Abstract
Immunoglobulin superfamily (IgSF) proteins are known for their ability to specifically recognize and adhere to other molecules, mediating cell surface reception and pathogen recognition. Mammalian IgSF proteins such as antibodies are among the best characterized molecules of the immune system; in contrast, the involvement of invertebrate IgSF members in immunity has not been broadly studied. Analysis of the predicted Anopheles gambiae transcriptome identified 138 proteins that have at least one immunoglobulin domain. Challenge with Plasmodium, Gram-negative or Gram-positive bacteria resulted in significant regulation of 85 IgSF genes, indicating potential roles for these molecules in infection responses and immunity. Based on sequence and expression data, six infection-responsive with immunoglobulin domain (IRID 1-6) genes were chosen and functionally characterized with regard to their role in innate immunity. Reverse-genetic gene-silencing assays showed IRID3, IRID5 and IRID6 contribute to viability upon bacterial infection while IRID4 and IRID6 are involved in limiting Plasmodium falciparum infection.
Keywords: Immunoglobulin domain, IgSF, Anopheles gambiae, innate immunity, Plasmodium, malaria
1. Introduction
The immunoglobulin superfamily (IgSF) is composed of genes encoding at least one immunoglobulin (Ig) domain. The structure of an Ig domain is ideal for ligand binding, accommodating broad amino acid sequence variability without changing the conserved structure, thus allowing a high degree of interaction specificity and diversity [1-3]. These properties make IgSF proteins candidates for processes requiring specific adhesion and recognition, especially when the domain is frequently repeated or found next to other highly interactive domains [3]. Such processes include cell-cell adhesion, cell-surface recognition and pathogen recognition [4 and 5].
In mammals, IgSF members are known primarily as immune molecules. In many cases, Ig-containing proteins are responsible for recognizing non-self entities and promoting their elimination. Although lower organisms do not have antibodies or other mammalian IgSF immune proteins, functional and structural parallels exist between these Ig molecules and invertebrate pathogen recognition proteins. Invertebrate immune surveillance is generally undertaken by pattern recognition receptor (PRR) molecules that, like mammalian Ig proteins, recognize specific foreign motifs and help initiate a molecular cascade resulting in activation of effector molecules or mechanisms. Like vertebrate Ig molecules, PRRs have adhesive domains that are capable of binding to pathogen-associated molecular patterns. In some cases, they also have catalytic or signaling domains that initiate an effector response upon pathogen detection. Each of these characteristics could theoretically be applied to insect IgSF members, thus making this superfamily a highly potent source of PRRs.
To our knowledge, the IgSF as a superfamily has never been thoroughly screened as a source of immune genes in insects; however, several individual Ig proteins have been linked to invertebrate immunity. Dscam of Anopheles gambiae has been shown to be involved in defense against bacteria and Plasmodium parasites [6]. Fibrinogen-related proteins (FREPs) of Biomphalaria glabrata [7, 8] (which are also present in the A. gambiae and D. melanogaster genomes [9]) and molluscan defense molecule (MDM) of Lymnaea stagnalis [10] are Ig domain-containing proteins that are either induced or repressed in response to parasitic infection. Peroxinectin, described mainly in crayfish and black shrimp, combines Ig domains with a peroxidase region and has been proposed to act as an opsonin that enhances encapsulation and phagocytosis [11, 12]. Hemolin binds bacteria [13], and triggers phagocytosis and the phenoloxidase cascade in Hyalophora cecropia [14, 15] and prevents hemocyte aggregation in Manduca sexta [16]. This was the first insect IgSF molecule to be considered a PRR, since it has broad specificity for lipopolysaccharide and lipotechoic acid [17, 18] and is a major player during Gram-negative bacteria infections [19-21]. These proteins are intriguing and raise questions about the extent of immunity roles throughout the superfamily.
Studies of the IgSF in Drosophila and C. elegans determined that IgSF members carry out diverse functions in multiple physiological systems [22-25] which we also expect in mosquitoes. The specific and diverse molecular recognition mechanisms required by the innate immune system are analogous to those required in other physiological systems, particularly the nervous system [26]. In fact, phylogenetic studies that incorporate IgSF members have suggested that invertebrate immune molecules are more closely related to invertebrate and vertebrate neuronal molecules than to vertebrate immune molecules [27]. Co-expression of IgSF members in immune and nervous systems has long been observed in vertebrates [28], and dual roles in the immune and nervous systems have recently been documented for two insect IgSF members. Dscam, an IgSF member studied in both Drosophila and Anopheles, has been shown to mediate axon guidance and pathogen recognition [6, 29-30]. Similarly, a Manduca sexta molecule, neuroglian, is implicated in axon sorting and extension in the nervous system and in hemocyte aggregation in the immune system [31-32]. Molecular integration of the nervous and immune systems is not uncommon in insects. The Toll receptor, Dif, cactus and several peptides have all been assigned dual roles in these systems [33-35] while general immune responses have been implicated in learning and memory of honeybees [36].
Here we provide evidence of the extensive involvement of the A. gambiae IgSF in the mosquito’s innate immune system.
2. Materials and methods
2.1 Determining IgSF members
Proteins predicted by the Ensembl A. gambiae genome browser (http://www.ensembl.org/Anopheles_gambiae/) to contain at least one immunoglobulin domain were considered. Ensembl defines likely domains using the Prosite, Prints, Pfam and UnitPro/SwissProt databases. Seven different Interpro identifiers were returned for immunoglobulin domains. Of these, two identified domains found in predicted Anopheles genes: Ig-MHC (Interpro domain IPR003006, Prosite accession number PS00290) and Ig-like (Interpro domain IPR007110, Pfam accession number PF00047, Prosite accession number PS50835). Amino acid sequences for proteins containing these domains were retrieved from Ensembl and entered into the Simple Modular Architecture Research Tool (SMART) database, version 4.0, http://smart.embl-heidelberg.de[37 and 38]) to confirm domain arrangement and to increase stringency. At the time of our search, the SMART library contained 667 hidden Markov models (HMMs). SMART uses HMMER searches by default, and within the SMART analysis we included additional domain support through the Pfam HMM set. We also included signal peptide and transmembrane region prediction through SMART access to Sigcleave and TMHMM2, respectively. Our final catalogue of IgSF genes contained only those that were identified by Ensembl and corroborated by SMART. Gene identifiers used here are either the catalogue name given by Ensembl or the name given by previous experimental investigators.
Ensembl predicts putative orthologs by reciprocal BLAST hits. Any D. melanogaster Ig domain gene that was returned as a unique best reciprocal hit (UBRH) to an A. gambiae Ig domain gene query was considered an ortholog. Similarity between orthologs was determined using BLAST to align Ensembl-predicted sequences of A. gambiae, D. melanogaster and Aedes aegypti. When more than one transcript was predicted, the sequence returning the highest percent similarity was used.
2.2 Mosquito rearing
A. gambiae Keele strain mosquitoes were maintained on sugar solution at 27°C and 70% humidity with a 12-h light/dark cycle according to standard procedures [39].
2.3 Bacterial challenge for microarray
For preparation of bacterial-challenged samples for microarray analyses, 4-day old female mosquitoes were first injected with approximately 20,000 heat-inactivated Salmonella typhimurium or Bacillis subtilis and approximately 20 whole mosquitoes were collected 4 hr after challenge [40].
2.4 Microarray analysis
For Escherichia coli, Staphylococcus aureus, Plasmodium berghei and Plasmodium falciparum challenges, comparison of IgSF gene expression between infected and naïve mosquitoes was derived from the data set generated and validated by Dong et al. [40]. For S. typhimurium and B. subtilis challenges, RNA was extracted from challenged whole mosquitoes using the RNeasy kit (QIAGEN), quantified using a Beckman DU640 spectrophotometer, and assessed for quality by RNA Nano LabChip analysis on an Agilent Bioanalyzer 2100. Probes were synthesized with 2-3 μg RNA using the Agilent Technologies low-input RNA labeling kit according to the manufacturer’s instructions. Hybridizations were done with the Agilent technologies in situ hybridization kit according to the manufacturer’s instructions. Microarray scanning was done with an AXON 4200AL scanner; laser power set to 100%, PMTs adjusted manually. Scan images were analyzed with Genepix 6.0 software and processed with the TIGR MIDAS software through the following steps: a) Spots with intensity lower than 100 or signal-to-background ratio lower than 2 in either Cy5 or Cy3 channel were excluded; b) spots flagged manually for poor quality were excluded; c) arrays were normalized according to a LOWESS normalization method. The TIGR MeV software was then used to Log2 transform the Cy5/Cy3 ratios and perform T-TESTs with the following parameters: mean value to be tested against = 0, Welsh approximation method for degree of freedom calculation, P-values based on t-distribution, the overall alpha (critical p-value) 0.05, no p-value correction. Gene expression values are the average of 3 biological replicas and are considered significant if p < .05 and display regulation of at least 1.7 fold [41].
2.5 RNAi gene silencing
Sense and antisense RNAs were synthesized from PCR-amplified gene fragments using the T7 Megascript kit (Ambion). PCR-amplified fragments were each ∼300bp; the primers used are described in Supplementary Material. About 69nl dsRNAs (3μg/μl) in water was introduced into the thorax of cold-anesthetized 4-day-old female mosquitoes by using a nano-injector (Nanoject, Drummond) with glass capillary needles according to established methodology [42].
2.6 Challenge with bacteria and Plasmodium
Gram-positive S. aureus and Gram-negative E. coli DH5α dwere cultured in LB broth overnight, washed three times with phosphate-buffered saline (PBS) and resuspended in PBS. Four days after dsRNA injections for each tested gene and the control, approximately 20,000 bacteria of each species were injected in a 69-nl PBS suspension into the hemolymph of 50 cold-anesthetized female mosquitoes,. Injections were done using a microcapillary Nanoject II injector (Drummond). Concentrations of bacteria were estimated based on OD and counting with light microscopy. Control-challenged mosquitoes were injected with 69 nl sterile PBS. Dead mosquitoes were counted and removed daily over a 7-day period. The results shown here are representative of at least three independent experiments per tested gene, 80 mosquitoes per treatment. Two-way ANOVA analysis was used to determine the significance of treatment at each time point.
Plasmodium infections were carried out according to standard procedure [43]. For P. berghei infection, mosquitoes were fed on the same infected mouse 4 days after dsRNA treatment. For P. falciparum, mosquitoes were fed on NK54 gametocytes in human blood through a membrane feeder 4 days after dsRNA treatment. Unfed mosquitoes were removed within 24 h and the rest were maintained at 21°C for 13 days or 27°C for 7 days for P. berghei and P. falciparum infections, respectively. Mosquito midguts were dissected and stained with mercurochrome, and oocyst numbers were recorded using a light microscope (Olympus). Each assay was done with at least 20 mosquitoes, and the data represent three independent experiments.
2.7 Real-time quantitative PCR
Samples were treated with Turbo DNase (Ambion) and reverse-transcribed using Superscript III (Invitrogen) with random hexamers. Real-time quantification was performed using the QuantiTect SYBR Green PCR Kit (Qiagen) and ABI Detection System ABI Prism 7000. Primer sequences were the same as used for generating dsRNA templates. All PCR reactions were performed in triplicate; to check for specificity of the PCR reactions, melting curves were analyzed for each data point. For the tissue- and stage-specific analyses, the relative levels of expression of IRID genes were estimated by normalizing cDNAs using ribosomal protein S7 gene.
2.8 Hemolymph perfusion and colony forming unit assays
Adult female mosquitoes were subject to gene silencing as described above and incubated under normal insectary conditions for 4 days. They were dipped in 70% EtOH for surface sterilization and washed with sterile PBS. Each mosquito underwent three rounds of sterilization. Sterile PBS (2 uL) was injected at the anterior end of each thorax using a microcapillary needle, and the PBS/hemolymph perfusion was collected from the posterior end of the thorax using a second microcapillary needle. The perfusion was serially diluted in PBS, then plated onto LB agar and incubated at 27° C overnight. Colony forming units (CFU) were scored for morphology, counted and streaked for isolation. Morphologically distinct colonies were subject to PCR amplification of the 16S ribosomal gene, which was used for sequence analysis. Analytic data for each gene are representative of three biologically independent replicas.
3. Results
3.1 The Anopheles IgSF
The A. gambiae genome contains 138 predicted IgSF genes. The smallest of these has only a single predicted Ig domain, while others have up to 35. Thirty-six predicted IgSF transcripts (26% of the total IgSF) begin with a methionine residue, and the remaining transcripts are incompletely annotated. Currently, 11.5% of the predicted genes contain signal peptide regions (Table 1) and therefore represent putative secreted proteins that can participate in interactive processes. Twenty-nine percent (40 genes) of the IgSF members have putative transmembrane domains (Table 1), suggesting a localization at the cell surface. Forty-two percent (58 genes) have modular structures with additional functional domains, many of which have known roles in innate immunity. The next most abundant domain is fibronectin III (FnIII), which is encoded by 36 IgSF genes (26% of the superfamily) and is known for its ligand-binding capabilities [44, 45]. Additional binding domains include collagen, leucine-rich repeats, EGF and C-type lectin and 10 IgSF transcripts (7%) contain signaling domains such as kinase, phosphatase or SH3 domains (Table 1). Like AgDscam [6], several members exhibit repeating Ig domains or repeating units of an Ig domain plus other domains.
Table 1.
Domain representation within the A. gambiae IgSF. Domains encoded by genes of the IgSF are listed in the left column. The number of genes containing each domain and the corresponding percentage are given in the right column. Percentages refer to the number of proteins containing one or more of the indicated domain, divided by the total number of IgSF proteins. (A) Signal peptide and transmembrane domains. (B) Functional domains
| A | |
|---|---|
| Feature | # genes (%) |
| Transmembrane | 34 (24.6) |
| Signal Peptide | 10 (7.2) |
| Both | 6 (4.3) |
| Neither | 88 (63.8) |
| B | |
|---|---|
| Domain | # genes (%) |
| Immunoglobulin Only | 80 (57.9) |
| FibronectinIII | 36 (26.1) |
| Leucine-rich repeat | 6 (4.3) |
| Kinase | 5 (3.6) |
| Epidermal growth factor | 4 (2.9) |
| Thrombospondin | 4 (2.9) |
| Phosphatase | 3 (2.2) |
| Low-density lipoprotein | 2 (1.4) |
| Laminin | 2 (1.4) |
| vonWillebrand | 2 (1.4) |
| Kunitz | 1 (0.8) |
| Src homology 3 | 1 (0.8) |
| Pleckstrin homology | 1 (0.8) |
| Histone deacetylase | 1 (0.8) |
| C-type lectin | 1 (0.8) |
| Peroxidase | 1 (0.8) |
| Domain of unknown function | 1 (0.8) |
| Extensin | 1 (0.8) |
| Collagen | 1 (0.8) |
| Genes with single non-Ig | 45 (32.6) |
| Genes with 2-4 non-Ig | 10 (7.2) |
| Genes with 5+ non-Ig | 3 (2.2) |
The IgSF includes a small set of Anopheles-specific transcripts encoding peptides with single Ig domains. These genes have no described function and may represent a unique class of genes or be artifacts of incomplete sequencing or annotation. It is noteworthy that none of these single Ig-domains peptides share significant sequence similarity with any other Anopheles protein, nor do they have orthologs in D. melanogaster or A. aegypti.
Because D. melanogaster has been widely used as a genetically tractable model, we used the Drosophila orthologs of our genes to consider possible roles of novel A. gambiae IgSF members. Also, the availability of the published A. aegypti genome sequence allowed us to include a second mosquito species in our comparative analysis. We found that 58% of Anopheles IgSF genes have orthologs in both of the other species, tending to share more sequence similarity with Aedes than with Drosophila (Fig. 1). Although 10 genes show homology between A. gambiae and D. melanogaster but not A. aegypti, 28 genes are orthologs only between the two mosquito species. Twenty-three IgSF genes are uniquely anopheline. Of these, two share similarity only with Apis mellifera, 18 encode short, Ig-only peptides/proteins consisting only of 1-4 Ig domains and 3 have a more complex composition. Twelve of the short Ig-only gene sequences are incomplete, so we cannot exclude the possibility that truncations did not allow for an ortholog match; however, eight are likely to be complete. Of the 87 Drosophila IgSF orthologs, most have experimentally described functions [22]. All three species have a similar number of unique genes (23 for A. gambiae, 20 for D. melanogaster and 24 for A. aegypti).
Figure 1.
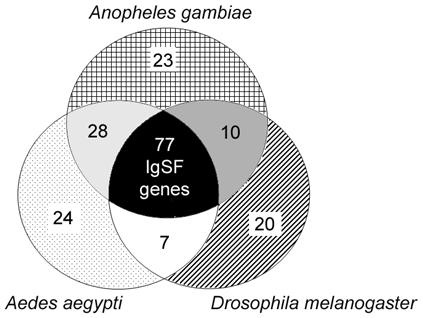
IgSF orthologs among A. gambiae, A. aegypti, and D. melanogaster. Numbers of genes displaying overlapping homology are partitioned as displayed in the different sections of the Venn diagram. Patterned regions indicate the number of IgSF genes unique to the organism indicated. Gray regions indicate number of genes with homologs in only two organisms, and the black center region indicates the number of genes common to A. gambiae, A. aegypti and D. melanogaster.
3.2 Infection-responsive gene expression
A high degree of correlation between transcript abundance and gene function allows predictions of a gene’s immune relevance by assessing gene regulation after microbial challenge. DNA microarray analysis was used to determine gene expression in response to bacterial and parasitic elicitors and thereby link IgSF members with putative immune functions. The array design incorporated probes for individual exons within the genes of the IgSF to extend the expression analysis beyond genetic regulation. By assessing the expression of each exon, immune-responsive regulation within each gene could be analyzed. This is of particular relevance for the IgSF, since at least one member (Dscam) is known to form alternative transcripts upon immune challenge [6]. Since the analysis is focused on genes with a common domain, the potential for cross-hybridization was minimized by exclusion of oligo probes with multiple predicted targets and validation of several exon-specific results via quantitative real-time PCR (data not shown).
IgSF exon transcript abundance in whole mosquitoes was determined after challenge with each of four bacterial species and in mosquito midguts and carcasses during challenge with invading ookinetes of either Plasmodium berghei or Plasmodium falciparum (Table 2) (40). Overall, 256 exons of 85 different Ig genes were differentially regulated between control and challenged mosquitoes (Supplementary Table 1 A and B). Bacterial infections elicited more prominent responses than did Plasmodium infection. E. coli caused the greatest enrichment of IgSF exons, while S. aureus caused a striking down-regulation. The other bacterial challenges (B. subtilis and S. typhimurium) elicited a greater induction than repression of IgSF genes (Table 2A and Supplementary Table 1A).
Table 2.
Summary of IgSF gene regulation during immune challenge. Numbers indicate the IgSF exons and genes significantly regulated during the indicated challenge. Upward and downward arrows indicate that, in the challenged mosquitoes, transcript levels of those exons/genes are increased or decreased, respectively. The number of alternatively regulated genes indicates the number of genes that, for the indicate challenge, showed at least one exon significantly up-regulated and one exon significantly down-regulated. (A.) Expression in response to bacterial challenge. (B.) Expression in response to Plasmodium challenge. CTRP, circumsporozoite- and TRAP-related protein, indicates parasites deficient in this protein which is essential for midgut entry by the ookinete
| A | ||||
|---|---|---|---|---|
| Challenge | E. coli | S typhimurium | S. aureus | B. subtilis |
| # regulated exons | 64↑ 34↓ | 44↑ 17↓ | 8↑ 136↓ | 48↑ 29↓ |
| # regulated genes | 45↑ 27↓ | 28↑ 9↓ | 8↑ 54↓ | 32↑ 22↓ |
| # alternatively regulated genes | 14 | 6 | 3 | 11 |
| B | ||||||
|---|---|---|---|---|---|---|
| Challenge | P.f. gut | P.f. carcass | P.b. gut | P.b. carcass | P.f. CTRP gut | P.f. CTRP carcass |
| # regulated exons | 6↑ 2↓ | 0↑ 3↓ | 6↑ 15↓ | 2↑ 0↓ | 7↑ 5↓ | 6↑ 12↓ |
| # regulated genes | 4↑ 2↓ | 0↑ 3↓ | 5↑ 10↓ | 2↑ 0↓ | 6↑ 5↓ | 5↑ 10↓ |
| # alternatively regulated genes | 0 | 0 | 0 | 0 | 1 | 1 |
The IgSF response to Plasmodium has been categorized first according to tissue and then according to parasite stage, facilitated by the occurrence of circumsporozoite- and TRAP-related (CTRP) knockout mutant strains of both P. falciparum and P. berghei [46]. CTRP-parasites are unable to invade the midgut epithelium, allowing distinction between gene expression triggered by malaria-infected blood and gene expression triggered by actively invading parasites. In the midgut, P. berghei ookinete invasion triggered down-regulation of 15 exons from 10 Ig-containing genes, while P. falciparum invasion caused repression of only 2 exons, each from a different gene (Table 2B). In the carcasses of those mosquitoes, very few IgSF genes were differentially regulated in response to invading ookinetes of either parasite species. Malaria-infected blood induced 7 exons of 6 IgSF genes in the gut and 6 exons of 5 genes in the carcass. The same treatment caused repression of 5 genes in the gut and 10 genes in the carcass (Table 2B).
In general, some genes responded to only one elicitor, while others responded to multiple microbes and no exon responded similarly to all challenges. Individual exons can be differentially regulated within the same gene, such that, for a specific challenge, one (or more) displays increased transcription, while at least one other display decreased transcription. Such alternatively regulated genes were few in number when compared to the total number of regulated genes (Tables 2A and 2B, third rows) and may represent alternative splicing events.
3.3 Selecting candidates for immune relevance
Sequence and expression data were used to select immune-related candidates for a more focused reverse-genetic functional analysis. Only genes significantly regulated in response to at least two pathogens were considered. Also, selection was biased toward genes predicted to have multiple Ig domains and/or additional domains of known implication in immunity. Accordingly, six candidates were chosen for further analysis (Fig. 2) and are herein referred to as infection responsive with immunoglobulin domain (IRID) 1-6.
Figure 2.
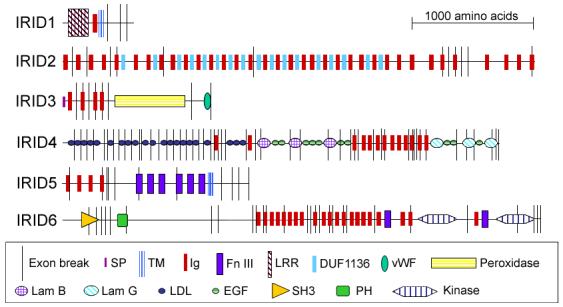
Domain architectures of six IRID proteins. For each protein, the black horizontal line represents the sequence length, black vertical lines represent the predicted exon boundaries, red bars represent Ig domains and other symbols represent predicted domains as defined in the key. In all cases, the N- terminus is at the left. IRID1, ENSANGG00000004060; IRID2, ENSANGG00000007024; IRID3, ENSANGG00000007168; IRID4, ENSANGG00000017722; IRID5, ENSANGG00000017864; and IRID6, ENSANGG00000019232. Domain abbreviations: SP, signal peptide; TM, transmembrane region; Ig, immunoglobulin; FnIII, fibronectin III; LRR, leucine-rich repeat; DUF1136, domain of unknown function 1136 (Interpro ID); vWF, vonWillebrand factor; Lam, laminin; LDL, low density lipoprotein; EGF, epidermal growth factor; SH3, src homology 3; PH, pleckstrin homology.
The shortest candidate, IRID1 (Fig. 2), is the ortholog of Drosophila kekkon1 [47, 48]. Midgut invasion by P. falciparum significantly induced expression of this kekkon1-like gene (Supplementary Table 1B).
The longest candidate, IRID2 (Fig. 2), is an ortholog of the D-Titin allele sallimus [49]. Ig domains are arranged in tandem repeating units with DUF1136 (IPR010939), a domain of unknown function often found adjacent to Ig domains, noted by Interpro annotation. Exons from this gene are up-regulated in response to P. falciparum but down-regulated in response to P. berghei and infected blood (Supplementary Table 1B).
IRID3 (Fig. 2) is an ortholog of peroxidasin, with the peroxidase domains sharing the most similarity as well as sharing similarity to mammalian neutrophil and eosinophil peroxidases. Of the 18 A. gambiae proteins containing peroxidase domains, IRID3 is the only one that also incorporates repeating putative binding domains. The Drosophila peroxidasin is about 222 amino acids longer than the current Anopheles prediction and contains leucine-rich repeats (LRRs) upstream of the Ig domains. Sequence analysis of the genomic regions upstream of IRID3 revealed that LRR domains that could be encoded by IRID3 remain unidentified because of a potential annotation error. Drosophila peroxidasin is made by hemocytes and is incorporated into basement membranes during early development but may have implications for phagocytosis later in life [50]. In other invertebrates, proteins such as peroxinectin with both adhesive and peroxidase domains have been shown to be produced by hemocytes [11] and are associated with activation of the prophenyloxidase system [12, 51], melanotic encapsulation [52], and possibly phagocytosis. These characteristics make a secreted adhesive peroxidase a good opsonic candidate. IRID3 is highly induced during both E. coli and B. subtilis challenge (Supplementary Table 1A) and is not one of the peroxidases described as having anti-malaria activity [53].
Another candidate, IRID4 (Fig. 2), encodes an ortholog of the fruit fly trol (perlecan) [54]. IRID4 has the most domain variety of all Anopheles IgSF members and is predicted by Ensembl to have alternative transcripts. This trol-like gene has 48 predicted exons, the largest number in any protein in the superfamily, and has six of the most infection-responsive exons. Some exons display similar regulation, whereas others are regulated differently, with some exons elevated and others repressed during infection with a single pathogen species (Supplementary Table 1 A and B).
IRID5 (Fig. 2) is the ortholog of frazzled [55, 56] and is induced by several bacteria but is strongly repressed by P. falciparum-infected blood (Supplementary Table 1 A and B).
IRID6 (Fig. 2), encodes a complex protein which, though fly homolog has unknown function, shares some identity with C. elegans UNC-89 [57]. IRID6 is down-regulated to varying degrees during the three bacterial infections and is strongly down-regulated in the gut during P. berghei ookinete invasion (Supplementary Table 1 A and B).
3.3 Tissue-specific and stage-specific expression of IRID genes
The developmental and tissue specificity of gene expression can reveal important features of a gene’s function. Expression of selected IgSF members was assayed throughout development and in the adult female head, thorax, abdomen and midgut tissues. Developmental IRID expression was assayed in total RNA extracts from eggs, larvae, pupae, female adults and male adults and normalized against the expression of the ribosomal S7 protein gene in each sample to give a relative level of abundance (Fig. 3A). Expression of IRID1 displayed a steady decrease during the progression from egg to adult. IRID2 showed similar levels of expression at all stages except the pupa, when it exhibited a sharp increase. IRID3 expression was elevated in the egg, then maintained a lower level in larva and pupa and decreased further in adulthood, with the depression more pronounced in males. IRID4 and IRID6 showed a gradual increase in expression from the egg through the pupal stage, then dropped off in adults of both sexes. IRID5 showed decreasing expression through development and remained low in adult females, but increased in adult males. IRID3 and IRID4 also showed differential expression in males and females.
Figure 3.
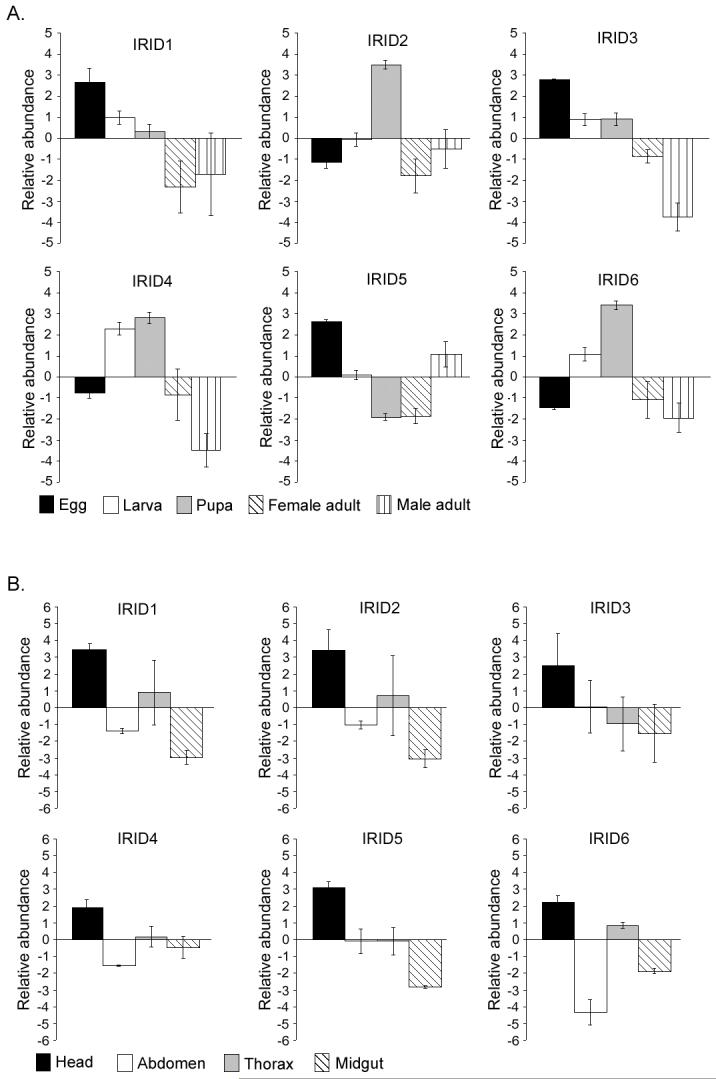
Stage- and tissue-specific IRID expression under naïve conditions. (A) Bars represent the average abundance of the indicated IRID transcript in eggs (black), larvae (open), pupae (gray), adult males (diagonally striped) and adult females (vertically striped) as compared to a pool of equal amounts of RNA from each stage.
(B) Bars represent the average abundance of the indicated IRID transcript in females heads (black), abdomens (open), thoraces (gray) and midguts (diagonally striped), as compared to expression in the whole adult female.
Tissue specificity was more uniform among the IRID genes, with all of them showing a preferential expression in the head tissue and being reduced to varying degrees in the abdomen and midgut (Fig. 3B). Expression in the thorax varied among the genes. A high level of expression in the head was observed consistently in repeated independent assays.
3.4 Silencing of four IgSF genes decreases survival after challenge
A standard RNA interference (RNAi) gene-silencing approach was used to determine whether these six candidate genes are essential for defending the mosquito against bacteria. Each gene was knocked down, and mosquitoes were infected with either E. coli or S. aureus and then monitored for survival; control mosquitoes were treated with dsRNA against GFP to activate RNAi mechanisms without interfering with mosquito-derived transcripts. The average silencing efficiency ranged from 75 to 99% for all six genes (Supplementary Fig. 1A). To ensure that any fluctuations in survival were related to infection and not to a change in overall health, survival of mosquitoes after RNAi treatment was monitored in a non-infectious context, and survival was found to be comparable in all groups (Supplementary Fig. 1B).
Treatment with dsRNA against IRID3, IRID5 and IRID6 caused a significant reduction in viability during bacterial infection (Fig. 4A). Also, mosquitoes lacking IRID1 show a significant (p<.01) decrease in survival at several time points during E. coli infection, but the phenotype was not observed throughout the survival time course. IRID3-silenced mosquitoes experienced a drastic decrease in survival during the first 2 days after infection with either E. coli or S. aureus (p<.01 for both infections at all time points). Less than 10% of the infected IRID3-silenced mosquitoes survived to 7 days after infection with S. aureus, less than 20% with E. coli. In contrast, IRID5- and IRID6-KD mosquitoes experience a more gradual, consistent decrease in survival through day 7. The dsIRID5-treated mosquitoes survived S. aureus infection somewhat better than did those treated with the positive control, the anti-microbial peptide dsGambicin (data not shown). In contrast, the dsIRID3-treated mosquitoes were somewhat (but not significantly, p=.118) less able to survive S. aureus infection than were dsGambicin-treated mosquitoes (data not shown).
Figure 4.
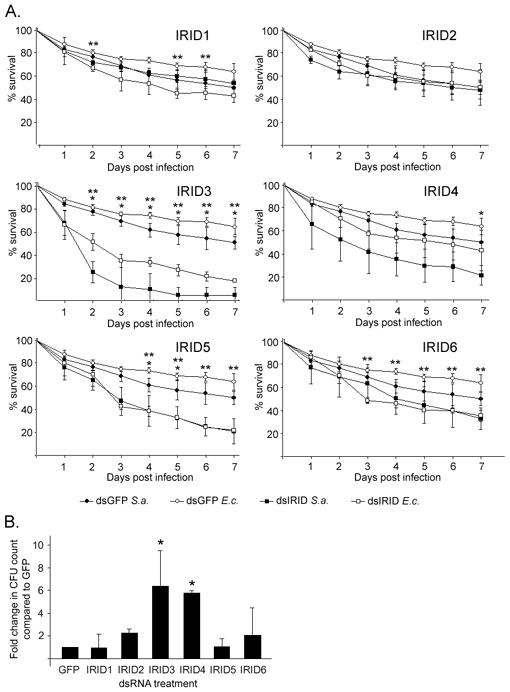
Lack of IRIDs causes increased sensitivity to bacteria. (A) IRID-silenced mosquitoes show reduced viability after immune challenge. Mosquitoes were treated with the indicated dsIRID (squares) or dsGFP (circles) and subsequently challenged with either S. aureus (S.a., closed symbols) or E. coli (E.c., open symbols). Points represent the average percent survival of three experiments consisting of 80 female mosquitoes each; error bars represent standard error. *, p< .01 for S. aureus infection; **, p< .01 for E. coli infection. For both infections, the p-values generated for dsIRID3 treatment 2 days post-infecton through 7 days post-infection were < .001. (B) IRID-silenced mosquitoes experience altered immune homeostasis. Bars represent the -fold increase in hemolymph colony forming units as calculated against a GFP value normalized to 1. The error bars indicate standard deviation of three experiments, assaying six mosquitoes per gene. *, p < .05.
3.5 Silencing of IRID3 and IRID4 causes a deficiency in immune homeostasis
Mosquitoes are continuously exposed to bacterial pathogens, and the immune system is therefore required to maintain immune homeostasis throughout the insect’s lifetime. Gene silencing assays using established methodology [6] were designed to assess the involvement of IRID in this immune homeostasis. In sum, the number of colony-forming units isolated from hemolymph of IRID3- or IRID4-silenced mosquitoes was, respectively, 6.3 and 5.6 times greater than the number of CFUs isolated from control mosquitoes (Fig. 3B), supporting a role for IRID3 in immune homeostasis and general bacterial clearance. Other IRIDs showed a more modest effect, being increased by about 2.5-fold for bacteria. Sequence analysis of isolated colonies showed that IRID-deficient mosquitoes allowed the proliferation of Novosphingobium, Escherichia, Pseudomonas and other species.
3.6 IRIDs are implicated in anti-Plasmodium defense
In order to assess the involvement of IRIDs in anti-Plasmodium defense, mosquitoes were fed on P. falciparum- or P. berghei-infected blood after IRID gene silencing. An increase in the number of oocysts harbored in the midgut of IRID gene-silenced mosquitoes would support a role for the IRID protein in limiting the malaria parasite in the mosquito. The P. falciparum oocyst load increased in mosquitoes treated with RNAi against IRID4, producing infection levels almost twice those observed in control mosquitoes (p<.01). Oocyst counts following ingestion of both P. falciparum- (Fig 4, left panel) and P. berghei- (right panel) infected blood showed that a lack of the IRID6 transcript caused the infection intensity to increase by more than 2-fold over non-silenced levels (p<.001 for P. falciparum, p<.05 for P. berghei). The oocyst levels observed in mosquitoes depleted of IRID6 transcripts were comparable to those observed in mosquitoes depleted of the highly potent anti-Plasmodium factor TEP1 [40, 58]. Silencing of the other four IRID genes had no significant effect on oocyst levels, and therefore these genes do not appear to play a role in the defense against ookinete-stage Plasmodium. Data for each experiment is outlined in the Supplementary Table 2 A and B.
4. Discussion
Because the immunoglobulin domain is capable of facilitating molecular interactions and is an essential component of mammalian immunity, proteins containing these domains are effective candidates for immune effector molecules in invertebrates. We present here a comprehensive sequence and gene expression analysis of all the IgSF members of A. gambiae. The sequence data indicate that multiple members of this family have other immune-relevant domains and the potential for alternative splicing. Some of these domains, such as C-type lectin and FnIII, are adhesive, supporting a potential role in pathogen interaction. The sequences of other immune-relevant domains, such as kinase and SH3, suggest that IgSF members are involved in transducing signals, an essential component of innate immune reactions. The modular structures of some members suggest that they may undergo alternative splicing, which can contribute to diversity and specificity of interaction; however, without experimental validation of each gene, a definitive quantification of the IgSF members capable of alternative splicing cannot yet be made.
As expected, a comparative analysis of the IgSFs of A. gambiae, D. melanogaster and A. aegypti revealed that the two mosquito species have a greater number of shared orthologs than either mosquito shares with the fruit fly. We also observed that each species has ∼20 unique IgSF genes. Because the nature of the proteins encoding many of these genes is unknown, it is possible that the molecules lack significant sequence homology yet perform analogous functions.
Transcript abundance data have shown that infection with a variety of microbes elicits transcriptional responses from many Ig genes. While the responses to bacteria were robust, responses to infected blood and invading ookinetes were restricted to just a few genes; however, this reactivity was proportional to the total number of genes responding to either of these conditions [38]. This quantitative disparity could reflect differences in the intensity of infection (millions of bacteria, as compared to a few or even a single Plasmodium) or location of infection (hemocoel, as compared to midgut) or could reflect tolerance inflicted by the close evolutionary relationship between Anopheles and Plasmodium. The fact that some genes did show regulation after malarial infection indicates that IgSF genes are regulated by the mosquito during infection with either P. falciparum or P. berghei and that they are responding at one or both stages of malarial infection: the detection of infected blood by the mosquito and the ookinete invasion of the midgut epithelium.
We also observed that no Ig-containing molecules were ubiquitously regulated in response to the entry of a foreign microbe. Thus, the regulation of individual IgSF transcript abundance exhibited specificity, the degree of which differed among IgSF members. The breadth of this response to microbes within one superfamily is not surprising, since the Ig domain is capable of accommodate many binding partners, and Ig domains in mammals bind to all types of pathogens. It is not known which foreign moieties are being bound or otherwise detected by IgSF members, nor do we know how specific this detection is.
Probing for specific exons showed that exon usage was not uniform across a transcript from the same gene. In some cases, we saw a certain pattern of exon expression in response to one elicitor and a completely different pattern in response to another elicitor. In other cases we observed enrichment, depletion and non-use of exons encoded by the same gene in response to the same elicitor. In particular, one gene, ENSANGG00000017722, had many regulated exons, with at least one induced exon and at least one repressed exon in the expression profile generated by each of the four bacterial elicitors. This pattern of response is a likely indication of alternative splicing, making it possible for an infection to elicit increased usage of particular alternative transcripts. While this exon expression patterning was most striking in ENSANGG00000017722, it was also exhibited by ENSANGG00000015823, ENSANGG0000000-7823, ENSANGG00000018890, ENSANGG-00000017503 and others (Supplementary Tables 1 and 2). These data suggest that although a single gene may be important for immunity, different exons specifically respond to different classes of pathogen. This mechanism is at present only speculative, since alternative splicing has only definitively been shown for the IgSF member Dscam [6, 29-30], and it is unknown whether any such splicing events are dependent on challenge.
This gene expression analysis has provided a solid basis for the selection of potential immune molecules to be functionally analyzed in greater detail. In the present study, we have conducted a focused, detailed analysis of six of these potential immune molecules, several of which proved to be critical players in immune mechanisms.
An analysis of IRID expression throughout development (Fig. 3A) showed that the expression of IRID2 was specifically increased in the pupal stage, potentially reflecting its involvement in metamorphosis. IRID3 could be involved in embryonic developments, since it displayed a high level of expression in the egg, which waned as development continued. Other IRIDs showed varying patterns of stage-specific expression. It may be that certain developmental stages have a greater need for protective immunity, and these molecules respond accordingly. It is also likely that, as is true for other known immune molecules, the IRID genes are important for development as well as immunity, and they are thus expressed discontinuously until adulthood.
All six IRIDs were abundantly expressed in the head (Fig. 3B), an observation that may reflect the prevalence of Ig proteins in neural functions. The similarities between the nervous and immune systems allow for a dual role, which has already been reported for the Ig proteins Dscam [29, 30] and neuroglian [59]. The expression profiles of other, non-IgSF immune genes assayed using the same mRNA samples did not show a ubiquitous elevation in the head (data not shown).
Knock-down of three genes, IRID3, IRID5 and IRID6, affected the way A. gambiae female mosquitoes were able to combat Gram+ (S. aureus) and Gram- (E. coli) bacteria. dsIRID3-treated mosquitoes exhibited severe die-off within a few days, while a lack of the other IRIDs caused a gradual yet prolonged increase in the death rate. The contrasting survival curves may indicate that these molecules are involved in different immune mechanisms or that IRID3 has a required antimicrobial role but that there is redundancy in the role of IRID5. The IRID3 protein contains not only Ig domains but also a peroxidase domain, which shows homology to mammalian neutrophil and eosinophil peroxidase. It may be that this Anopheles peroxidase has a function similar to that of mammalian peroxidases and is somehow involved in generating toxic oxygen species that quickly and non-specifically target bacterial cells. This is similar to the model put forth for peroxinectin [11, 12]. The more general composition of IRID5 suggests an ability to bind at the cell surface, but offers few extra insights. dsIRID6 was the only treatment that induced a sustained, significant decrease in viability during infection with one bacterial species and not the other. It is possible that IRID6 may have specific activity against E. coli or Gram-negative bacteria.
Mosquitoes lacking IRID3 and IRID4 are unable to clear bacteria from the hemolymph. It is probable that some hemolymph-dwelling bacteria were undetected by the media and growth conditions we used; however, this assay serves as a proof of principle that silencing of IgSF members does influence the proliferation of naturally acquired microorganisms. Additionally, the temperature and humidity at which colonies were incubated are identical to mosquito rearing conditions, making the bacteria that we detected particularly relevant to mosquito immune homeostasis.
While significant increases in bacteria were observed in the hemolymph 4 days after dsRNA treatment, survival was still uncompromised. This continued survival could have been observed because the species of bacteria naturally acquired under laboratory conditions are innocuous, whereas injected bacteria are virulent. The species that best matched the ribosomal sequence from the hemolymph-isolated bacteria were all Gram-negative. This result could have occurred if IRID3 is specifically active against these types of bacteria or if it is indiscriminate in its anti-bacterial activity, but the environmental and physiological conditions favor Gram-negative species. In any case, the inability of the IRID3- and IRID4-silenced mosquitoes to clear ambient pathogens from the hemolymph supports a role for these genes in bacterial clearance.
Our data also demonstrate that IRID6 and IRID4 play a role in limiting Plasmodium infection. The observation that the contribution of IRID6 was relatively equal during infections with both Plasmodium species is interesting, since many parasite antagonists show species specificity [60, 61]. At present, we are unable to determine whether the increase in infection in the IRID4- and IRID6-KD mosquitoes was due to an active immune response against the parasite or to secondary mechanisms that indirectly affected the maturation or invasion events of the parasite. The IRID4 molecule is quite complex, having many domains of multiple types and a composition and gene expression profile that would support alternative splicing. The presence of EGF and laminin domains, together with the Ig domains, would suggest a role in adhesion and attachment. The IRID6 molecule also displays appreciable complexity. In addition to 20 Ig domains, it has an SH3 domain and a Pleckstrin domain toward the N-terminus and two protein kinase domains at the C-terminus; these features point to a potential role for this gene in signal transduction. Participation in an immune signaling pathway could explain why IRID6 affects viability during E. coli infection as well as the intensity of Plasmodium infection.
It should be noted that the infection phenotyping addressed in this study only considered IRID involvement up to the oocyst stage. It is possible that IRID molecules are acting against other stages of malarial parasites in the mosquito. We are currently assessing this possibility and have raised antibodies against some of the IRIDs to allow us to begin studying the mechanisms by which IgSF proteins contribute to immunity.
Understanding the IgSF as a whole will be advantageous not only for translating knowledge from other invertebrate studies to Anopheles but also for identifying and characterizing genes unique to disease-transmitting invertebrates. With the present analysis, we broaden the scope of innate immune recognition to include additional Ig domain-containing proteins, and we provide evidence for the importance of novel IgSF superfamily members in defending the mosquito against different classes of microbes. Considering that we found 85 IgSF genes to be infection-responsive and four out of the six chosen candidate genes to have an impact on mosquito viability during infection or infection intensity, there are probably many more IgSF members involved in the mosquito innate immune system.
In summary, our data demonstrate that IgSF members in A. gambiae are capable of reacting to pathogen challenge and of controlling events that contribute to the mosquito’s defense against infection.
Supplementary Material
Figure S1: (A) Silencing efficiencies of IRID and GFP dsRNA treatment. Bars indicate % KD (percent knock-down) of the indicated gene, as determined by qRT-PCR. (B) Survival of IRID-silenced naïve mosquitoes is uncompromised. Post-silencing viability was determined at day 4 post-silencing in all cohorts of mosquitoes receiving dsRNA treatment. Bars represent the average percent survival of mosquitoes receiving each treatment.
Table S1: IgSF infection-responsive gene expression. The degree of differential gene expression in challenged adult female mosquitoes is given as the log2-transformed -fold change in transcript level, as compared to PBS-injected control mosquitoes. Gene IDs given are derivatives of ENSANGG000000xxxxxx IDs assigned by Ensembl. For visual simplification, we have deleted “ENSANG” and “000000” from the IDs.
(A) Transcriptional responses of IgSF exons to bacterial (E. coli [E.c.], S. typhimurium [S.t.], S. aureus [S.a.] and B. subtilis [B.s.]) challenge.
(B) Transcriptional responses of IgSF exons to malarial challenge (invading ookinetes of P. berghei [Pb IO]) and P. falciparum [Pf IO]) and infected blood containing invasion-incapable P. falciparum ookinetes (Pf IB).
Table S2: Oocyst counts for P. falciparum and P. berghei infections in IRID-silenced mosquitoes. Rows are labeled according to dsRNA treatment; columns are labeled according to the type of data and replica number. Included are data on the incidence of infection, mean oocyst load, range of infection intensities and -fold change in infection intensity (when compared to GFP-treated controls) for each of three independent assays, as well as the average -fold change and standard deviations between the assays. (A) P. falciparum infection (B) P. berghei infection.
Figure 5.
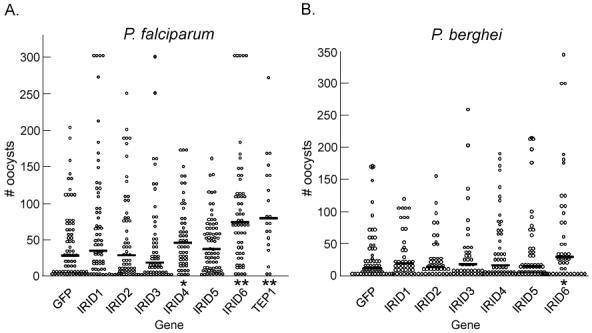
Plasmodium oocyst intensity increases in IRID-silenced mosquitoes. Points indicate the absolute oocyst counts in individual mosquitoes treated with the dsRNA indicated on the x-axis. Horizontal black bars in each column represent the average number of oocysts observed in each treatment group from three independent assays. P-values were calculated using a Mann-Whitney analysis. Incidence of infection, average infection intensity from each replica and standard deviations are given in Supplemental Tables S2 A and B.
(A) Oocyst counts of P. falciparum-infected midguts (n = 45-60) *, p <.05; **, p <.01.
(B) Oocyst counts of P. berghei-infected midguts (n = 45-60). An asterisk denotes p < .05.
Acknowledgments
We are grateful to Dr. Yuemei Dong for sharing gene expression data. We thank the microarray core facility, the parasitology core facility and the insectary personnel at the Johns Hopkins Malaria Research Institute for assistance with microarray, P. falciparum culture and mosquito rearing. We also thank Dr. Deborah McClellan at the Editing Referral Service, William H. Welch Medical Library, Johns Hopkins University School of Medicine. This material is based upon work supported under a National Science Foundation Graduate Research Fellowship (LG). This work is also supported by the National Institutes of Health/National Institute for Allergy and Infectious Disease 1R01AI061576-01A1, United Nations Development Program/World Bank/World Health Organization Special Program for Research and Training in Tropical Diseases, The Ellison Medical Foundation, Johns Hopkins School of Public Health, and Johns Hopkins Malaria Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Amit AG, Mariuzza RA, Phillips SEV, Poljak RJ. Three-dimensional structure of an antibody-antigen complex at 2.8 A resolution. Science. 1986;233:747–753. doi: 10.1126/science.2426778. [DOI] [PubMed] [Google Scholar]
- [2].Williams AF. A year in the life of the Immunglobulin Superfamily. Immunol Today. 1987;8:298–303. doi: 10.1016/0167-5699(87)90016-8. [DOI] [PubMed] [Google Scholar]
- [3].Willams AF, Barclay AN. The Immunoglobulin Superfamily—domains for cell surface recognition. Ann Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- [4].Hynes RO, Zhao Q. The evolution of cell adhesion. J Cell Biol. 2000;150(2):F89–96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- [5].Hutter H, Vogel BE, Plenefisch JD, Norris CR, Proenca RB, Spieth J, Guo C, Mastwal S, Zhu X, Scheel J, Hedgecock EM. Conservation and novelty in the evolution of cell adhesion and extracellular matrix genes. Science. 2000;287:989–994. doi: 10.1126/science.287.5455.989. [DOI] [PubMed] [Google Scholar]
- [6].Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the Anopheles gambiae innate immune system. PLoS Biol. 2006;4(7):e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94(16):8691–6. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang SM, Leonard PM, Adema CM, Loker ES. Parasite-responsive IgSF members in the snail Biomphalaria galabrata: characterization of novel genes with tandemly arranged IgSF domains and a fibrinogen domain. Immunogenetics. 2001;53(8):684–94. doi: 10.1007/s00251-001-0386-8. [DOI] [PubMed] [Google Scholar]
- [9].Wang X, Xiao Q, Christensen BM. Identification and characterization of the fibrinogen-like domain of fibrinogen-related proteins in the mosquito, Anopheles gambiae, and the fruitfly, Drosophila melanogaster, genomes. BMC Genomics. 2005;6:114. doi: 10.1186/1471-2164-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hoek RM, Smit AB, Frings H, Vink AM, de Jong-Brink M, Geraerts WP. A new Ig-superfamily member, molluscan defence molecule (MDM) from Lymnaea stagnalis, is down-regulated during parasitosis. Eur J Immunol. 1996;26(4):939–44. doi: 10.1002/eji.1830260433. [DOI] [PubMed] [Google Scholar]
- [11].Johansson MW, Lind MI, Holmblad T, Thornqvist PO, Soderhall K. Peroxinectin, a novel cell adhesion protein from crayfish blood. Biochem Biophys Res Commun. 1995;216(3):1079–87. doi: 10.1006/bbrc.1995.2731. [DOI] [PubMed] [Google Scholar]
- [12].Sritunyalucksana K, Wongsuebsantati K, Johansson MW, Soderhall K. Peroxinectin, a cell adhesive protein associated with the proPO system from the black tiger shrimp, Penaeus monodon. Dev Comp Immunol. 2001;25(56):353–63. doi: 10.1016/s0145-305x(01)00009-x. [DOI] [PubMed] [Google Scholar]
- [13].Sun SC, Lindstrom SI, Boman HG, Faye I, Schmidt O. Hemolin: an insect-immune protein belonging to the immunoglobulin superfamily. Science. 1990;250(4988):1729–32. doi: 10.1126/science.2270488. [DOI] [PubMed] [Google Scholar]
- [14].Lanz-Mendoza H, Bettencourt R, Fabbri M, Faye I. Regulation of the insect immune response: the effect of hemolin on cellular immune mechanisms. Cell Immunol. 1996;169(1):47–54. doi: 10.1006/cimm.1996.0089. [DOI] [PubMed] [Google Scholar]
- [15].Terenius O, Bettencourt R, Lee SY, Li W, Soderhall K, Faye I. RNA interference of Hemolin causes depletion of phenoloxidase activity in Hyalophora cecropia. Dev Comp Immunol. 2006;31(6):571–5. doi: 10.1016/j.dci.2006.09.006. [DOI] [PubMed] [Google Scholar]
- [16].Ladendoff NE, Kanost MR. Bacteria-induced protein P4 (hemolin) from Manduca sexta: a member of the immunoglobulin superfamily which can inhibit hemocyte aggregation. Arch Insect Biochem Physiol. 1991;18(4):285–300. doi: 10.1002/arch.940180410. [DOI] [PubMed] [Google Scholar]
- [17].Daffre S, Faye I. Lipopolysaccharide interaction with hemolin, an insect member of the Ig-superfamily. FEBS Lett. 1997;408(2):127–30. doi: 10.1016/s0014-5793(97)00397-9. [DOI] [PubMed] [Google Scholar]
- [18].Yu XQ, Kanost MR. Binding of hemolin to bacterial lipopolysaccharide and lipoteichoic acid. An immunoglobulin superfamily member from insects as a pattern-recognition receptor. Eur J Biochem. 2002;269(7):1827–34. doi: 10.1046/j.1432-1033.2002.02830.x. [DOI] [PubMed] [Google Scholar]
- [19].Eleftherianos I, Millichap PJ, ffrench-Constant RH, Reynolds SE. RNAi suppression of recognition protein mediated immune responses in the tobacco hornworm Manduca sexta causes increased susceptibility to the insect pathogen Photorhabdus. Dev Comp Immunol. 2006;30(12):1099–107. doi: 10.1016/j.dci.2006.02.008. [DOI] [PubMed] [Google Scholar]
- [20].Eleftherianos I, Marokahazi J, Millichap PJ, Hodgkinson KJ, Sriboonlert A, ffrench-Constant RH, Reynolds SE. Prior infection of Manduca sexta with non-pathogenic Escherichia coli elicits immunity to pathogenic Photorhabdus luminescens: roles of immune-related proteins shown by RNA interference. Insect Biochem Mol Bio. 2006;36(6):517–25. doi: 10.1016/j.ibmb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- [21].Eleftherianos I, Gokcen F, Felfoldi G, Millichap PJ, Trenczek TE, ffrench-Constant RE, Reynolds SE. The immunoglobulin family protein Hemolin mediates cellular immune responses to bacteria in the insect Manduca sexta. Cell Microbiol. 2007;9(5):1137–1147. doi: 10.1111/j.1462-5822.2006.00855.x. [DOI] [PubMed] [Google Scholar]
- [22].Vogel C, Teichmann SA, Chothia C. The immunoglobulin superfamily in Drosophila melanogaster and Caenorhabditis elegans and evolution of complexity. Development. 2003;130(25):6317–6328. doi: 10.1242/dev.00848. [DOI] [PubMed] [Google Scholar]
- [23].Teichmann SA, Chothia C. Immunoglobulin superfamily proteins in Caenorhabditis elegans. J Mol Biol. 2000;296:1367–1383. doi: 10.1006/jmbi.1999.3497. [DOI] [PubMed] [Google Scholar]
- [24].Rougon G, Hobert O. New insights into the diversity and function of neuronal immunoglobulin superfamily molecules. Annu Rev Neurosci. 2003;26:207–238. doi: 10.1146/annurev.neuro.26.041002.131014. [DOI] [PubMed] [Google Scholar]
- [25].Lanz-Mendoza H, Faye I. Physiological aspects of the immunoglobulin superfamily in invertebrates. Dev Comp Immunol. 1999;23:359–374. doi: 10.1016/s0145-305x(99)00017-8. [DOI] [PubMed] [Google Scholar]
- [26].Yoshihara Y, Oka S, Ikeda J, Mori K. Immunoglobulin superfamily molecules in the nervous system. Neurosci Res. 1991;10(2):83–105. doi: 10.1016/0168-0102(91)90033-u. [DOI] [PubMed] [Google Scholar]
- [27].Hughes AL. Protein phylogenies provide evidence of a radical discontinuity between arthropod and vertebrate immune systems. Immunogenetics. 1998;47(4):283–96. doi: 10.1007/s002510050360. [DOI] [PubMed] [Google Scholar]
- [28].Parnes JR, Hunkapillar T. L3T4 and the immunoglobulin gene superfamily: new relationships between the immune system and the nervous system. Immunol Rev. 1987;100:109–27. doi: 10.1111/j.1600-065x.1987.tb00529.x. [DOI] [PubMed] [Google Scholar]
- [29].Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101(6):671–84. doi: 10.1016/s0092-8674(00)80878-8. [DOI] [PubMed] [Google Scholar]
- [30].Watson FL, Puttman-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel DI, Schmucker D. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309(5742):1874–1878. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- [31].Gibson NJ, Tolbert LP. Activation of epidermal growth factor receptor mediates receptor axon sorting and extension in the developing olfactory system of the moth Manduca sexta. J Comp Neurol. 2006;495(5):554–72. doi: 10.1002/cne.20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nardi JB, Zhuang S, Pilas B, Bee CM, Kanost MR. Clustering of adhesion receptors following exposure of insect blood cells to foreign surfaces. J Insect Physiol. 2005;51(5):555–64. doi: 10.1016/j.jinsphys.2005.02.005. [DOI] [PubMed] [Google Scholar]
- [33].Imler JL, Hoffmann JA. Toll receptors in Drosophila: a family of molecules regulating development and immunity. Curr Top Microbiol Immunol. 2002;270:63–79. doi: 10.1007/978-3-642-59430-4_4. [DOI] [PubMed] [Google Scholar]
- [34].Cantera R, Roos E, Engstrom Y. Dif and cactus are colocalized in the larval nervous system of Drosophila melanogaster. J Neurobiol. 1999;38(1):16–26. [PubMed] [Google Scholar]
- [35].Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32(10):1295–309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- [36].Mellon EB, Brockmann A, Schmid-Hempel P. Immune response inhibits associative learning in insects. Proc Biol Sci. 2003;270(1532):2471–3. doi: 10.1098/rspb.2003.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28(1):231–4. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32(Database issue):D142–4. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Benedict MQ. Care and maintenance of anopheline mosquitoes. In: Crampton JM, Beard CB, Louis C, editors. The molecular biology of disease vectors: A methods manual. Champman and Hall; London: 1997. pp. 3–12. [Google Scholar]
- [40].Dong Y, Aguilar A, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune response to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2(6):e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yang IV, Chen E, Hasseman JP, Liang W, Frank BC, Wang S, Sharov V, Saeed AI, White J, Li J, Lee NH, Yeatman TJ, Quackenbush J. Within the fold: assessing differential expression measures and reproducibility in microarrays. Genome Biol. 2002;3(11) doi: 10.1186/gb-2002-3-11-research0062. research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Blandin S, Moita LF, Kocher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Rep. 2002;3(9):852–6. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Vlachou D, Schlegelmilch T, Christophides GK, Kafatos FC. Functional genomic analysis of midgut epithelial responses in Anopheles during Plasmodium invasion. Curr Biol. 2005;15:1185–1195. doi: 10.1016/j.cub.2005.06.044. [DOI] [PubMed] [Google Scholar]
- [44].Sekiguchi K, Hakomori S. Functional domain structure of fibronectin. Proc Natl Acad Sci USA. 1980;77(5):2661–5. doi: 10.1073/pnas.77.5.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Akiyama AK, Yamada KM, Hayashi M. The structure of fibronectin and its role in cellular adhesion. J Supramol Struct Cell Biochem. 1981;16(4):345–8. doi: 10.1002/jsscb.1981.380160405. [DOI] [PubMed] [Google Scholar]
- [46].Dessens JT, Beetsma AL, Dimopoulos G, Wengelnik K, Crisanti A, Kafatos FC, Sinden RE. CTRP is essential for mosquito infection by malaria ookinetes. EMBO J. 1999;18(22):6221–7. doi: 10.1093/emboj/18.22.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mussachio M, Perrimon N. The Drosophila kekkon genes: novel members of both the leucine-rich repeat and immunoglobulin superfamilies expressed in the CNS. Dev Biol. 1997;178(1):63–76. doi: 10.1006/dbio.1996.0198. [DOI] [PubMed] [Google Scholar]
- [48].Ghiglione C, Amundadottir L, Andresdottir M, Bilder D, Diamonti JA, Noselli S, Perrimon N, Caraway KL., III Mechanism of inhibition of the Drosophila and mammalian EGF receptors by the transmembrane protein Kekkon 1. Development. 2003;130(18):4483–93. doi: 10.1242/dev.00617. [DOI] [PubMed] [Google Scholar]
- [49].Machado C, Andrew DL. D-Titin: a giant protein with dual roles in chromosomes and muscles. J Cell Biol. 2000;151(3):639–52. doi: 10.1083/jcb.151.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nelson RE, Fessler LI, Takagi Y, Blumberg B, Keene DR, Olsen PF, Parker CG, Fessler FE. Peroxidasin: a novel enzyme-matrix protein of Drosophila development. EMBO J. 1994;13(15):3438–47. doi: 10.1002/j.1460-2075.1994.tb06649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Johansson MW, Holmblad T, Thornqvist PO, Cammarata M, Parrinello N, Soderhall K. A cell-surface superoxide dismutase is a binding protein for peroxinectin, a cell-adhesive peroxidase in crayfish. J Cell Sci. 1999;112(Pt 6):917–25. doi: 10.1242/jcs.112.6.917. [DOI] [PubMed] [Google Scholar]
- [52].Kobayashi M, Johansson MW, Soderhall K. The 76 kD cell-adhesion factor from crayfish hemocytes promotes encapsulation in vitro. Cell Tissue Res. 1990;260:13–18. [Google Scholar]
- [53].Kumar S, Gupta L, Han YS, Barillas-Mury C. Inducible peroxidases mediate nitration of anopheles midgut cells undergoing apoptosis in response to Plasmodium invasion. J Biol Chem. 2004;279(51):53475–82. doi: 10.1074/jbc.M409905200. [DOI] [PubMed] [Google Scholar]
- [54].Datta S, Kankel DR. l(1)trol and l(1)devl, loci affecting the development of the adult central nervous system in Drosophila melanogaster. Genetics. 1992;130(3):523–37. doi: 10.1093/genetics/130.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kolodziej PA, Timpe LC, Mitchell KJ, Fried SR, Goodman CS, Jan LY, Jan YN. frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell. 1996;87(2):197–204. doi: 10.1016/s0092-8674(00)81338-0. [DOI] [PubMed] [Google Scholar]
- [56].Hiramoto M, Hiromi Y, Giniger E, Hotta Y. The Drosophila Netrin receptor Frazzled guides axons by controlling Netrin distribution. Nature. 2000;406(6798):886–9. doi: 10.1038/35022571. [DOI] [PubMed] [Google Scholar]
- [57].Benian GM, Tinsley TL, Tang X, Borodovsky M. The Caenorhabditis elegans gene unc-89, required for muscle M-line assembly, encodes a giant modular protein composed of Ig and signal transduction domains. J Cell Biol. 1996;132(5):835–48. doi: 10.1083/jcb.132.5.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104(5):709–18. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- [59].Nardi JB, Pilas B, Bee CM, Zhuang S, Garsha K, Kanost MR. Neuroglian-positive plasmatocytes of Manduca sexta and the initiation of hemocyte attachment to foreign surfaces. Dev Comp Immunol. 2006;30(5):447–62. doi: 10.1016/j.dci.2005.06.026. [DOI] [PubMed] [Google Scholar]
- [60].Cohuet A, Osta MA, Morlais I, Awono-Ambene PH, Michel K, Simard F, Christophides GK, Fontanille D, Kafatos FC. Anopheles and Plasmodium: from laboratory models to natural systems in the field. EMBO Rep. 2006;7(12):1285–9. doi: 10.1038/sj.embor.7400831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Michel K, Suwanchaichinda C, Morlais I, Lambrechts L, Cohuet A, Awono-Ambene PH, Simard F, Fontanille D, Kanost MR, Kafatos FC. Increased melanizing activity in Anopheles gambiae does not affect development of Plasmodium falciparum. Proc Natl Acad Sci USA. 2006;103(45):16858–63. doi: 10.1073/pnas.0608033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: (A) Silencing efficiencies of IRID and GFP dsRNA treatment. Bars indicate % KD (percent knock-down) of the indicated gene, as determined by qRT-PCR. (B) Survival of IRID-silenced naïve mosquitoes is uncompromised. Post-silencing viability was determined at day 4 post-silencing in all cohorts of mosquitoes receiving dsRNA treatment. Bars represent the average percent survival of mosquitoes receiving each treatment.
Table S1: IgSF infection-responsive gene expression. The degree of differential gene expression in challenged adult female mosquitoes is given as the log2-transformed -fold change in transcript level, as compared to PBS-injected control mosquitoes. Gene IDs given are derivatives of ENSANGG000000xxxxxx IDs assigned by Ensembl. For visual simplification, we have deleted “ENSANG” and “000000” from the IDs.
(A) Transcriptional responses of IgSF exons to bacterial (E. coli [E.c.], S. typhimurium [S.t.], S. aureus [S.a.] and B. subtilis [B.s.]) challenge.
(B) Transcriptional responses of IgSF exons to malarial challenge (invading ookinetes of P. berghei [Pb IO]) and P. falciparum [Pf IO]) and infected blood containing invasion-incapable P. falciparum ookinetes (Pf IB).
Table S2: Oocyst counts for P. falciparum and P. berghei infections in IRID-silenced mosquitoes. Rows are labeled according to dsRNA treatment; columns are labeled according to the type of data and replica number. Included are data on the incidence of infection, mean oocyst load, range of infection intensities and -fold change in infection intensity (when compared to GFP-treated controls) for each of three independent assays, as well as the average -fold change and standard deviations between the assays. (A) P. falciparum infection (B) P. berghei infection.


