is used for eye development and crystallin gene expression in invertebrates as it is in vertebrates (15–22).
MATERIALS AND METHODS
Adult jellyfish T. cystophora were collected from sea water under mangroves in La Parguera with the permission of Department of Natural Resources and Agriculture of Puerto Rico.
Cloning jRXR Gene and cDNA.
Cloning was initiated by PCR using degenerate primers designed according to conserved regions in DBD of NHRs as described (23). A single 130-bp fragment was obtained from genomic DNA prepared from male sperm as well as from cDNA prepared by reverse transcription of total RNA extracted from larval forms obtained by dissection of female jellyfish. The fragment was used as a probe for screening in a genomic library made with a Stratagene λ-Zap construction kit. This yielded two clones containing part of the gene (Fig. 1A). PCR strategy was used to clone cDNA. Nested primers were designed in a fragment cloned originally. Region 3′ to DBD was cloned with a reverse transcription primed with a poly-T oligomer coupled on its 5′ position to a short sequence designed to raise the annealing temperature for subsequent PCR (5′GACTCGAGTCGAGGTGGAT17). The amplified fragment was cloned with a Clone-Amp System (Life Technologies, Gaithersburg, MD). The 5′ end of the cDNA was cloned using a 5′rapid amplification of cDNA ends (RACE) System (Life Technologies). 5′ RACE products were separated by agarose gel electrophoresis, blotted on a membrane (Gene Screen Plus, NEN) and screened by hybridization using an internal probe labeled by 32P-dATP. 5′ RACE was made in duplicate exploiting two different primers separated by a known distance. Autoradiography resulted in number of labeled fragments from which only those that differed by expected size in duplicates were considered correct. A duplicate gel prepared at the beginning of the procedure was positioned over the autoradiogram and gel from areas revealed by the radioactive probe (but not visible in UV light) was removed and used for a PCR amplification and cloning. A radioactive probe used in this procedure was prepared by using PCR: A portion of the known sequence of the cDNA not containing the region covered by primers used for 5′ RACE reverse transcription was amplified by using internal primers and a fragment was harvested from an agarose gel after an electrophoretic separation. The radioactive probe (300 bp long) was prepared by another round of amplification by PCR (eight cycles) with 32P-dATP (5 μl of 3,000 Ci/mmol, Amersham; 1 Ci = 37 GBq) in reaction mixture (in volume 50 μl). Hybridization was performed at 65°C in 10 ml of 6× standard saline citrate, 5× Denhardt’s solution, 0.5% (wt/vol) SDS, 100 μg/ml denatured salmon DNA, and 2 × 107 cpm of radioactive probe. The membrane was washed at low stringency conditions. Southern analysis of genomic DNA was performed similarly except that the wash was done at high stringency (24).
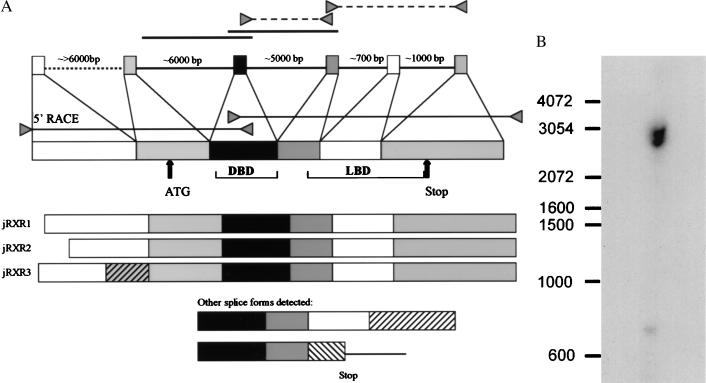
Figure 1.
(A) Organization of the jRXR gene and cDNA isoforms. A full length cDNA was obtained by PCR with primers based on the previously determined 5′ and 3′ ends of the cDNA. Three isoforms of cDNA alternatively spliced in the region 5′ to the DBD were sequenced. Two genomic clones indicated by solid lines were obtained by screening in a genomic library. Regions indicated by triangles connected by dashed lines were amplified by PCR from genomic DNA prepared from male gonads. The size of the transcript was confirmed by RNA primer extension. The position of intron 1 was derived from cDNA isoforms, although we failed to amplify this intron from genomic DNA by PCR. The presence of intron 4 and 5 is determined by PCR from genomic DNA. (B) Southern blot analysis of genomic DNA prepared from gonads of 21 male adult jellyfish. DNA restricted by EcoRI and probed by cDNA probe covering exon 3 yielding two bands of 2.5 and 3 kb.
Prepration of (His)6jRXR Fusion Protein, Anti-jRXR Antibody and Electrophoresis Mobility Shift Analysis (EMSA).
jRXR fusion protein was generated by cloning the full-length jRXR cDNA into the pRSET B vector (Invitrogen) and expressing in BL 21 (LysS) cells after induction for 2 hr. Cells were lysed in denaturing or native conditions and (His)6jRXR fusion protein was purified by affinity chromatography on Ni-Sepharose columns. Protein obtained in denaturing conditions was used for immunizations of rabbits (Hazelton, VA). Protein preparations obtained in both denaturing and native conditions were used for EMSA, the former only after overnight incubation in renaturation solution (50 mM Tris⋅HCl, pH 7.4/1 mM EDTA/5 mM DTT/8% glycerol and protease inhibitors; Boehringer Mannheim).
For EMSA, DNA oligomers were synthesized, end labeled with T4 Polynucleotide Kinase (Life Technologies), annealed and double-stranded DNA probes separated by PAGE. Probes used in this experiment were:
DR5: TCGACGACCAGGTCACCGGAAGGTCACGTTCTAG; DR1: TCGACGACCAGGTCAAAGGTCACGTTCTAG; DR4: AGCTTCAGGTCAGAGGAGGTCAGAGAGAGCT; J1BRE: -106 TAAATGAAAAAGGTCATTTACCGTTACAAT -135; and NBRE: GAGTTTTAAAAGGTCATGCTCAATTTGGAT.
EMSA was performed in a final volume of 20 μl containing 100–400 ng of jRXR fusion protein alone and/or 5 μl of in vitro translated jRXR, mouse RXRb, or Xenopus TR beta. The total volume of reticulocyte lysate was also 5 μl in cases where two in vitro translated proteins were used. The incubation buffer consisted of 20 mM Hepes (pH 7.9) (N-2-hydroxyethylpiperazine-N′-2ethanesulfonic acid), 1 mM DTT, 2 mM MgCl2, 10% (vol/vol) glycerol, 0.1 mg/ml BSA, and contained 104 cpm of a labeled probe and 1 μg of a nonspecific competitor poly(dI-dC) (Sigma) per reaction. For competition reactions, 100× molar excess of unlabeled probes was added for 10 min before the labeled probes. Binding was performed on wet ice for 30 min and products were analyzed by PAGE at nondenaturing conditions after at least of 2 hr of pre-run at 100 V. Results were analyzed by autoradiography after exposition from 1–5 days at −70°C with intensification screens.
Laboratory Culture of T. cystophora.
Planulae obtained by dissection of female jellyfish were collected and incubated in natural sea water (filtered samples obtained in open sea 1 Km from T. cystophora habitat) or in artificial sea water (Instant Ocean, Aquarium Systems, Mentor, OH) with specific gravity 1.021 g/ml at 21°C. Polyps and ephydrae were fed in intervals from 5 to 10 days with newly hatched brine shrimp (Artemia cysts originated from Great Salt Lake, UT, distributed by Inve, Grantsville, UT).
For histology, polyps or ephydrae were fixed in 4% (vol/wt) paraformaldehyde in 50 mM phosphate buffer (pH 7.4) and 0.6 M NaCl.
Western Blot Analysis.
Non motile or swimming planulae were collected and frozen in aliquots of 20–50 μl on dry ice. Primary and secondary polyps, metamorphosing polyps and freshly released ephydrae were harvested from laboratory culture and samples containing 20 individual animals were frozen in about 5 μl of artificial seawater. Upon thawing, 5× Laemmli buffer was added together with protease inhibitors (0.1 mM polymethylsulfonyl fluoride/10 μg/ml of antipain and leupeptin; Sigma) and volume adjusted with deionized water. Samples were boiled for 8 min, and 20 μg of total protein were loaded on a minigel and electrophoresed (Mini-Protean II, Bio-Rad). Separated proteins were electrotransfered to a nitrocellulose membrane. Membranes were pre-incubated in PBS containing 0.02% (vol/vol) of Tween 20 and 5% (wt/vol) of dried low fat milk for 2 hr at room temperature or overnight at 4°C. Primary antibody was added to the preincubation solution in dilution 1:104 and incubated for 2 hr at room temperature. Membranes were washed three times in PBS containing 0.02% (vol/vol) of Tween 20 at room temperature for a total of 2 hr and incubated with secondary antibody (goat anti-rabbit IgG coupled to peroxidase (Sigma). Chemoluminiscent detection system was used for peroxidase visualization. In some experiments, the classical Laemmli buffer was exchanged for a KBO buffer [20 mM n-octylglucoside (Boehringer Mannheim)/0.5% (vol/vol) Triton X/0.3 M NaCl/0.025 M sodium phosphate, pH 7.4/0.02% (wt/vol) NaN3]. Protein concentration was estimated using the Lowry method (Bio-Rad DC Protein Assay) with BSA as a standard.
Retinoic Acid Binding Studies.
9-cis-[3H]retinoic acid and all-trans [3H]retinoic acid were purchased from DuPont/NEN. Either 10 or 20 ng of (His)6jRXR fusion protein was incubated on ice for 3 hr with labeled retinoids at variable concentration with or without a 100 fold excess of unlabeled competitors in binding buffer [50 mM Tris⋅HCl, pH 7.4/1 mM EDTA/5 mM dithiotreitol/120 mM KCl, 8% (vol/vol) glycerol/0.5% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propane sulfonate/1 mM phenylmethylsulfonyl fluoride/10 ng/ml soybean trypsin inhibitor/2 μM E64/1.6 mM benzamidine/1 μM pepstatin A/5 μg/ml of leupeptin (all inhibitors from Boehringer Mannheim)] in final volumes of 500 μl. Proteins were separated from free ligands with a hydroxyapatite slurry (Ceramic Hydroxyapatite, Bio-Rad) as described (25) and washed 3 times in 1 ml of ice cold wash solution [50 mM Tris⋅HCl, pH 7.4/1.5 mM EDTA/120 mM KCl/0.5% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate with intervening centrifugation (5 min at 1,100 × g). Total radioactivity was estimated by scintillation beta detector (Beckman) from aliquots of samples taken during the incubation, bound radioactivity was measured by transferring the final pelleted hydroxyapatite-protein fraction resuspended in 400 μl of ethanol to scintillation solution and free ligands were estimated from collected wash fractions. Data were subjected to Scatchard analysis as described (26).
RESULTS AND DISCUSSION
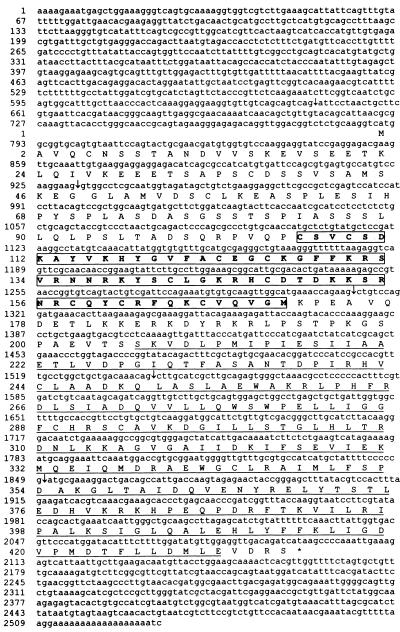
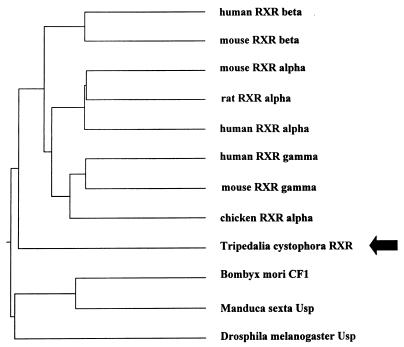
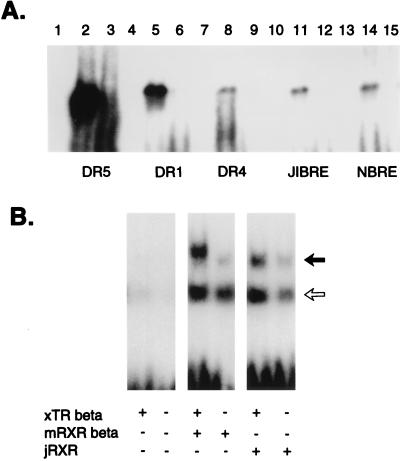
Screening for sequences homologous to known NHRs by PCR with degenerate primers covering the most conserved region of DBD resulted in a sequence showing 75% to 45% identity to a variety of known nuclear hormone receptors. Cloning of cDNA was done by using a PCR strategy employing this sequence and the polyadenylation region or the 5′ RACE method (Life Technologies). Three cDNA isoforms of an mRNA alternatively spliced in the 5′ untranslated region and two short fragments resembling incompletely spliced transcripts were sequenced. Two different clones containing part of the cDNA were obtained by screening a genomic library (Fig. 1A). The gene that we term jRXR was found to be >20 kb in length; three large introns were not sequenced (Fig. 1A). The deduced protein is 435 amino acids long with a molecular mass of 48 kDa. There are five introns, intron 3 being conserved in many mammalian NHRs including the RXRs (27, 28). Southern blot analysis of jRXR showed two bands of equal intensity (Fig. 1B). It is not clear whether this is due to the presence of two hybridizing genes or a polymorphism. Contrary to mammalian RXRs however, the DBD of jRXR is not interrupted by an intron. Although Northern analysis was unsuccessful, RNA primer extension showed a single RNA species (not shown). The predicted amino acid sequence (Fig. 2) showed 61% identity to human RXR alpha in 71 amino acids comprising the DBD and an astonishing 68% identity in the C-terminal 101 amino acids. pileup analysis (Wisconsin Package, version 9.1, Genetics Computer Group) shows less homology to ultraspiracle, the RXR homologue found in Drosophila (Fig. 3). The jRXR protein was prepared as a fusion protein (His)6 and expressed in bacteria or translated in vitro. In either case, the protein bound the core sequence AGGTCA as a monomer (Fig. 4A). The (His)6jRXR fusion protein also bound to promoter sites from the crystallin gene JIB (and JIA, not shown) cloned previously (29) from T. cystophora (Fig. 4A, lane 11). These binding sites were identified by the findpattern Program (Wisconsin Package, version 9.1, Genetics Computer Group). Binding was less than that to the AGGTCA motif separated by either 1 or 5 bases (Fig. 4A). Similar to mammalian RXRs, jRXR heterodimerized with Xenopus TR beta on DR4 (Fig. 4B). Also like mammalian RXRs, the jRXR fusion protein bound 9-cis-retinoic acid with a kd = 0.4 nM (Fig. 5), somewhat greater than values reported for vertebrate RXRs (9, 25). This is in contrast to a recent suggestion that primitive NHRs developed as orphan receptors (13). Binding of all-trans-retinoic acid to the jRXR protein was little greater than nonspecific binding. To study the expression of jRXR during various stages of development, we cultured T. cystophora as described earlier (30) carrying the polyps through the stage of metamorphosis during which the rhopalia with camera eyes are formed (Fig. 6). Western blot analysis with a rabbit antibody raised against the (His)6jRXR fusion protein revealed widely different levels of expression during early nonmotile embryonic stages as well as in swimming planulae, small polyps, and polyps undergoing metamorphosis to medusae. The expression of jRXR with Mr = 48 kDa disappeared sharply (<1 day) after the young ephydra were released. At this stage, a shorter protein was recognized by the antibody. The antibody failed to recognize any proteins in a lysate prepared from another jellyfish Aurelia aurita (Fig. 7), or from brine shrimp used as food for T. cystophora (not shown).
Figure 2.
cDNA of jRXR1 and predicted amino acid sequence. Splicing junctions are indicated by arrows. DBD is boxed. LBD is underlined. Sequence of jRXR2 has deleted nucleotides 528–599; jRXR3 has nucleotides 483–643 exchanged for the sequence: actaggaggaaggtgttgtcagcagtcagatgatcgccgtctgtacatcaacgttcatgtttccatcattactatatcttctcatatggctgcggagatcggacttcaggccagcagctgatttaca.
Figure 3.
Phylogenetic tree of members of NHR family showing branching of jRXR before the differentiation of vertebrate RXRs but distinct from Usp.
Figure 4.
(A) EMSA. DR5, DR1, DR4, J1BRE, a sequence identified in jellyfish J1B crystallin promoter (29), and Nurr1RE (NBRE) (35), bind jRXR expressed in bacteria as a fusion protein. Lanes 2, 5, 8, 11, and 14 are probes (indicated at bottom) incubated with 400 ng of (His)6jRXR; lanes 1, 4, 7, 10, and 13 are probes incubated without the (His)6jRXR; lanes 3, 6, 9, 12, and 15 are probes incubated with (His)6jRXR in presence of 100× in excess of unlabeled probe. (His)6jRXR also bound a sequence found in the promoter of J1A crystallin (29):-195 AAGGAAACAATTTAGAGGTCATGCTACATTGTTGGACGC–158 (data not shown). Motif PuGGTCA was also identified in the promoter of J1C crystallin: -83 TACCAGTAATAAGACTTGGGTCATGTCATCTAAACAGATGCAT-40. (B) Heterodimerization of jRXR fusion protein expressed in bacteria or mouse RXR beta with Xenopus laevis TR beta on DR4. Xenopus TR beta and mouse RXR beta were expressed using TNT Reticulocyte Lysate System (Promega). The solid arrow corresponds to a heterodimer and the open arrow to the monomer position. The faint bands at the heterodimer position are probably due to the presence of another heterodimerization partner present in the lysate.
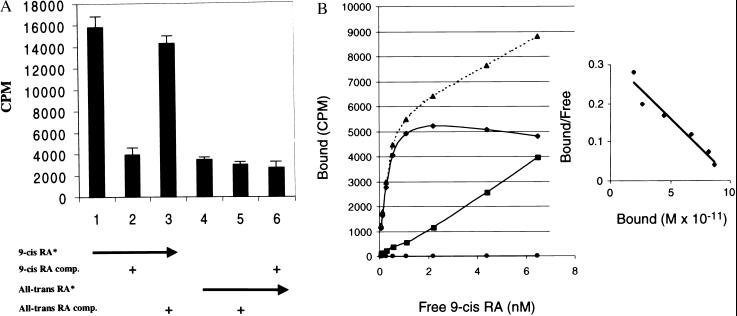
Figure 5.
Binding of 3H 9-cis-retinoic acid or 3H All-trans-retinoic acid to (His)6jRXR. (A) 10 ng of fusion protein was incubated with 3H 9-cis-retinoic acid or 3H-all-trans-retinoic acid (purchased from DuPont/NEN) and competed or not with a hundred fold excess of unlabeled 9-cis-retinoic acid or all-trans-retinoic acid (Sigma) as indicated. A representative of five independent experiments done in triplicates is shown. SED is indicated. (B) A kinetic analysis of binding; insert shows the Scatchard analysis. Kd = 0.4 nM. ▴ are total binding, ■ are nonspecific binding and ♦ are specific binding.
Figure 6.
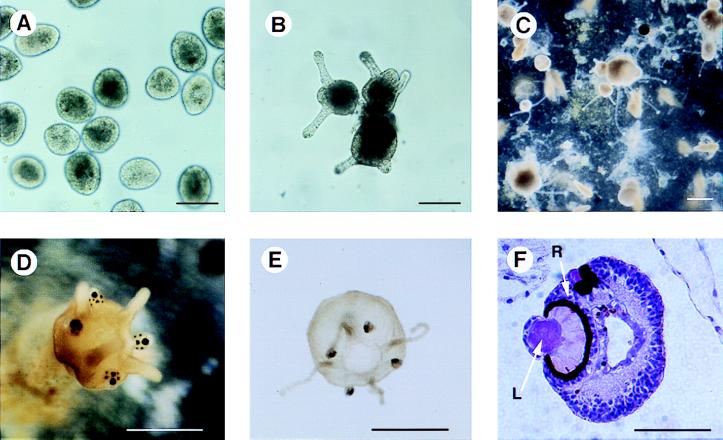
Laboratory culture of T. cystophora. (A) planulae obtained from adult female jellyfish after one day of culture in artificial sea water. (B) the formation of attached polyps with two or three tentacles 48 hr after incubation at 21°C. (C) Primary polyps with attached daughter polyps. (D) A polyp undergoing metamorphosis in process of formation of rhopalia containing the eyes. (E) Young ephydra released about an hour previously from metamorphosing polyp. (F) Rhopalia in paraffin embedded section stained by hematoxylin/eosin. Note the retina (R) and the lens (L). Bar = 100 μm (A and B), 200 μm (C), 500 μm (D), 800 μm (E), and 60 μm (F).
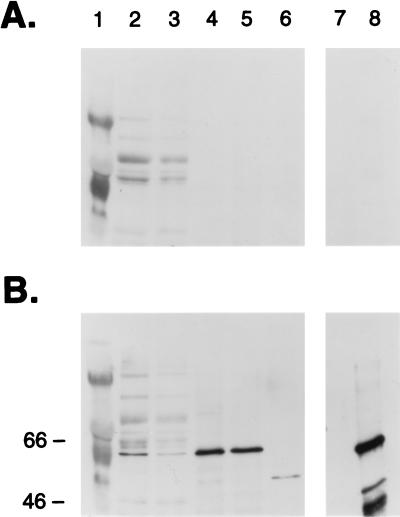
Figure 7.
Immunoblots of extracts from cultured stages of T. cystophora, and from polyps of Aurelia. A is probed with preimmune serum, B with rabbit polyclonal antibodies prepared by immunization with (His)6jRXR. Protein extract prepared from non motile embryos, swimming larvae, small polyps, polyps undergoing metamorphosis (with rhopalia), ephydrae <24 hr old, extract from polyps of Aurelia aurita and 10 ng of (His)6 fusion protein are in lanes 2–8, respectively. Lane 1 is molecular markers.
Our data establishing that the Cnidarian jellyfish, T. cystophora, has at least one NHR, has evolutionary implications. Many views place the Cnidaria as a primitive phylum with no links to other phyla (31, 32). The presence in a Cnidarian of an NHR homologous to a vertebrate NHR suggests, but does not prove that this phylum is ancestral to all other phyla and is not an offshoot without descendents. This conclusion is consistent with the presence of ancestral homeobox (33) and Pax (34) genes in Cnidaria. A major challenge resulting from the present finding is to determine the functional significance of retinoid signaling in Cnidaria and other primitive species. Initial evidence that jRXR plays a number of unknown developmental roles comes from the western immunoblots with proteins from different stages. The gel shift experiments showing that jRXR can bind a consensus DNA sequence as a monomer and as a heterodimer in the presence of TR, as well as its strong affinity for 9-cis-retinoic acid suggest that jellyfish, like vertebrates, use retinoid signaling to express specific genes. Among the potential target genes of jRXR are those encoding the soluble crystallins in the cellular lenses of the camera-type eye of T. cystophora (14) (a role already noted for RAR with crystallin genes in vertebrates; refs. 16, 18–22). We show here that the promoters of J1A and J1B crystallin genes (29) have sequences that bind recombinant jRXR, consistent with it having a functional role in the expression of these genes. Thus the present data raise the possibility that jRXR contributes to the regulation of jellyfish crystallin genes in the lens that, if true suggests that retinoid signaling is a conserved pathway for crystallin gene expression throughout the animal kingdom. In this connection, it is interesting to consider that regulation by retinoic acid isomers might have evolved together with vision.
Acknowledgments
Authors thank Drs. Donald D. Brown for xTR beta, Keiko Ozato for RXR beta, Hans Cahnmann for discussion and advice on retinoic acid isomers, Dame Margaret Clagett for providing the protocol for retinoic acid binding study before publication, and Josef Lazar for advice on EMSA analysis. We also thank George Poy for DNA oligomers and automated sequencing. We are grateful for support and help of Dr. Jeff Holmquist, University of Puerto Rico, Mayaguez, and members of his laboratory.
ABBREVIATIONS
- NHR
nclear hormone receptors
- RXR
retinoic acid X receptor
- jRXR
retinoid receptor for T. cystophora
- DBD
DNA binding domain
- EMSA
electrophoretic mobility shift assay
- RACE
rapid amplification of cDNA ends
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF091121).
To whom reprint requests should be addressed at: National Institute of Diabetes, and Digestive and Kidney Diseases, National Institutes of Health, Building 10, Room 6S201, 9000 Rockville Pike, Bethesda, MD 20892-1829. e-mail: Joseph_Rall@nih.gov.
References
- 1.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blumberg B, Kang H, Bolado J, Jr, Chen H, Craig A G, Moreno T A, Umesono K, Perlmann T, De Robertis E M, Evans R M. Genes Dev. 1998;12:1269–1277. doi: 10.1101/gad.12.9.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forman B M, Goode E, Chen J, Oro A E, Bradley D J, Perlmann T, Noonan D J, Burka L T, McMorris T, Lamph W W, et al. Cell. 1995;81:687–693. doi: 10.1016/0092-8674(95)90530-8. [DOI] [PubMed] [Google Scholar]
- 4.Lala D S, Syka P M, Lazarchik S B, Mangelsdorf D J, Parker K L, Heyman R A. Proc Natl Acad Sci USA. 1997;94:4895–4900. doi: 10.1073/pnas.94.10.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forman B M, Ruan B, Chen J, Schroepfer G J, Jr, Evans R M. Proc Natl Acad Sci USA. 1997;94:10588–10593. doi: 10.1073/pnas.94.20.10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laudet V, Hanni C, Coll J, Catzeflis F, Stehelin D. EMBO J. 1992;11:1003–1013. doi: 10.1002/j.1460-2075.1992.tb05139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laudet V. J Mol Endocrinol. 1997;19:207–226. doi: 10.1677/jme.0.0190207. [DOI] [PubMed] [Google Scholar]
- 8.Kliewer A, Umesono K, Mangelsdorf D J, Evans R M. Nature (London) 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin A A, Sturzenbecker L J, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, et al. Nature (London) 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]
- 10.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 11.Oro A E, McKeown M, Evans R M. Nature (London) 1990;347:298–301. doi: 10.1038/347298a0. [DOI] [PubMed] [Google Scholar]
- 12.Jones G, Sharp P A. Proc Natl Acad Sci USA. 1997;94:13499–13503. doi: 10.1073/pnas.94.25.13499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Escriva H, Safi R, Hanni C, Langlois M C, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R, Laudet V. Proc Natl Acad Sci USA. 1997;94:6803–6808. doi: 10.1073/pnas.94.13.6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piatigorsky J, Horwitz J, Kuwabara T, Cutress C E. J Comp Physiol A. 1989;164:577–587. doi: 10.1007/BF00614500. [DOI] [PubMed] [Google Scholar]
- 15.Cvekl A, Piatigorsky J. BioEssays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- 16.Gopal-Srivastava R, Cvekl A, Piatigorsky J. J Biol Chem. 1998;273:17954–17961. doi: 10.1074/jbc.273.28.17954. [DOI] [PubMed] [Google Scholar]
- 17.Kastner P, Grondona J M, Mark M, Gansmuller A, LeMeur M, Decimo D, Vonesch J L, Dolle P, Chambon P. Cell. 1994;78:987–1003. doi: 10.1016/0092-8674(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 18.Patek C E, Clayton R M. Exp Eye Res. 1990;50:345–354. doi: 10.1016/0014-4835(90)90135-h. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Cvekl A, Bassnett S, Piatigorsky J. Dev Genet. 1997;20:258–266. doi: 10.1002/(SICI)1520-6408(1997)20:3<258::AID-DVG8>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Tini M, Otulakowski G, Breitman M L, Tsui L C, Giguere V. Genes Dev. 1993;7:295–307. doi: 10.1101/gad.7.2.295. [DOI] [PubMed] [Google Scholar]
- 21.Tini M, Tsui L C, Giguere V. Mol Endocrinol. 1994;8:1494–1506. doi: 10.1210/mend.8.11.7877618. [DOI] [PubMed] [Google Scholar]
- 22.Tini M, Fraser R A, Giguere V. J Biol Chem. 1995;270:20156–20161. doi: 10.1074/jbc.270.34.20156. [DOI] [PubMed] [Google Scholar]
- 23.Kostrouch Z, Kostrouchova M, Rall J E. Proc Natl Acad Sci USA. 1995;92:156–159. doi: 10.1073/pnas.92.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Clagett-Dame M, Repa J J. Methods Enzymol. 1997;282:13–24. doi: 10.1016/s0076-6879(97)82092-7. [DOI] [PubMed] [Google Scholar]
- 26.Bylund D B, Yamamura H I. In: Methods in Neurotransmitter Receptor Analysis. Yamamura H I, Enna S J, Kuhar M J, editors. New York: Raven; 1990. pp. 1–35. [Google Scholar]
- 27.Liu Q, Linney E. Mol Endocrinol. 1993;7:651–658. doi: 10.1210/mend.7.5.8391126. [DOI] [PubMed] [Google Scholar]
- 28.Nagata T, Kanno Y, Ozato K, Taketo M. Gene. 1994;142:183–189. doi: 10.1016/0378-1119(94)90259-3. [DOI] [PubMed] [Google Scholar]
- 29.Piatigorsky J, Horwitz J, Norman B L. J Biol Chem. 1993;268:11894–11901. [PubMed] [Google Scholar]
- 30.Werner B, Cutress C E, Studebacher J P. Nature (London) 1971;232:582–583. doi: 10.1038/232582a0. [DOI] [PubMed] [Google Scholar]
- 31.Willmer P. Invertebrate Relationships. Cambridge: Cambridge University Press; 1990. [Google Scholar]
- 32.Valentine J W. Proc Natl Acad Sci USA. 1997;94:8001–8005. doi: 10.1073/pnas.94.15.8001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez D E, Bridge D, Masuda-Nakagawa L M, Cartwright P. Nature (London) 1998;393:748–749. doi: 10.1038/31616. [DOI] [PubMed] [Google Scholar]
- 34.Sun H, Rodin A, Zhou Y, Dickinson D P, Harper D E, Hewett-Emmett D, Li W H. Proc Natl Acad Sci USA. 1997;94:5156–5161. doi: 10.1073/pnas.94.10.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson T E, Fahrner T J, Johnston M, Milbrandt J. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]