Abstract
The oligomerization of activated d- and l- and racemic guanosine-5′-phosphoro-2-methylimidazole on short templates containing d- and l-deoxycytidylate has been studied. Results obtained with d-oligo(dC)s as templates are similar to those previously reported for experiments with a poly(C) template. When one l-dC or two consecutive l-dCs are introduced into a d-template, regiospecific synthesis of 3′-5′ oligo(G)s proceeds to the end of the template, but three consecutive l-dCs block synthesis. Alternating d-,l-oligomers do not facilitate oligomerization of the d-, l-, and racemic 2-guanosine-5′-phosphoro-2-methylimidazole. We suggest that once a “predominately d-metabolism” existed, occasional l-residues in a template would not have led to the termination of self-replication.
Keywords: self-replicating system/enantiomeric cross-inhibition/l-nucleotides/alternating d-l-oligonucleotides
All living organisms use the d-enantiomers of ribo- and deoxynucleotides as building blocks for their nucleic acids. Any abiotic synthesis of a nucleotide would yield equal amounts of the d- and l- isomers. It is not clear why only d-nucleotides were chosen during evolution (1–4). In the present study, we explore the way in which l-nucleotides in a prebiotic soup would effect a self-replicating system of d-nucleotides early in the development of nucleic acid replication. As a model system, we used the highly efficient template-directed oligomerization of activated guanosine mononucleotides on oligo(dC) templates (5). We report here on the oligomerization of activated d- and l-guanosine mononucleotides on templates containing d- and l-deoxycytidylate residues (Fig. 1).
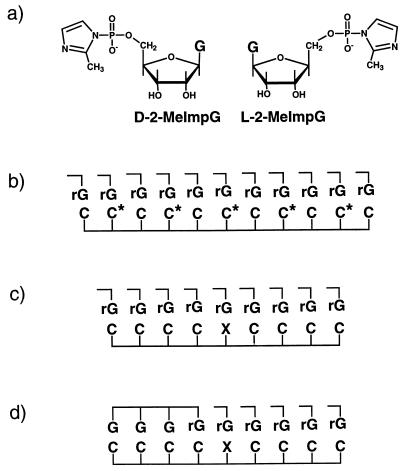
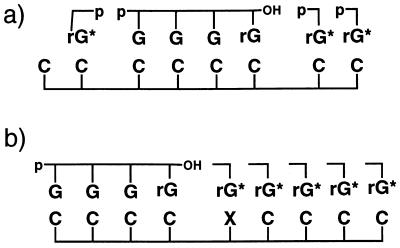
Figure 1.
(a) Structures of d-2-MeImpG and l-2-MeImpG. (b) Schematic representation of the template-directed oligomerization of 2-MeImpG on d-l mixed oligodeoxynucleotide template d(CC*)5dC, where dC* represents l-deoxycytidine. (c) Schematic representation of the template-directed oligomerization of 2-MeImpG and primer (dG)3G extension reaction with 2-MeImpG on d-l mixed oligodeoxynucleotide templates that have structure d(C4XC4) where X can be dC*; d(C*)2, d(C*)3, d(C*CC*), or d(C*CC*CC*). (d) Schematic representation of the primer (dG)3G extension reaction with 2-MeImpG on d-l mixed oligodeoxynucleotide templates that have structure d(C4XC4), where X can be dC*, (dC*)2, (dC*)3, d(C*CC*), or d(C*CC*CC*). In b–d, C is d-dC, C* is l-dC, and rG is d-2-MeImpG or l-2-MeImpG.
MATERIALS AND METHODS
Unless otherwise noted, all chemicals were reagent grade, were purchased from commercial sources, and were used without further purification. d-Guanosine 5′-monophosphate was obtained from Sigma. l-Guanosine 5′-monophosphate was synthesized from N3-isobutyryl-9-(2′,3′-di-O-benzoyl-β-l-ribofuranosyl)guanosine (6) by phosphitylation with bis(2-cyanoethyl)-N,N-diisopropyl phosphoramidite, followed by oxidation and aminolysis (85% yield). The enantiomers of guanosine 5′-phosphoro-2-methylimidazolide (2-MeImpG) were synthesized by a published procedure (7) with yields of 95%. β-l-Deoxycytidine(N-bz)β-cyanoethyl N,N-diisopropyl phosphoramidite was purchased from Chem-Genes (Waltham, MA). Oligodeoxyribonucleotide templates were synthesized on a 391A DNA synthesizer (Applied Biosystems), were deprotected in concentrated ammonia at 55°C, were purified by 20% PAGE, and were desalted on a Nensorb column (DuPont/NEN). The (dG)3G primer is an oligodeoxynucleotide terminated by a single ribonucleotide residue at the 3′ terminus. It was obtained as described above but by using a ribo controlled pore glass GAc column (Glen Research, Sterling, VA) to introduce the 3′-ribonucleotide. Nuclease P1 was obtained from Pharmacia. The primer was labeled with [γ32P]-ATP and T4 polynucleotide kinase (New England Biolabs) as described in ref. 8 and was purified on a Nensorb column. To determine the position of Gpp(dG)3G in PAGE it was synthesized as described in ref. 9.
All PAGE separations were run on a denaturing (8 M urea) 20% polyacrylamide gel. The elution buffer was 50 mM Tris⋅borate (pH 8.3) containing 1 mM EDTA. Loading buffer was prepared by mixing 900 μl of deionized formamide, 25 μl of xylene cyanol (2%), 25 μl of bromophenol blue (2%), and 50 μl of 10× Tris⋅borate, EDTA buffer.
Reaction conditions for the polymerization of 2-MeImpG on various templates were chosen to permit comparison with earlier published work (5, 10, 11). The reactions were run for 7 days at 0°C in 0.2 M 2,6-lutidine buffer (pH 7.9 at 25°C) containing 1.2 M NaCl, 0.2 M MgCl2, 0.5 mM template, and 0.1 M 2-MeImpG. When a mixture of d- and l- isomers was used, the concentration of each isomer was 0.1 M. The volume of the reaction mixture was 3 μl. To prepare the reaction mixture, the appropriate amounts of stock solutions of NaCl, MgCl2, and a template were mixed in an Eppendorf tube and were evaporated to dryness. The residue was redissolved in 3 μl of fresh, prepared solution of 2-MeImpG in 0.2 M 2,6-lutidine buffer to start the reaction. HPLC analyses of the reaction mixtures were performed on an RPC5 column as described (7). Reaction products were eluted with a linear gradient of NaClO4 (pH 12, 0–0.06 M in 60 min) and were monitored by UV absorption at 254 nm.
Reaction conditions for p(dG)3G primer extension with 2-MeImpG on different templates were chosen again to permit comparison with earlier published work (12, 13). The reactions were run for 5 days at 0°C in 0.2 M 2,6-lutidine buffer (pH 7.9 at 25°C) containing 1.2 M NaCl, 0.2 M MgCl2, 20 μM template, 20 nM primer, and 50 mM 2-MeImpG. When a mixture of d- and l-isomers was used, concentration of each isomer was 25 mM. The volume of the reaction mixture was 6 μl. To prepare the reaction mixture, appropriate amounts of stock solutions of NaCl, MgCl2, primer, and a template were mixed in an Eppendorf tube and were evaporated to dryness. The residue was redissolved in 3 μl of 0.2 M 2,6-lutidine buffer. The solution was heated to 95°C and was cooled to 25°C for 1 hour, then kept at 0°C for 20 min. Then, fresh prepared 2-MeImpG solution in 0.2 M 2,6-lutidine buffer (3 μl) was added to start the reaction. Reaction mixtures were analyzed by 20% PAGE.
RESULTS
Oligomerization of the Enantiomers of 2-MeImpG on a d(C10) Template.
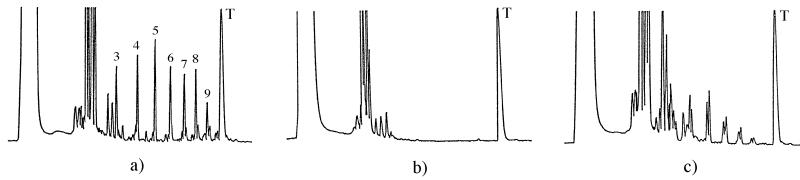
In the absence of a template, oligomerization of the d- and l- enantiomers of 2-MeImpG yields identical patterns of products; only dimers and smaller amounts of trimers are formed (data are not shown). The results that we obtained by using a d(C10) template and enantiomers of 2-MeImpG as substrates (Fig. 2 a, b, and c) are analogous to those reported for a poly(C) template (10). The presence of the d(C10) template leads to efficient oligomerization of d-2-MeImpG (Fig. 2a). The major peaks on the HPLC profile correspond to all 3′-5′-linked oligo(G)ns ranging in length from the dimer to 9-mer (5, 14–16). The addition of the 10th nucleotide to the 9-mer, as anticipated, leads to formation of a mixture of 2′-5′-linked and 3′-5′-linked products in low yield (5). In striking contrast to these results, a d(C10) template has no effect on the oligomerization of l-2-MeImpG (Fig. 2b).
Figure 2.
Oligomerization of d-2-MeImpG and l-2-MeImpG on d(C10) template. (a) d-2-MeImpG. (b) l-2-MeImpG. (c) A mixture of d-2-MeImpG and l-2-MeImpG. The numbers of the peaks represent the length of the oligo(G)n products; T represents the position of the template.
The HPLC profile in Fig. 2c shows the product distribution obtained in the template-directed polymerization of an equimolar mixture of d- and l-2-MeImpG. The single peaks of the 3′-5′-linked products of the template-directed reaction of d-2-MeImpG are replaced by a shorter series of complex groups of peaks. A similar effect has been reported by Joyce et al. (10) for oligomerization of d-2-MeImpG on a poly(C) template and was designated as enantiomeric cross-inhibition. It was shown that, on a poly(C) template, l-2-MeImpG attaches to the 2′(3′) terminus of d-oligo(G)n by means of either a 2′-5′ or a 3′-5′ internucleotide bond and also to the 5′ terminus of d-oligo(G)n by means of a pyrophosphate bond. The presence of these adducts, together with those formed with the d-isomer, explains the complexity of the elution profile.
Oligomerization of d-2-MeImpG on Templates Containing d- and l-dC.
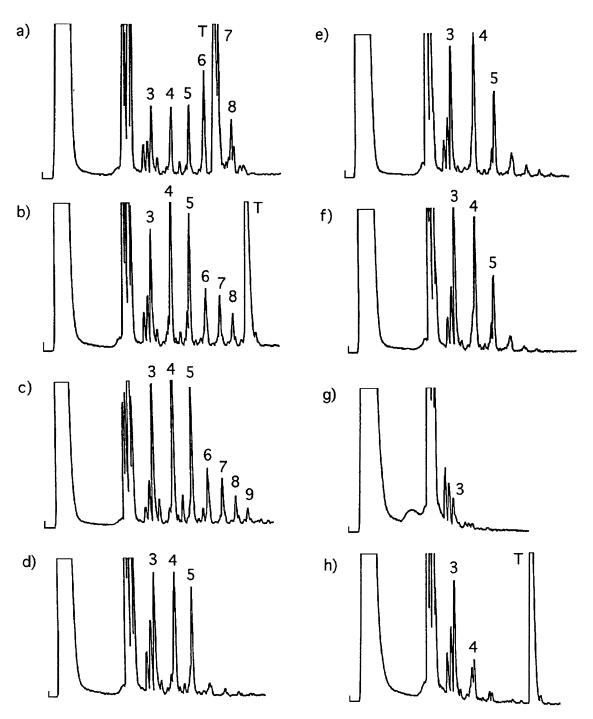
We studied the oligomerization of d-2-MeImpG on “mixed” d-l-oligo(dC) templates, which have the sequence d(C4XC4), where X can be dC*, d(C*)2, d(C*)3, d(C*CC*), or d(C*CC*CC*) and dC* represents l-dC (Fig. 1c). The results obtained are shown in Fig. 3. The presence of the d(C8) template leads to efficient oligomerization of d-2-MeImpG (Fig. 3a). The major peaks on the HPLC profile correspond to all 3′-5′-linked oligo(G)n ranging in length from the dimer to the 7-mer. The insertion of one or more l-dC residues into the d(C8) template leads to a significant decrease in the oligomerization efficiency (Fig. 3 b–f). The largest peaks on the HPLC profiles (Fig. 3 b–f) in every case correspond to all 3′-5′-linked oligo(G)n ranging in length from the dimer to the 5-mer. The presence of one or two l-dC residues in the middle of a d(C8) template does not stop extension of oligo(G)ns completely (Fig. 3 b and c). All oligo(G)ns from the dimer to the 8-mer or the 9-mer are detected in the HPLC profiles, although the yields of oligomers are somewhat reduced compared with those obtained with a d(C8) template. These products were shown to be exclusively 3′-5′-linked by cochromatography with corresponding oligo(G)ns synthesized in a reaction on a d(C8) template. It also was shown that purified oligomers from G3 to G7 are hydrolyzed completely by nuclease P1, an enzyme that is specific for 3′-5′ linkages in RNA (17). The oligomerization of d-2-MeImpG yields very little product larger than G5 when templates d(C4C*C*C*C4), d(C4C*CC*C4), and d(C4C*CC*CC*C4) are used (Fig. 3 d–f). Control experiments showed that a d(C4) (Fig. 3g,) or a d(C4AC4) (Fig. 3h) template does not facilitate efficient oligomerization of d-2-MeImpG.
Figure 3.
(a–f) Oligomerization of d-2-MeImpG on d-l mixed oligo(dC) templates. (a) d(C8). (b) d(C4C*C4). (c) d(C4C*C*C4). (d) d(C4C*C*C*C4). (e) d(C4C*CC*C4). (f) d(C4C*CC*CC*C4). (g and h) Control templates. (g) d(C4). (h) d(C4AC4). The numbers of the peaks represent the length of the oligo(G)n products; T represents position of the template. The positions of the template on diagrams c–f are not shown.
Extension of p(dG)3G Primer with d-2-MeImpG or l-2-MeImpG on d-l Mixed Oligo(dC) Templates.
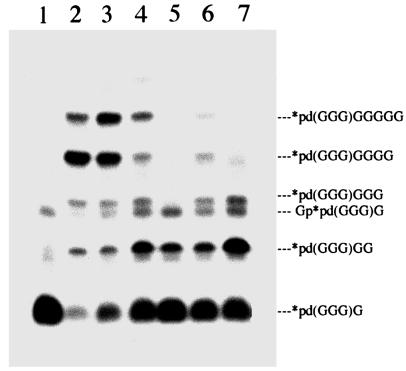
The products distribution in reaction of 32P-labeled p(dG)3G with d-2-MeImpG on mixed d-l oligo(dC) templates (Fig. 1d) are shown in Fig. 4. The results obtained confirm the results for the oligomerization of d-2-MeImpG on these templates. A d(C8) template directs extension of the primer up to p(dG)3G5. The presence of one or two l-dC residues does not stop the extension of the primer whereas the presence of three l-dC residues prevents the attachment of more then one nucleotide. The presence of alternating sequences d(C*CC*) or d(C*CC*CC*) in the middle of template allows the first nucleotide to be attached with high efficiency, and then only small amounts of second and third nucleotides.
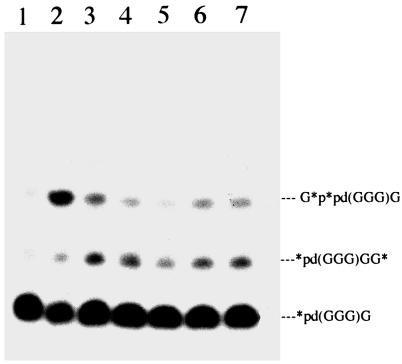
Figure 4.
Extension of p(dG)3G primer with d-2-MeImpG on d-l mixed oligo(dC) templates. Lanes: 1, no template; 2, d(C8); 3, d(C4C*C4); 4, d(C4C*C*C4); 5, d(C4C*C*C*C4); 6, d(C4C*CC*C4); and 7, d(C4C*CC*CC*C4). *p(dG)3G is 32p(dG)3G (5′-32P-labeled primer).
The product distribution in reaction of a p(dG)3G primer with l-2-MeImpG on different mixed d-l templates is shown in Fig. 5. The d(C8) template directs attachment of one l nucleotide to the 5′ terminus of the primer through a pyrophosphate bond to give G*pp(dG)3G, where G* is l-G. A single l-nucleotide also adds to the 2′(3′) terminus of the primer p(dG)3G, but less efficiently, to produce a small amount of p(dG)3GG*. Templates containing one or more l-dC residue behave differently. Attachment of one l-nucleotide to the 2′(3′) terminus of the primer is the main reaction, and the formation of G*pp(dG)3G is somewhat less efficient. These differences are attributable to differences in the ways that primer and substrate can be accommodated on the various templates (see Discussion).
Figure 5.
Extension of p(dG)3G primer with l-2-MeImpG on d-l mixed oligo(dC) templates. Lanes: 1, no template; 2, d(C8); 3, d(C4C*C4); 4, d(C4C*C*C4); 5, d(C4C*C*C*C4); 6, d(C4C*CC*C4); and 7, d(C4C*CC*CC*C4). G* is l-G. *p(dG)3G is 32p(dG)3G (5′-32P-labeled primer).
The extension of a p(dG)3G primer with a mixture of d- and l-2-MeImpG on the mixed oligo(dC) templates gives a complicated mixture of products. A d(C8) template directs the formation of a mixture of p(dG)3Gn and Gpp(dG)3Gn products containing both d- and l-G. Templates containing one or more l-dC residues allow attachment of one mononucleotide to the 2′(3′) terminus of the primer and also direct attachment of one mononucleotide to the 5′ terminus of the primer through a pyrophosphate bond (data not shown).
Oligomerization of the Enantiomers of 2-MeImpG on an Alternating d-l Template.
The presence of an alternating d-l d(CC*)5dC template in which dC* represents l-dC has no effect on the oligomerization of d-2-MeImpG, l-2-MeImpG, or a racemic mixture of d- and l-2-MeImpG (data not shown).
DISCUSSION
The template-directed reactions of d-2-MeImpG on a variety of oligo(C) and oligo(dC) templates, and of racemic 2-MeImpG on poly(C), have been studied extensively (5, 7, 10, 14–16, 18, 19). Our results on the oligomerization of d-, l- and racemic 2-MeImpG on d(C10) are consistent with previous findings. d-2-MeImpG yields 3′-5′-ligated oligomers up to the 9-mer rapidly and the 10-mer more slowly and less regiospecifically. l-2-MeImpG does not oligomerize significantly on d(C10). Finally, racemic 2-MeImpG oligomerizes less efficiently than d-2-MeImpG on d(C10) and yields a complex mixture of products because of enantiomeric cross-inhibition (10).
Although l-2-MeImpG inhibits the synthesis of long d-G oligomers on poly(C), it seemed possible that an oligo(dC) containing alternating d- and l- residues might act as an efficient template when incubated with racemic 2-MeImpG. Our experiments show that alternating the d-,l-template does not catalyze polymerization of d- or l-2-MeImpG or the racemate.
The oligomerization of d-2-MeImpG and the elongation of a p(dG)3G primer on templates of the form d(C4XC4), where C represents a d-dC residue and X represents a sequence initiated by an l-dC residue, are unexpected. In every case, 3′-5′-linked-oligomers up to G5 are obtained in substantial yield. When X is l-dC or (l-dC)2, synthesis continues to the end of the template strand, but when X is (l-dC)3, synthesis stops at G5. In control experiments, we found that d(C4) and d(C4AC4) do not support template-directed synthesis in the same way. They yield only G3 and small amounts of G4. Many other published experiments indicate that a mismatch between template and substrate prevent efficient primer extension, except when wobble pairing between T and G is possible (18, 20–22).
The results presented above indicate that d-2-MeImpG interacts specifically with an l-dC residue in the template in such a way as to permit elongation of an all d-primer. It seems almost certain that base pairing between l-dC and d-2-MeImpG occurs in the normal way with the formation of three hydrogen bonds. Then, because the syn-conformation of pyrimidine nucleotides is inaccessible, one possible way of bringing the 5′-phosphate of d-2-MeImpG into contact with the 3′-OH of the primer is by holding the d-2-MeImpG in the syn-configuration. It is also possible that d-G could form a Watson–Crick pair with l-dC in the manner suggested by Urata and coworkers (23).
Attempts to extend a p(dG)3G primer with l-2-MeImpG on the templates used in our experiments were unsuccessful. When the template contained one or more l-dC residues, small amounts of the primer extended at the 3′-end by one residue were the major products, together with lesser amounts of the 5′-capped pyrophosphate. On a d(C8) template, pyrophosphate formation was the main reaction, presumably because the primer and the l-2-MeImpG residue could be accommodated on a continuous sequence of d-residues of the template (Fig. 6). A similar pyrophosphate-capping reaction on poly(C) was reported by Joyce et al. and was discussed by them in detail (10). The attachment of an l-nucleotide to the 5′ terminus of the d-primer through a pyrophosphate bond would not contribute to the enantiomeric cross-inhibition of primer extension at the 3′ terminus.
Figure 6.
Schematic representation of the extension of a p(dG)3G primer by reaction with l-2-MeImpG (G*). (a) On a d(C8) template. Pyrophosphate formation is the main reaction, presumably because the primer and the l-2-MeImpG residue can be accommodated on a continuous sequence of d-residues of the template. (b) On d-l mixed oligo(dC) templates that have sequences d(C4XC4), where X is a sequence beginning with l-dC; once the primer occupies its most stable position on the template, there is no place to accommodate an additional l-2-MeImpG residue at the 3′ terminus of the template.
It is striking, and possibly relevant to the origin of the RNA world, that primer elongation with d-2MeImpG continues, even if at a somewhat slower rate, past one or two l-dC residues in the template. If the RNA world was the first organized biological world, as often has been proposed, it must have arisen in an environment containing racemic nucleotides. Our findings do not overcome the problems presented by enantiomeric cross inhibition because none of our templates are copied efficiently when the monomeric substrate is racemic. However, our results do suggest that once a “predominantly d-metabolism” was in place, a small proportion of l-monomers in the template or the substrate would not lead to the termination of replication. Nonenzymatic replication may be more resistant to poisoning by the incorrect enantiomer than previously seemed likely.
Acknowledgments
We thank Aubrey R. Hill, Jr. for technical assistance and Bernice Walker for manuscript preparation. This work was supported by National Aeronautics and Space Administration Grant NAG5-4118 and National Aeronautics and Space Administration Specialized Center of Research and Training/Exobiology Grant NAG5-4546.
ABBREVIATION
- 2-MeImpG
guanosine-5′-phosphoro-2-methylimidazole
References
- 1.Mason S F. Nature (London) 1984;311:19–23. doi: 10.1038/311019a0. [DOI] [PubMed] [Google Scholar]
- 2.Miller S L, Orgel L E. The Origins of Life on the Earth. Englewood Cliffs, NJ: Prentice–Hall; 1974. [Google Scholar]
- 3.Joyce G F. Nature (London) 1989;338:217–224. doi: 10.1038/338217a0. [DOI] [PubMed] [Google Scholar]
- 4.Joyce G F, Orgel L E. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 1–25. [Google Scholar]
- 5.Chen C-H B, Inoue T, Orgel L E. J Mol Biol. 1985;181:271–279. doi: 10.1016/0022-2836(85)90091-9. [DOI] [PubMed] [Google Scholar]
- 6.Pitsch S. Helv Chim Acta. 1997;80:2286–2312. [Google Scholar]
- 7.Joyce G F, Inoue T, Orgel L E. J Mol Biol. 1984;176:279–306. doi: 10.1016/0022-2836(84)90425-x. [DOI] [PubMed] [Google Scholar]
- 8.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 9.Chu B C F, Orgel L E. Biochim Biophys Acta. 1984;782:103–105. doi: 10.1016/0167-4781(84)90111-8. [DOI] [PubMed] [Google Scholar]
- 10.Joyce G F, Visser G M, van Boeckel C A A, van Boom J H, Orgel L E, van Westrenen J. Nature (London) 1984;310:602–604. doi: 10.1038/310602a0. [DOI] [PubMed] [Google Scholar]
- 11.Schmidt J G, Nielsen P E, Orgel L E. J Am Chem Soc. 1997;119:1494–1495. doi: 10.1021/ja963563c. [DOI] [PubMed] [Google Scholar]
- 12.Böhler C, Nielsen P E, Orgel L E. Nature (London) 1995;376:578–581. doi: 10.1038/376578a0. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt J G, Nielsen P E, Orgel L E. Nucleic Acids Res. 1997;25:4797–4802. doi: 10.1093/nar/25.23.4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bridson P K, Orgel L E. J Mol Evol. 1980;144:567–577. doi: 10.1016/0022-2836(80)90337-x. [DOI] [PubMed] [Google Scholar]
- 15.Inoue T, Orgel L E. J Am Chem Soc. 1981;103:7666–7667. [Google Scholar]
- 16.Inoue T, Orgel L E. J Mol Biol. 1982;162:201–217. doi: 10.1016/0022-2836(82)90169-3. [DOI] [PubMed] [Google Scholar]
- 17.Sawai H, Totsuka S, Yamamoto K, Ozaki H. Nucleic Acids Res. 1998;26:2995–3000. doi: 10.1093/nar/26.12.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue T, Orgel L E. Science. 1983;219:859–862. doi: 10.1126/science.6186026. [DOI] [PubMed] [Google Scholar]
- 19.Inoue T, Joyce G F, Grzeskowiak K, Orgel L E, Brown J M, Reese C B. J Mol Biol. 1984;178:669–676. doi: 10.1016/0022-2836(84)90244-4. [DOI] [PubMed] [Google Scholar]
- 20.Hill A R, Jr, Orgel L E, Wu T. Origins Life Evol Biosphere. 1993;23:285–290. doi: 10.1007/BF01582078. [DOI] [PubMed] [Google Scholar]
- 21.Wu T, Orgel L E. J Am Chem Soc. 1992;114:5496–5501. doi: 10.1021/ja00040a002. [DOI] [PubMed] [Google Scholar]
- 22.Wu T, Orgel L E. J Am Chem Soc. 1992;114:7963–7969. doi: 10.1021/ja00047a001. [DOI] [PubMed] [Google Scholar]
- 23.Urata H, Ueda Y, Suhara H, Nishioka E, Akagi M. J Am Chem Soc. 1993;115:9852–9853. [Google Scholar]