Abstract
Background
The INSIG2 rs7566605 and PFKP rs6602024 polymorphisms have been identified as obesity gene variants in genome-wide association (GWA) studies. However, replication has been contradictory for both variants. The aims of this study were to validate these obesity-associations through case-control studies and analyses of obesity-related quantitative traits. Moreover, since environmental and genetic factors may modulate the impact of a genetic variant, we wanted to perform such interaction analyses. We focused on physical activity as an environmental risk factor, and on the GWA identified obesity variants in FTO (rs9939609) and near MC4R (rs17782313) as genetic risk factors.
Materials and Methods
The four variants were genotyped in a combined study sample comprising a total of 18,014 subject ascertained from, the population-based Inter99 cohort (n = 6,514), the ADDITION screening cohort (n = 8,662), a population-based study sample (n = 680) and a type 2 diabetic patient group (n = 2,158) from Steno Diabetes Center.
Results
No association with overweight, obesity or obesity-related measures was shown for either the INSIG2 rs7566605 or the PFKP rs6602024 variants. However, an interaction between the INSIG2 rs7566605 variant and the level of self-reported physical activity (p Int = 0.004) was observed. A BMI difference of 0.53 (SE 0.42) kg/m2 was found when comparing physically passive homozygous C-allele carriers with physically passive G-allele carriers. No interactions between the two variants and FTO rs9939609 and MC4R rs17782313 were observed.
Conclusions
The INSIG2 rs7566605 and PFKP rs6602024 polymorphisms play no apparent role in the development of common forms of obesity in the Danish population. However, if replicated, the INSIG2 rs7566605 may influence the level of BMI in combination with the level of physical activity.
Introduction
The identification of genetic variants contributing to common forms of obesity has been challenging using traditional strategies for selection of candidate genes. However, within the last few years rapid advancements have made genome-wide association (GWA) studies feasible and these studies have provided new insight into the pathogenesis of a number of complex polygenic disorders. Several GWA studies have been performed on obesity, and have resulted in the identification of new potential obesity susceptibility gene variants. The rs7566605 located 10 kb upstream of the insulin induced gene 2 (INSIG2), was the first common obesity-related gene variant to be identified through a GWA approach, including 694 individuals [1]. Approximately 10% of the study population carried the risk CC-genotype, and were on average 1 body mass index (BMI) unit heavier than heterozygous or homozygous G-allele carriers. The initial finding was validated in four out of five replication samples, involving a total of 9,881 individuals, with a combined odds ratio (OR) of 1.22 (1.05–1.42) under a recessive model [1]. Moreover, an independent GWA scan, comprising 1,000 unrelated U.S. Caucasians, has observed an association between the INSIG2 rs7566605 variant and obesity [2]. The association of INSIG2 rs7566605 with BMI has further been observed in five out of nine independent large cohorts from eight populations across multiple ethnicities, and a meta-analysis of all nine cohorts, comprising nearly 17,000 individuals also confirmed the association [3]. Several other independent studies have, however, failed to validate the originally proposed association between rs7566605 and obesity when examining a total of 28,043 Caucasian [4]–[9], 2,292 Indian [9], [10] or 6,477 Asian individuals [11]–[13]. INSIG2 is the predominant protein isoform in adipocytes [14], and regulates the transcription of genes promoting fatty acid synthesis and adipogenesis [15], [16], making INSIG2 a good biological candidate gene for obesity.
The rs6602024 in the platelet type phosphofructokinase (PFKP) gene was identified in a GWA scan of obesity-related traits in 4,741 individuals, from a genetically isolated population of Sardinia, to associate strongly with BMI, body weight and hip circumference under an additive model [17]. The association was, however, not validated in independent replication cohorts, including a total of 3,467 subjects, even though homozygous carriers of the minor A-allele on average were ∼1–3 BMI units heavier than homozygous carriers of the G-allele [17].
PFKP encodes a phosphofructokinase acting as a key regulatory enzyme in the glycolysis. Enhanced activity of PFKP might alter the balance between glycolysis and glycogen production, increasing the glucose utilization and thereby lipogenesis, ultimately leading to obesity [17]–[19].
GWA studies have already proved successful and reported novel, unanticipated genetic variants contributing to disease risk. A now well-established link to obesity includes variants within the first intron of the fat mass and obesity associated gene (FTO) [17], [20]–[22]. The consistency of replication across several samples of Caucasian origin [23]–[26] and lately in other ethnic samples [27], [28] has considerably strengthened the important contribution of FTO to obesity. FTO has upon identification been found to encode a 2-oxoglutarate-dependent nucleic acid demethylase [29], [30], however, the link between this enzyme and the development of obesity remains to be elucidated. We have previously validated the association between the rs9939609 variant within FTO and obesity measures, and interestingly, an interaction with physical activity was demonstrated to differentiate the degree of body fat accumulation between genotype groups [23].
A large-scale meta-analysis of GWA data available for 16,876 individuals identified a variant (rs17782313) mapping 188 kb downstream of the gene encoding melanocortin-4 receptor (MC4R) to influence on BMI and the finding were replicated in large-scale studies of 77,228 individuals, both adults and children, with a combined OR for obesity of 1.12 (1.08–1.16), p = 5.2×10−9 [31]. Although mapping hundreds of kilo bases from the coding sequence of MC4R the identified variant presumable disrupt the transcriptional control of MC4R. MC4R is involved in appetite regulation and represent a compelling biological candidate, as rare coding mutations in the gene are a cause of monogenic obesity in humans [32].
Statistically powered replications of GWA findings are essential to determine the true influence of novel gene variants in the pathogenesis of polygenic obesity. Therefore, the aims of the present study are to validate the obesity-associations with the GWA identified INSIG2 rs7566605 and PFKP rs6602024 variants, through case-control studies and analyses of obesity-related quantitative traits. Further, since gene-environment and gene-gene interactions may modulate the effect the variants exert on BMI, analyses are extended to investigate such interactions, focusing on physical activity as an environmental risk factor and on GWA identified obesity variants, reaching a stringent genome-wide significance threshold, (FTO rs9939609 and MC4R rs17782313) as genetic risk factors.
Materials and Methods
Study samples
The INSIG2 rs7566605 and PFKP rs6602024 variants were genotyped in 18,014 subjects ascertained from four different study groups; 1) the Inter99 cohort, which is a population-based, randomized, non-pharmacological intervention study of middle-aged subjects for the prevention of ischemic heart disease (n = 6,514), conducted at the Research Centre for Prevention and Health in Glostrup, Copenhagen (ClinicalTrials.gov ID-no: NCT00289237) [33]; 2) the ADDITION screening cohort, Denmark (Anglo–Danish–Dutch Study of Intensive Treatment in People with Screen-Detected Diabetes in Primary Care) (ClinicalTrials.gov ID-no: NCT00237548) [34], which is a population-based, high-risk screening and intervention study for type 2 diabetes in general practice (n = 8,662); 3) a population-based group of unrelated middle-aged subjects (n = 680) examined at Steno Diabetes Center; and 4) unrelated type 2 diabetic patients (n = 2,158) sampled through the out-patient clinic at Steno Diabetes Center. The combined study sample refers to the combination of these four study groups. In the combined study sample 1,914 had screen-detected and 2,302 had known type 2 diabetes, 5,512 were normal weight, 7,458 were overweight and 5,044 were obese. All participants in study group 1 and 3 underwent a standard 75 g oral glucose tolerance test. Type 2 diabetes was defined according to the World Health Organization [35]. Overweight and obesity were defined as 25≤BMI<30 and BMI≥30 kg/m2, respectively. All study participants were Danes by self-report. Informed written consent was obtained from all subjects before participation. The studies were approved by the regional Ethics Committees (ethics committee, Copenhagen County for study group 1,3 and 4 and ethics committee, Aarhus County for study group 2) and were in accordance with the principles of the Helsinki Declaration.
Biochemical and anthropometrical measurements
In all four study groups body weight and height were measured in light indoor clothes and without shoes. BMI was defined as weight in kilograms divided by height in meters squared (kg/m2). Waist circumference (cm) was measured with subjects in standing position midway between the iliac crest and the lower costal margin. In study group 1, 3 and 4 blood samples were drawn after a 12-hour overnight fast and serum triglycerides, total cholesterol and high density lipoprotein (HDL) -cholesterol were determined using enzymatic colourimetric methods (GPO-PAP and CHOD-PAP; Roche Molecular Biochemicals, Mannheim, Germany). Serum insulin levels excluding des(31,32)- and intact proinsulin were measured using the AutoDELFIA insulin kit (Perkin-Elmer, Wallac, Turku, Finland). Plasma glucose was analysed using a glucose oxidase method (Granutest; Merck, Darmstadt, Germany) [36]. The level of physical activity in study group 1 was self-reported by questionnaire and divided into five categories as; physically passive and light, medium, hard or very hard physically active [36]. These categories were combined in two ways in the analyses; three groups defined as physically passive, light or medium physically active, hard or very hard physically active, and in two groups defined as physically passive and physically active including all four levels of physical activity.
Genotyping
The INSIG2 rs7566605 and PFKP rs6602024 variants were genotyped in the combined study sample using Taqman allelic discrimination (KBioscience, Herts, UK). The discordances between 686 random duplicate samples were 0.3% and 0.0% respectively. Genotyping success rates were 96.8% for both variants, and the minor allele frequencies (MAF) were 32.9% and 10.8%, respectively. The FTO rs9939609 and MC4R rs17782313 variants used in the gene-gene interaction analyses were genotyped using the same technique. The discordances between 1,464 and 721 random duplicate samples were 0.3% and 0.0% respectively. Genotyping success rates were 97.4% and 95.8%, and the MAF were 41.5% and 25.2%, respectively. All genotype groups obeyed Hardy-Weinberg equilibrium.
Statistical analyses
In case-control studies of overweight and obesity, logistic regression was applied to examine differences in genotype distributions between affected and unaffected subjects, applying a recessive model for the INSIG2 rs7566605 variant, and an additive model for the PFKP rs6602024 variant. Adjustments for age, sex and study group were introduced analysing the combined study sample, and for age and sex when analysing the population-based Inter99 study sample. A general linear model was used to test quantitative traits for differences between genotype groups applying the prior stated genetic model for each variant. Adjustment for sex, age, study group and BMI was applied when appropriate. Linear models extended with environmental parameters were used to test for gene-environment interactions using an analysis of variance (ANOVA) test, treating physical activity as a categorical variable. Likewise, additional genetic parameters were added to a linear model, when analysing gene-gene interactions with GWA identified obesity variants. All analyses were performed in RGui version 2.6.2 [37]. p-values<0.05 were considered significant. A test for homogeneity between the Inter99 cohort, the ADDITION cohort, the population-based and type 2 diabetic patient group from Steno Diabetes Center, was performed by means of the Mantel-Haenszel method (fixed effects model), revealing no significant heterogeneity between the study groups (p = 0.5). Statistical power was determined using the CaTS power calculator version 0.0.2. To get robust p-values for the gene-environment and gene-gene interactions, we used a permutation procedure where the phenotype labels where randomly permuted between individuals. The lowest p-value from all iterations (n = 1,000) were saved, and used as an empirical distribution to obtain robust p-values which are corrected for multiple testing.
Results
Using the population-based Inter99 cohort as reference, we found that the prevalence of overweight and obesity was 39% and 17%, respectively, in the Danish population. This gives us a statistical power of 100% observing association between a variant with a MAF of 10% and both overweight and obesity with a relative risk of 1.2, applying an additive model. For a variant with a MAF of 30%, we have a statistical power of 97% and 72% observing an association between overweight and obesity respectively with a relative risk of 1.2, when applying a recessive model.
Potential associations of the INSIG2 rs7566605 C-allele and the PFKP rs6602024 A-allele with overweight and obesity were evaluated by performing case-control studies in the combined study sample and further in the population-based Inter99 cohort, to elucidate the effect on a population-based level, however, no association was observed (Table 1).
Table 1. Case-control study of the INSIG2 rs7566605 and PFKP rs6602024 variants in relation to overweight and obesity.
| INSIG2 rs7566605 | n (men/women) | Genotype distribution n GG/GC/CC (%) | MAF (95% CI) | p rec | ORrec (95% CI) |
| Combined study sample* ( n = 16,781) | |||||
| Controls | 5,106 (2,113/2,993) | 2,264/2,274/568 (44/45/11) | 33.4 (32.5–34.3) | ||
| Overweight cases | 6,973 (4,332/2,641) | 3,162/3,056/755 (45/44/11) | 32.7 (32.0–33.5) | 0.7 | 0.98 (0.87–1.10) |
| Obese cases | 4,702 (2,425/2,277) | 2,118/2,077/507 (45/44/11) | 32.9 (31.9–33.8) | 0.8 | 1.02 (0.89–1.16) |
| Population-based Inter99 cohort ** ( n = 6,158) | |||||
| Controls | 2,708 (1,038/1,670) | 1,176/1,225/307 (44/45/11) | 34.0 (32.7–35.2) | ||
| Overweight cases | 2,391 (1,449/942) | 1,079/1,060/252 (45/44/11) | 32.7 (31.4–34.1) | 0.4 | 0.92 (0.77–1.10) |
| Obese cases | 1,059 (524/535) | 472/454/133 (44/43/13) | 34.0 (32.0–36.1) | 0.2 | 1.15 (0.93–1.44) |
Data are number of subjects, divided into genotype groups (% in each group), frequencies of the minor allele (MAF) in percentages (95% CI) and odds ratio (OR) for the applied genetic model (95% CI). Differences in genotype distribution was evaluated using logistic regression, applying a recessive model (p rec) for the INSIG2 rs7566605 variant (GG/GC vs. CC) and an additive model (p add) for the PFKP rs6602024 variant (GG vs. GA vs. AA). *In the combined study sample (where all four study groups, the Inter99 cohort, the ADDITION cohort, the SDC population-based and type 2 diabetes sample were included) p-values were adjusted for age, sex, and study group, whereas p-values in the **population-based Inter99 cohort were adjusted for age and sex. Controls were defined as BMI<25, overweight cases as 25≤BMI<30 and obese cases as BMI≥30 kg/m2 respectively.
The two variants were furthermore investigated for influence on quantitative obesity-related traits in the combined study sample and in the population-based Inter99 cohort excluding known type 2 diabetic patients. No association of obesity risk alleles with BMI, body weight or waist circumference could be shown (Table 2).
Table 2. Quantitative obesity-related traits in the combined study sample and the population-based Inter99 cohort.
| INSIG2 rs7566605 | PFKP rs6602024 | |||||||
| GG | GC | CC | p rec | GG | GA | AA | p add | |
| Combined study sample* | ||||||||
| n (men/women) | 6,658 (3,482/3,176) | 6,552 (3,467/3,085) | 1,619 (836/783) | 11,806 (6,177/5,629) | 2,840 (1,513/1,327) | 150 (71/79) | ||
| Age (years) | 55±10 | 55±10 | 54±10 | 54±10 | 55±10 | 55±10 | ||
| BMI (kg/m2) | 27.6±4.8 | 27.5±4.9 | 27.5±5.0 | 0.9 | 27.5±4.9 | 27.6±4.9 | 27.6±5.3 | 0.8 |
| Body weight (kg) | 81.1±16.2 | 81.1±16.2 | 80.9±16.7 | 0.9 | 81.0±16.2 | 81.3±16.6 | 79.4±15.3 | 1 |
| Waist (cm) | 92.7±14.4 | 92.5±14.1 | 92.3±14.6 | 0.9 | 92.5±14.2 | 93.0±14.4 | 91.7±15.5 | 0.6 |
| Population-based Inter99 cohort** | ||||||||
| n (men/women) | 2,559 (1,271/1,288) | 2,559 (1,282/1,277) | 652 (317/335) | 4,642 (2,303/2,339) | 1,056 (532/524) | 60 (25/35) | ||
| Age (years) | 46±8 | 46±8 | 46±8 | 46±8 | 46±8 | 46±8 | ||
| BMI (kg/m2) | 26.2±4.5 | 26.1±4.5 | 26.4±4.5 | 0.2 | 26.2±4.5 | 26.3±4.7 | 25.9±4.2 | 0.9 |
| Body weight (kg) | 78.0±15.7 | 78.0±15.9 | 78.3±17.2 | 0.4 | 78.1±15.9 | 78.3±16.4 | 75.1±13.4 | 0.9 |
| Waist (cm) | 86.5±13.2 | 86.4±13.1 | 86.7±13.9 | 0.4 | 86.4±13.2 | 87.0±13.3 | 84.4±12.4 | 0.6 |
Data are means±standard deviation. p-values were calculated assuming a recessive model (p rec) for INSIG2 rs7566605 variant (GG/GC vs. CC) and an additive model (p add) for PFKP rs6602024 variant (GG vs. GA vs. AA) in the combined study sample (where all four study groups, the Inter99 cohort, the ADDITION cohort, the SDC population-based and type 2 diabetes sample were included) and the population-based Inter99 cohort. Known type 2 diabetic patients were excluded from the analyses. *Adjustments for the effect of age, sex and study group was introduced for the combined study sample, and for age and sex in the **population-based Inter99 cohort.
Since the INSIG2 rs7566605 C-allele has been proposed to exert a larger effect in individuals already predisposed to obesity, we investigated the effect of the variant in the obese subgroup of the combined study sample (n = 3,878), with a mean BMI of 32.8 kg/m2, however, no genotype effect was found (data not shown).
In the population-based Inter99 cohort, serum insulin and plasma glucose measures during an oral glucose tolerance test were available, together with fasting levels of serum triglyceride and HDL-cholesterol. However, none of these measures were associated with INSIG2 rs7566605 or PFKP rs6602024 genotypes (data not shown).
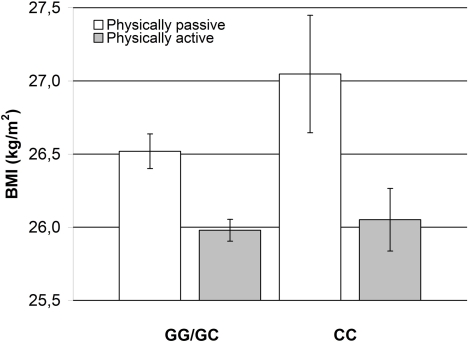
In analyses of gene-environment interactions comprising a total of 5,604 individuals from the population-based Inter99 cohort, excluding known type 2 diabetes patients, we found an interaction between INSIG2 rs7566605 genotype and the level of self-reported physical activity. When dividing subjects into three groups categorised as physically passive, light or medium physically active and hard or very hard physically active, a difference in BMI level was only observed between the group of physically passive homozygous C-allele carriers and physically passive G-allele carriers (p Int = 0.004). No difference in BMI was observed between genotype groups for light or medium physically active or hard or very hard physically active. Therefore, we tested physically passive against physically active (light to very hard physically active) (p Int = 0.004), and found that physically passive homozygous C-allele carriers had a BMI 1.00 (SE 0.46) kg/m2 higher than physically active homozygous C-allele carriers whereas physically passive G-allele carriers only had a BMI 0.54 (SE 0.14) kg/m2 higher than physically active G-allele carriers (Figure 1). This is also reflected in a difference of 0.53 (SE 0.42) kg/m2 between physically passive homozygous C-allele carriers and physically passive G-allele carriers. The robustness of the p-value was verified by the empirical value obtained by the permutation procedure (p = 0.005). No interaction between the PFKP rs6602024 variant and physical activity was demonstrated.
Figure 1. Effect of physical activity on the impact of the INSIG2 rs7566605 genotype on BMI.
Participants (n = 5,604) from the population-based Inter99 cohort, were divided according to self-reported physical activity categorised as physically passive and physically active and stratified according to INSIG2 rs7566605 genotype applying a recessive model. Bars indicate mean BMI, and error bars indicate standard error. The number of participants (physically passive/physically active) are (1,712/3,259) for G-allele carriers and (202/431) for homozygous C-allele carriers. We tested for interaction effects using linear models, with or without, interaction parameters for physical activity compared by an ANOVA test (p Int = 0.004).
We further investigated potential interactions between the rs7566605 and rs66020245 variants and GWA identified obesity variants reaching the stringent significance threshold; rs9939609 located in FTO and rs17782313 located downstream from MC4R, however, no interactions were observed (data not shown).
Discussion
Analysing 18,014 Danish individuals, we failed to demonstrate a correlation between the INSIG2 rs7566605 C-allele and the PFKP rs6602024 A-allele and excessive body fat accumulation both in case-control studies of overweight and obesity, and when analysing quantitative obesity-related traits. The obesity-association of PFKP rs6602024 was not successfully replicated in the initial GWA study, which was suggested to be due to the relatively low risk allele frequency of the variant resulting in only a small fraction of the population being affected. However, the variant exerted a relatively large effect size on BMI in the replication cohorts, with homozygous A-allele carriers having a BMI ∼3 units higher than G-allele carriers, and therefore it was proposed that larger population samples might be needed in order to reach statistical significance [17]. We did, however, not replicate the association between PFKP rs6602024 and obesity measures, either using an additive or recessive model. On the contrary we observed slightly lower BMI, body weight and waist circumference among homozygous PFKP rs6602024 A-allele carriers. The failure to replicate the association between PFKP rs6602024 and different measures of obesity in the present study could be due to differences in linkage disequilibrium patterns between the population in which the variant was first identified [17] and our study material. An independent GWA study in unrelated U.S. Caucasians has reported an association between three other polymorphisms in PFKP and excessive body fat accumulation [2]. Hence, PFKP could in theory be a true obesity susceptibility gene, with rs6602024 failing to be a marker for the functional variant in our population.
Several studies have failed to validate the initial GWA finding of the INSIG2 rs7566605 C-allele contributing to the pathogenesis of obesity. Despite the large-scale study samples included in our study, we were not able to confirm the proposed association, in neither case-control nor quantitative settings. The effect of the INSIG2 rs7566605 variant on BMI has been proposed to be predominant in already obese subjects, but neither in an obese subgroup did we observe an association. However, the effect of some genetic variants is only exerted in combination with environmental or other genetic risk factors [38]. When taking the level of physical activity into account, we detected an INSIG2 rs7566605 genotype effect on the level of BMI. Physical inactivity results in an increased BMI level in both G-allele carriers and homozygous C-allele carriers. In the group of G-allele carriers the effect of physical inactivity on BMI level is 0.54 (SE 0.14) kg/m2, whereas the effect is more pronounced in homozygous C-allele carriers increasing the BMI level by 1.00 (SE 0.46) kg/m2. This indicates an interaction between the INSIG2 rs7566605 variant and physical activity. This is supported by the observation that the BMI level in the subgroup of physically passive is 0.53 (SE 0.42) kg/m2 higher in homozygous C-allele carries compared with physically passive G-allele carriers. However, this interaction analysis is merely explorative, since it is based on self-reported measures, and replication in statistically well-powered populations with more precise physiological physical activity measures is imperative.
We can thus conclude that common variation in INSIG2 and PFKP, both candidate genes arising from GWA studies, do not play a significant role in the pathogenesis of obesity in the Danish population. Still, if replicated common variation in INSIG2 might, contribute to common forms obesity through interaction with a low level of physical activity.
Acknowledgments
The authors wish to thank Annemette Forman, Inge-Lise Wantzin and Marianne Stendal for technical assistance and Grete Lademann for secretarial support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This study was supported by the Danish Medical Research Council, the Danish Diabetes Association, the Gerda and Aage Haensch Foundation, the A.P. Møller Foundation for the Advancement of Medical Science, University of Copenhagen, and the Velux Foundation. This work is part of the project “Hepatic and adipose tissue and functions in the metabolic syndrome” (HEPADIP www.hepadip.org), which is supported by the European Commission as an integrated project under the 6th Framework Programme (LSHM-CT-2005-018734). The study also received support from The Danish Obesity Research centre (DanORC; www.danorc.dk), which is supported by The Danish Council for Strategic Research (Grant No 2101-06-0005).
References
- 1.Herbert A, Gerry NP, McQueen MB, Heid IM, Pfeufer A, et al. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]
- 2.Liu YJ, Liu XG, Wang L, Dina C, Yan H, et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet in press. 2008 doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lyon HN, Emilsson V, Hinney A, Heid IM, Lasky-Su J, et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007;3:e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosskopf D, Bornhorst A, Rimmbach C, Schwahn C, Kayser A, et al. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1130571. [DOI] [PubMed] [Google Scholar]
- 5.Loos RJ, Barroso I, O'rahilly S, Wareham NJ. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dina C, Meyre D, Samson C, Tichet J, Marre M, et al. Comment on “A common genetic variant is associated with adult and childhood obesity”. Science. 2007;315:187. doi: 10.1126/science.1129402. [DOI] [PubMed] [Google Scholar]
- 7.Hall DH, Rahman T, Avery PJ, Keavney B. INSIG-2 promoter polymorphism and obesity related phenotypes: association study in 1428 members of 248 families. BMC Med Genet. 2006;7:83. doi: 10.1186/1471-2350-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boes E, Kollerits B, Heid IM, Hunt SC, Pichler M, et al. INSIG2 Polymorphism is neither associated with BMI nor with phenotypes of lipoprotein metabolism. Obesity (Silver Spring) in press. 2008 doi: 10.1038/oby.2007.132. [DOI] [PubMed] [Google Scholar]
- 9.Smith AJ, Cooper JA, Li LK, Humphries SE. INSIG2 gene polymorphism is not associated with obesity in Caucasian, Afro-Caribbean and Indian subjects. Int J Obes (Lond) 2007;31:1753–1755. doi: 10.1038/sj.ijo.0803645. [DOI] [PubMed] [Google Scholar]
- 10.Kumar J, Sunkishala RR, Karthikeyan G, Sengupta S. The common genetic variant upstream of INSIG2 gene is not associated with obesity in Indian population. Clin Genet. 2007;71:415–418. doi: 10.1111/j.1399-0004.2007.00795.x. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Dong H, Xiang Q, Hong X, Wilker E, et al. Lack of association between rs7566605 and obesity in a Chinese population. Hum Genet. 2007;120:743–745. doi: 10.1007/s00439-006-0258-2. [DOI] [PubMed] [Google Scholar]
- 12.Kuzuya M, Ando F, Iguchi A, Shimokata H. No association between rs7566605 variant and being overweight in Japanese. Obesity (Silver Spring) 2007;15:2531–2534. doi: 10.1038/oby.2007.301. [DOI] [PubMed] [Google Scholar]
- 13.Tabara Y, Kawamoto R, Osawa H, Nakura J, Makino H, et al. No association between INSIG2 Gene rs7566605 polymorphism and being overweight in Japanese population. Obesity (Silver Spring) 2008;16:211–215. doi: 10.1038/oby.2007.25. [DOI] [PubMed] [Google Scholar]
- 14.Krapivner S, Popov S, Chernogubova E, Hellenius ML, Fisher RM, Hamsten A, et al. Insulin-induced gene 2 (INSIG2) involvement in human adipocyte metabolism and body weight regulation. J Clin Endocrinol Metab in press. 2008 doi: 10.1210/jc.2007-1850. [DOI] [PubMed] [Google Scholar]
- 15.Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10:1096–1107. doi: 10.1101/gad.10.9.1096. [DOI] [PubMed] [Google Scholar]
- 16.Fajas L, Schoonjans K, Gelman L, Kim JB, Najib J, et al. Regulation of peroxisome proliferator-activated receptor gamma expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol Cell Biol. 1999;19:5495–5503. doi: 10.1128/mcb.19.8.5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scuteri A, Sanna S, Chen WM, Uda M, Albai G, et al. Genome-Wide Association Scan Shows Genetic Variants in the FTO Gene Are Associated with Obesity-Related Traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakajima H, Raben N, Hamaguchi T, Yamasaki T. Phosphofructokinase deficiency; past, present and future. Curr Mol Med. 2002;2:197–212. doi: 10.2174/1566524024605734. [DOI] [PubMed] [Google Scholar]
- 19.Belfiore F, Borzi V, Napoli, Rabuazzo AM. Enzymes related to lipogenesis in the adipose tissue of obese subjects. Metabolism. 1976;25:483–493. doi: 10.1016/0026-0495(76)90001-9. [DOI] [PubMed] [Google Scholar]
- 20.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinney A, Nguyen TT, Scherag A, Friedel S, Bronner G, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) Variants. PLoS ONE. 2007;2:e1361. doi: 10.1371/journal.pone.0001361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dina C, Meyre D, Gallina S, Durand E, Korner A, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 23.Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, et al. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57:95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- 24.Peeters A, Beckers S, Verrijken A, Roevens P, Peeters P, et al. Variants in the FTO gene are associated with common obesity in the Belgian population. Mol Genet Metab in press. 2007 doi: 10.1016/j.ymgme.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Price RA, Li WD, Zhao H. FTO gene SNPs associated with extreme obesity in cases, controls and extremely discordant sister pairs. BMC Med Genet. 2008;9:4. doi: 10.1186/1471-2350-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt SC, Stone S, Xin Y, Scherer CA, Magness CL, et al. Association of the FTO Gene With BMI. Obesity (Silver Spring) in press. 2008 doi: 10.1038/oby.2007.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha SW, Choi SM, Kim KS, Park BL, Kim JR, et al. Replication of genetic effects of FTO polymorphisms on BMI in a Korean population Obesity (Silver Spring) in press. 2008 doi: 10.1038/oby.2008.314. [DOI] [PubMed] [Google Scholar]
- 28.Chang YC, Liu PH, Lee WJ, Chang TJ, Jiang YD, et al. Common variants in the FTO gene confers risk of obesity and modulates body mass index in the Chinese population. Diabetes in press. 2008 doi: 10.2337/db08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanchez-Pulido L, Andrade-Navarro MA. The FTO (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:23. doi: 10.1186/1471-2091-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loos RJF, Lindgren CM, Li S, Wheeler E, Zhao JH, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farooqi IS, Yeo GS, Keogh JM, Aminian S, Jebb SA, et al. Dominant and recessive inheritance of morbid obesity associated with melanocortin 4 receptor deficiency. J Clin Invest. 2000;106:271–279. doi: 10.1172/JCI9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jørgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glümer C, et al. A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99. Eur J Cardiovasc Prev Rehabil. 2003;10:377–386. doi: 10.1097/01.hjr.0000096541.30533.82. [DOI] [PubMed] [Google Scholar]
- 34.Lauritzen T, Griffin S, Borch-Johnsen K, Wolffenbuttel BH, Rutten G. The ADDITION study: proposed trial of the cost-effectiveness of an intensive multifactorial intervention on morbidity and mortality among people with Type 2 diabetes detected by screening. Int J Obes Relat Metab Disord 24 Suppl. 2000;3:S6–11. doi: 10.1038/sj.ijo.0801420. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization Study Group: Definition, diagnosis and classification of diabetes mellitus and its complications; Part 1: Diagnosis and classification of diabetes mellitus. Tech Rep Ser WHO/NCD/NCS/99.2. Geneva, World Health Organization, 1999.
- 36.Glümer C, Jørgensen T, Borch-Johnsen K. Prevalences of diabetes and impaired glucose regulation in a Danish population: the Inter99 study. Diabetes Care. 2003;26:2335–2340. doi: 10.2337/diacare.26.8.2335. [DOI] [PubMed] [Google Scholar]
- 37.R Development Core Team. R: A language and environment for statistical computing, R Foundation for Statistical Computering, Vienna, Australia. 2008. ISBN 3-900051-01-0, URL http://www.R-project.org.
- 38.Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]