Abstract
The molecular switching mechanism governing skeletal and cardiac muscle contraction couples the binding of Ca2+ on troponin to the movement of tropomyosin on actin filaments. Despite years of investigation, this mechanism remains unclear because it has not yet been possible to directly assess the structural influence of troponin on tropomyosin that causes actin filaments, and hence myosin-crossbridge cycling and contraction, to switch on and off. A C-terminal domain of troponin I is thought to be intimately involved in inducing tropomyosin movement to an inhibitory position that blocks myosin-crossbridge interaction. Release of this regulatory, latching domain from actin after Ca2+-binding to TnC presumably allows tropomyosin movement away from the inhibitory position on actin, thus initiating contraction. However, the structural interactions of the regulatory domain of TnI with tropomyosin and actin that cause tropomyosin movement are unknown and thus the regulatory process is not well defined. Here, thin filaments were labeled with an engineered construct representing C-terminal TnI and then 3D-EM was used to resolve where troponin is anchored on actin-tropomyosin. EM-reconstruction showed how TnI-binding to both actin and tropomyosin at low-Ca2+ competes with tropomyosin for a common site on actin and drives tropomyosin movement to a constrained, relaxing position to inhibit myosin-crossbridge association. Thus the observations reported reveal the structural mechanism responsible for troponin-tropomyosin-mediated steric-interference of actin-myosin interaction that regulates muscle contraction.
Keywords: actin, troponin, tropomyosin, calcium, electron microscopy
Contraction of skeletal and cardiac muscles is switched on and off by the thin filament proteins troponin and tropomyosin reacting in concert to changes in intracellular Ca2+ levels. By shifting position along thin filaments in response to Ca2+ binding to troponin, tropomyosin is known to either block or expose myosin-binding sites on actin, hence regulating myosin-crossbridge cycling and consequently contraction1–5; however, the structural basis of troponin’s effect on tropomyosin remains obscure. Troponin, itself a three-component complex, consists of TnI, the “inhibitory” subunit, TnC, the Ca2+-sensor that relieves inhibition, and TnT, the element linking the complex to tropomyosin6,7. The interplay between troponin and tropomyosin, and, in particular, the role of the C-terminal segment of TnI8, considered the transducer controlling tropomyosin movement, is not defined structurally in models of troponin function. Binding studies show that the C-terminal TnI domain, while unstructured in solution9–13, interacts with actin in relaxed thin filaments but translocates to the N-terminal lobe of TnC following Ca2+-activation14,15. The TnI domain may thus constrain tropomyosin in a position that interferes with myosin binding on actin in the relaxed state. Its release from actin after Ca2+-activation would then allow tropomyosin to move away from its inhibitory position, initiating contraction. Electron microscopy and image reconstruction, well suited to examining helical macromolecular assemblies, have the potential to solve thin filament structure completely. However, the low stoichiometry of troponin on thin filaments complicates analysis9,16 and has lead to conflicting models of troponin behavior. Here, we have circumvented these shortcomings by performing electron microscopy and image reconstruction on filaments saturated with a construct representing the C-terminal TnI domain, which is readily resolved on such thin filaments. Reconstructions reveal that by competing with tropomyosin for a common binding site, TnI causes tropomyosin movement toward the blocking position on actin. Our results thus provide direct structural insights into the role of troponin in the steric regulation of muscle contraction.
3D-reconstruction of cTerm-TnI-decorated F-actin and F-actin-tropomyosin
A construct representing the C-terminal 80 amino acids of TnI (“cTerm-TnI”, human cardiac TnI residues 131 to 210) was expressed in E. coli for the structural studies described below. The construct bound to F-actin and strongly inhibited actin-activated myosin-S1 ATPase to 20 percent of its initial value at equimolar TnI:actin. These results indicated stoichiometric binding of cTerm-TnI to each actin subunit of thin filaments under saturating conditions. The inhibition was enhanced by tropomyosin at lower ratios of TnI:actin (see Supplementary data for details).
Negatively stained F-actin or F-actin-tropomyosin controls showed characteristic double-helical arrays of actin monomers. The actin subunit backbone was partially masked by addition of cTerm-TnI, and these filaments showed a noticeable increase in diameter (Fig. 1). Reconstructions generated from tropomyosin-free F-actin labeled with cTerm-TnI revealed densities in addition to the actin that were attributable to the construct. These emerge from subdomain 1 of each actin subunit and bridge to the azimuthally neighboring actin monomer on the opposite helical strand of the filament (Figs. 2b, 2c; cf. 2a). EM reconstructions of F-actin-tropomyosin decorated with the construct showed additional TnI density, indicating that the presence of tropomyosin further structures TnI, which now occupies more of the actin surface (Fig. 2d, 2e, Fig. 3a). Tropomyosin was localized in its “blocking” position on actin, as if intact troponin were present at low-Ca2+ (4,5). Fitting atomic models of F-actin and tropomyosin within the reconstructions helped to further define the position of cTerm-TnI. The TnI extension again is seen crossing the cleft between azimuthally neighboring actin monomers (Fig. 3a), bridging from subdomain 1 of actin0 to the lower edge of subdomain 4 of actin−1 (over actin residues 223–232; green, Fig. 3b). The elongated density continues to traverse the adjoining subdomain 3 toward tropomyosin on actin−1. Here part of the TnI terminates as a fist-like structure abutting tropomyosin and sitting over actin residues 309 to 330 (magenta, Fig. 3b) normally occupied by tropomyosin in the “closed”, high-Ca2+ state5, while the rest splays out to drape over tropomyosin (Fig. 2d, 2e, Fig. 3a–c). Statistical maps show that the densities noted above, which contribute to the TnI in the reconstructions, are significant at greater than the 99% confidence levels (see Supplementary data). The binding of cTerm-TnI at the low protein concentrations used for electron microscopy (1 µM actin, 2 µM cTerm-TnI) and the clear definition of the cTerm-TnI densities on actin-tropomyosin indicate that the TnI construct bound specifically to thin filaments. Atomic structures of cTerm-TnI are not available for fitting within the corresponding density envelope of our reconstructions; nevertheless, the volume encompassed by the TnI domain in the reconstructions is comparable in size to those in an array of NMR solutions for cTerm-TnI12.
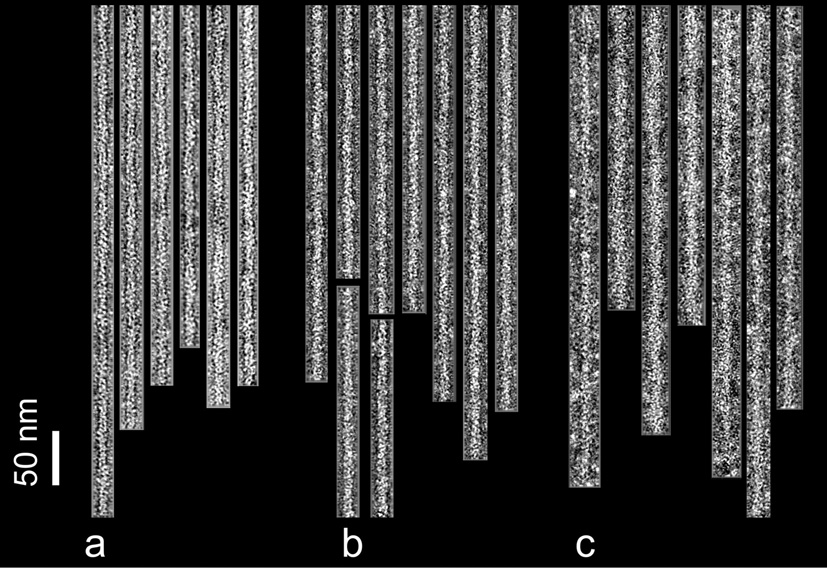
Fig. 1.
Electron micrographs of negatively stained filaments. F-actin control filaments (a), F-actin decorated with cTerm-TnI (b), F-actin-tropomyosin decorated with cTerm-TnI (c). Note the increase in diameter due to the TnI label, particularly in (c). Filaments are shown with their pointed ends facing up; polarity was determined by alignment tools in reference 22. Scale bar = 50 nm.
Preparation and assay of proteins: F-actin, cardiac troponin and tropomyosin were purified as previously20. cTerm-TnI was expressed and purified as follows: The cDNA for human cardiac TnI residues 131–210 was inserted into pET3d, transformed into Rosetta pLysS cells, and a single colony used to seed 1L of Overnight Express Media (CalBiochem/Novagen). Cells were lysed with sonication in 20mM Tris buffer (pH 8.0), 20% sucrose, 1mM EDTA, 5µg/ml TPCK, 5µg/ml TLCK and 0.3mM PMSF. The pellet fraction was resuspended and sonicated in 10mM MOPS buffer (pH 7.0), 5M urea, 1mM dithiothreitol, 5µg/ml each of TPCK and TLCK, 0.01% NaN3 and 0.3mM PMSF. The supernatant fraction was dialyzed against 10mM MOPS (pH 7. 0), 2mM dithiothreitol, 5µg/ml TPCK and TLCK, 0.01% NaN3 and 0.3mM PMSF, and purified by successive SP Sepharose and AKTA system-Resource S columns. Protein concentration was determined by quantitative amino acid analysis, and identify confirmed by MALDI-MS. Actin-activated S1 MgATPase rates were measured at 25 °C20 in the presence of 10mM MOPS (pH 7), 1mM dithiothreitol, 4mM MgCl2, 5mM KCl, 1.0mM ATP, 1mM phosphoenolpyruvate, 3mg/ml pyruvate kinase, 0.2 mg/ml lactate dehydrogenase, and the rate of NADH (0.3mM) oxidation monitored spectrophotometrically for 5 minutes.
EM and image processing: Filaments were prepared by mixing a two-fold molar excess of cTerm-TnI (40 µM) with F-actin or F-actin-tropomyosin (20 µM) to optimize binding in 100mM NaCl, 3mM MgCl2, 1mM NaN3, 0.2mM EGTA, 1mM dithiothreitol, 5mM sodium phosphate/5mM Pipes buffer (pH 7.0) at 25°C21. The mixture was diluted 20-fold, applied to carbon-coated grids and stained with 1% uranyl acetate21. EM was done on a Philips CM120 EM at a magnification of X45,000 under low dose conditions (~12 e−/Å). Helical reconstruction22 and single particle reconstruction23 were performed by standard methods as previously21. Both methods revealed comparable cTerm-TnI density. The significance of the densities contributing to cTerm-TnI, was evaluated by using a Student’s t-test24,25.
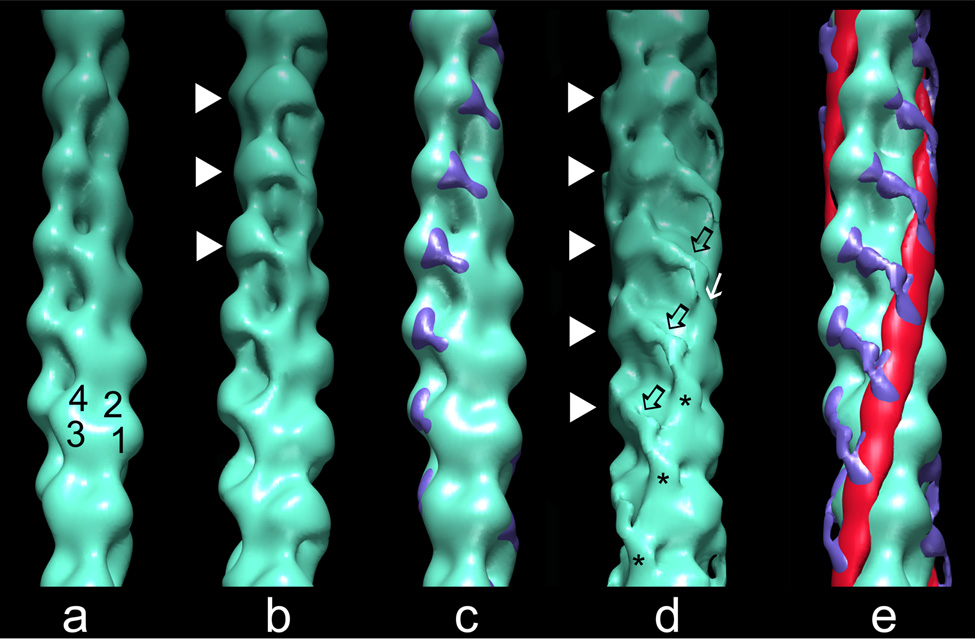
Fig. 2.
Surface views of thin filament reconstructions showing the position of cTerm-TnI on F-actin and F-actin-tropomyosin. Reconstructions of: (a) F-actin (subdomains noted). (b) F-actin decorated with cTerm-TnI; note density (white arrowheads) bridging between azimuthally neighboring actin subunits from subdomain 1 of one actin to subdomain 4 of the other. (c) Difference densities (blue), derived by subtracting F-actin from cTerm-TnI decorated F-actin, then shown superimposed on F-actin. (d) F-actin-tropomyosin decorated with cTerm-TnI; the TnI density is again seen bridging between azimuthally neighboring actin monomers (arrowheads), but the extra density here is more elongated and traverses obliquely over the lower actin (black open arrows). The TnI density ends as a finger-like mass (asterisk) on a clenched-hand that approaches and drapes over tropomyosin (white arrow). (e) Difference densities (blue for TnI and red for tropomyosin) between cTerm-TnI decorated F-actin-tropomyosin and F-actin, shown superimposed on F-actin. All of the reconstructions of thin filaments obtained were aligned to each other and are shown with filament pointed ends facing up.
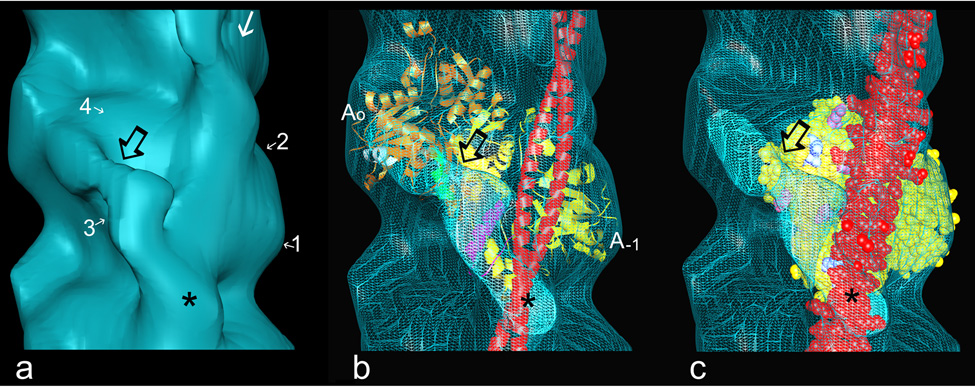
Fig. 3.
Fitting F-actin and tropomyosin models into reconstructions. Atomic models of F-actin26 and tropomyosin27 were docked into EM density envelopes, using coordinates previously determined28. The azimuthal location of tropomyosin is the same as that found previously for the blocked state5,16,21,28. (a) Reconstruction of F-actin-tropomyosin decorated with cTerm-TnI (enlargement of part of Fig. 2d, actin subdomains noted, other symbols the same as in Fig. 2); (b) ribbon models of two actin monomers (gold, yellow) and tropomyosin (red) fitted within the EM envelope shown in (a), displayed here in wire-mesh; TnI density is seen to originate near actin’s C-terminal residues 360 to 366 on subdomain 1 of actin0 (helix highlighted in white), to cross the cleft between actin subunits and over helix 223 to 232 (green) on subdomain 4 of actin−1, to buttress against tropomyosin at actin residues 309 to 330 (magenta) and to end over the tropomyosin strand (red); (c) same as (b) but now the fitting shown with one actin monomer (yellow space filling model) and tropomyosin (red space filling model), here highlighting amino acids on actin, which are thought to interact electrostatically with tropomyosin in the high-Ca2+ (“closed”) state27 (pink, acidic, blue, basic amino acids). Note that cTerm-TnI covers several of these amino acids clusters and hence sites occupied by C-state tropomyosin.
Electron tomography of thin filaments containing the intact troponin complex
Because the TnI construct in the above experiments was not tethered to the rest of troponin, it was free to bind to every actin monomer along thin filaments. In contrast, in muscle, the elongated N-terminal TnT ”tail” (TnT1) domain acts as a molecular spacer that is linked to defined regions along tropomyosin7. In muscle, this arrangement limits binding of the troponin core domain (the globular portion of troponin representing the confluence of TnI and TnC on the C-terminal end of the TnT10,11) and hence the C-terminal TnI domain, to every seventh actin along thin filaments, where the TnI domain may be targeted to bind to defined stretches of tropomyosin, insuring local cTerm-TnI – tropomyosin specificity. In fact, trace densities, possibly representing the domain, have been detected previously in single particle reconstructions of Ca2+-free thin filaments containing the entire troponin complex present at a native stoichiometry of 7 actins to 1 troponin and 1 tropomyosin9,12,16. While these densities were not well delineated, they appeared to localize between azimuthally adjacent actin monomers. To further investigate troponin organization in filaments with native stoichiometry, we carried out electron tomography of actin-tropomyosin filaments containing the whole troponin complex at the native ratio. Longitudinal sections through tomograms of single thin filaments revealed characteristic helically arranged actin monomers and troponin core domain “bulges”4,5 distributed regularly at 38.5 nm intervals (see Supplementary data). The signal-to-noise ratio of the component densities in tomograms of single filaments was too low to obtain details of molecular interactions, but averaging 38.5 nm long repeating units from the filament tomograms revealed key structural features. A tri-lobed mass, characteristic of the troponin core domain complex16, is easily identified in the averaged data, despite the relatively low resolution of the tomograms (Fig. 4a). In addition, a ridge of density originating from the core domain is seen extending between azimuthally related actin monomers (Fig. 4b). This density, derived from intact troponin, follows a path over the actin surface that is equivalent to that of cTerm-TnI shown above, corroborating those structural results (Fig. 4c). The near identity of the extension seen in tomograms of thin filaments containing intact troponin at native stoichiometry and the cTerm-TnI density observed at higher ratios on decorated filaments supports the contention that the two share common targets. Further, these results suggest that the effects of cTerm-TnI binding on actin-tropomyosin correspond to those of TnI from whole troponin.
Fig. 4.
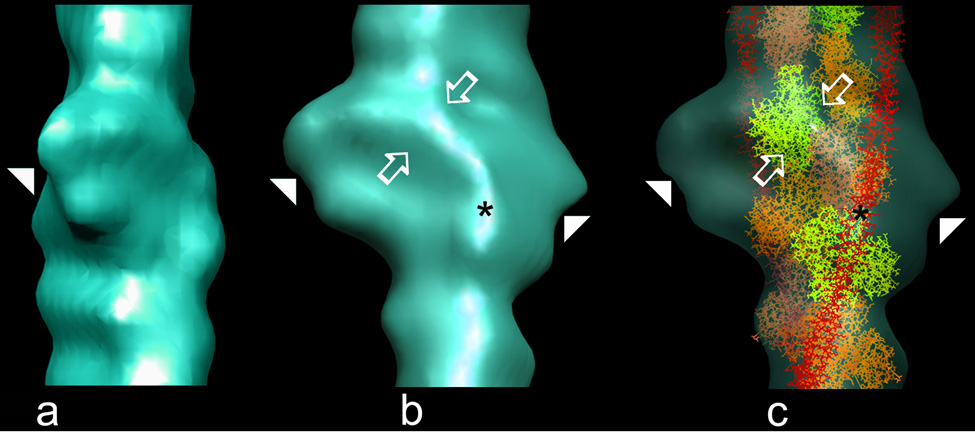
Electron tomography of thin filaments reconstituted from actin, tropomyosin and intact troponin. Surface views of averaged tomograms from 61 filaments maintained in low-Ca2+. (a) the core domain (arrowhead) of troponin is seen face on; note the trilobed density, where the upper two lobes match the near perpendicular orientation suggested for TnC relative to F-actin16,29,30 while the rest would correspond to the TnIT arm10,11. (b) orthogonal rotation of the tomogram shows a side view of the core domain from which a ridge of density (open arrows) emerges and crosses laterally over the surface of the filament. (c) fitting the atomic model of F-actin-tropomyosin within the tomogram, rotated as in (b) and made translucent (actin monomers alternately yellow, coral and orange; tropomyosin, red); note that the length of the ridge of density is sufficient to reach from the core domain to tropomyosin; further, this density follows a path comparable to that of cTerm-TnI in the reconstructions above and ends as a pad of density at a similar spot on the tropomyosin position as does the TnI domain (asterisks in (b) and (c)). Given the low resolution of the tomograms and possible uncertainties about domain positions in the low-Ca2+ crystal structure of the troponin core complex11, fitting these two structures to each other did not seem warranted. Moreover, the region of particular interest here, namely the C-terminal end of TnI, is not resolved in the low-Ca2+ crystal structure, since in the absence of its binding to actin, this region of TnI is likely to be disordered12,13.
Tomography protocols: Electron tomograms of thin filaments, prepared as previously21, were derived from EMs captured in a tilt series consisting of 1° increments between ±68° at a magnification of X31,000 (300 kV, 450 e−/Å total exposure) on a Technai F30 EM; tomograms of individual negatively stained F-actin troponin-tropomyosin filaments (see Supplementary data) were generated using Protomo software31. To improve the signal-to-noise ratio, tomograms of single thin filaments were divided into 40 nm long segments and aligned to a low-resolution model of the filament16. The aligned segments were placed within a Gaussian mask to reduce surrounding noise and the rotational and axial alignment refined using SPIDER32. Following final alignment, segments were averaged32.
A latching mechanism for TnI that induces tropomyosin movement to its blocking position
We conclude that the C-terminal domain of TnI is structured when bound on thin filaments, forming a defined extension that bridges adjacent actin monomers over the site normally occupied by tropomyosin at high-Ca2+ (Fig. 3c). Our results support the hypothesis that TnI binding and folding on actin-tropomyosin are intimately involved in the mechanism of muscle regulation13. The position of the TnI domain on azimuthally related actin monomers is common to several other actin-binding proteins, including caldesmon17 in smooth muscle thin filaments, which contain no troponin. By acting as a molecular tie straddling between the two long pitch strands of actin, troponin (and caldesmon) increase filament rigidity in the inhibited state (but not in the presence of calcium)18,19. This cross-strand communication involves no obvious change in F-actin conformation. While the mechanical effect of the troponin crosslink may have functional consequences, it is secondary to Ca2+-regulation per se, as filament stiffness can be manipulated experimentally (by phalloidin modification for example), without a noticeable change in the response to Ca2+ (20).
Our results directly relate the structural action of troponin to the movement of tropomyosin on actin. The interaction of the cTerm-TnI domain with actin subdomain 3 would be expected to dislodge tropomyosin from its high-Ca2+ position, inducing tropomyosin movement to the blocking position on actin, as is observed. The tip of the TnI domain that drapes over tropomyosin should further stabilize this blocked-state configuration. These steric effects would inhibit actin-myosin interaction and thus contraction in relaxed muscle. While we have not defined how the entire troponin complex controls tropomyosin position on actin, it appears that in intact filaments, blocked-state tropomyosin would be wedged between cTerm-TnI on one side and the tail of TnT and the troponin core domain on the other (Fig. 5). Thus each troponin complex appears to interact with tropomyosin molecules in the adjacent helical strands, suggesting a functional synergy between the two halves of the thin filament in the relaxed state and a configuration that has not before been fully appreciated. On the other hand during activation, Ca2+-saturated TnC will compete with actin for cTerm-TnI binding, thus favoring release of tropomyosin from the blocking position and leading to myosin-crossbridge interaction. In turn, myosin binding on actin will promote further movement of tropomyosin5 that would also likely displace actin-bound cTerm-TnI. Calcium binds more strongly to troponin than to TnC14, presumably because in troponin the TnI can stabilize the open state of the N-lobe of TnC, which binds Ca2+. By facilitating TnI dissociation from actin and thus favoring its association with TnC, myosin would indirectly cause an increase in TnC Ca2+affinity. This linkage between myosin binding, tropomyosin movement and TnI mobility would increase the Ca2+-sensitivity of the activation process. Our results on troponin-tropomyosin interactions presented here provide a framework for understanding the structural mechanics of the thin filament and a basis for defining regulatory protein mutants with phenotypes that lead to human cardiomyopathy.
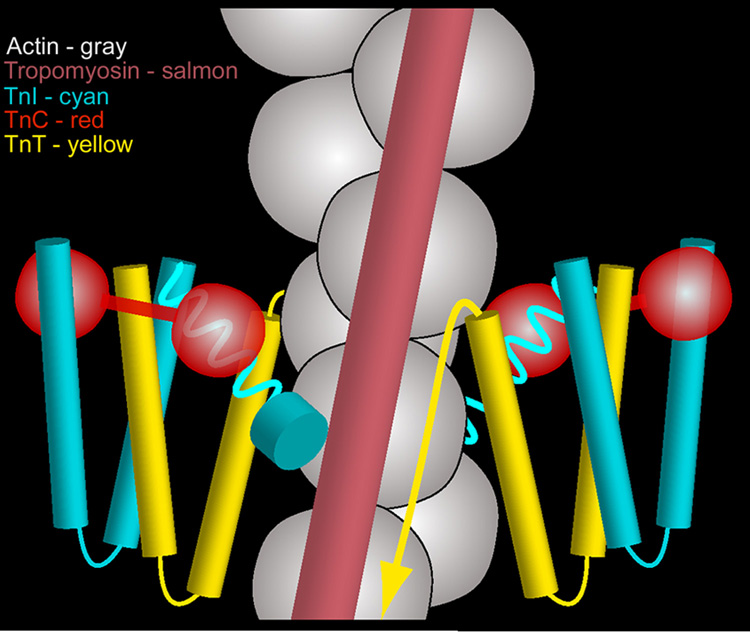
Fig. 5.
Cartoon representation of the organization of the thin filament at low-Ca2+. The troponin core domain complexes on either side of F-actin are depicted as W-shaped TnIT structures supporting dumbbell-shaped TnC13–15; actin, grey; tropomyosin, salmon; TnI, cyan; TnC, red; TnT, yellow. C-terminal TnI extensions are shown emerging from the core domain (as cork-screws), crossing the cleft between neighboring actin monomers and abutting tropomyosin (as a cyan disk). The TnT tail is depicted as an arrow running in parallel to tropomyosin. Tropomyosin is shown wedged in the blocking position between cTerm-TnI on one side and the troponin core domain and TnT on the other in a vise-like grip. No attempt was made to rationalize possible interactions between the troponin core domain and F-actin (here depicted as a double chain of beads for simplicity). Please note: Only one of two tropomyosin strands is shown. Also note: The tip of the TnI extension emanating from the troponin core on the right side of the diagram is hidden behind actin in back of the filament and not seen.
Supplementary Material
Supplementary data: To help in understanding the experimental data presented, we provide statistical evidence to indicate the boundaries of cTerm-TnI densities in our reconstructions (Item 1), images of single thin filaments used to generate averaged tomograms (Item 2), and a graph showing the inhibition of actin-activated S1 ATPase by cTerm-TnI (Item 3).
Supplementary data - Item 1. Statistical significance of densities contributing to cTerm-TnI in reconstructions shown in figure 2 and figure 3. Right: Five consecutive cross-sections covering an axial distance of 28 Å were made through the reconstruction of cTerm-TnI decorated actin-tropomyosin. (Since azimuthally related actin subunits are staggered by 27.5 Å, the density profiles of actin monomers (55 Å long) and actin-associated proteins are all represented over this distance). Left: The region of the reconstruction (displayed at 2 sigma over the mean density) that was sectioned is marked. Difference maps (not shown) comparing corresponding sections were calculated by subtracting densities in F-actin-tropomyosin controls from those in cTerm-TnI decorated F-actin-tropomyosin. The significance of the densities contributing to the difference, representing cTerm-TnI, was evaluated by using a Student’s t-test22,25. Significant differences at >99% confidence levels that are accounted for by cTerm-TnI are shown (in cyan) superimposed on maps of corresponding actin (gold) and tropomyosin (magenta). Note that the entire cTerm-TnI extension, including the parts of TnI that extend between azimuthally related actin subunits (arrowhead), and those that abut and drape over tropomyosin (arrow, asterisk) are significant. Similar analysis of differences between cTerm-TnI decorated F-actin (no tropomyosin) also show significant cTerm-TnI densities at these confidence levels (not shown).
Supplementary data - Item 2. Longitudinal sections through tomograms of negatively stained filaments reconstituted from actin, tropomyosin and intact troponin. Top: (a, b) Sections through tomograms of two individual thin filaments (prior to cutting filaments into ~40 nm segments for alignment and averaging, see Fig. 4). Bottom (c, d) An enlargement of the central sections in the top panel. Arrows indicate positions of troponin core domain bulges and arrowhead, actin subunits in the longitudinal slices.
Supplementary data - Item 3. Inhibitory effect of cTerm-TnI on actin-activated S1 MgATPase rate. Increasing concentrations of cTerm-TnI were added to samples containing myosin S1 (1 µM) and either 3 µM actin (solid line), or 3 µM actin with saturating concentrations of tropomyosin (2 µM, dashed line). The ATPase inhibition (in the absence of tropomyosin) is consistent with a cTerm-TnI - actin affinity of 3 × 106 M−1, assuming inhibition is proportional to fractional saturation of actin with the fragment. ATPase rates were normalized to those in the absence of the TnI construct, which under these conditions are also higher in the absence of tropomyosin.
Acknowledgements
We thank Ms. Victoria Hatch for technical assistance during the initiation of these studies and Ms. Karen Moore (Moore Design) for artwork in Figure 5. This research was supported by grants from the National Institutes of Health to W.L. (HL36153, HL86655), L.S.T. (HL38834, HL63774) and R.C (AR34711, RR08426).
Footnotes
Supplementary Data Supplementary data associated with this article can be found in the online version, at …
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Huxley HE. Structural changes in actin- and myosin-containing filaments during contraction. Cold Spring Harbor Symp. Quant. Biol. 1972;37:361–376. [Google Scholar]
- 2.Haselgrove JC. X-ray evidence for a conformational change in actin-containing filaments of vertebrate striated muscle. Cold Spring Harbor Symp. Quant. Biol. 1972;37:341–352. [Google Scholar]
- 3.Parry DAD, Squire JM. Structural role of tropomyosin in muscle regulation: Analysis of the X-ray patterns from relaxed and contracting muscles. J. Mol. Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- 4.Lehman W, Craig R, Vibert P. Ca2+-induced tropomyosin movement in Limulus thin filaments revealed by three dimensional reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 5.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J. Mol. Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 6.Greaser M, Gergely J. Reconstitution of troponin activity from three protein components. J. Biol. Chem. 1971;246:4226–4233. [PubMed] [Google Scholar]
- 7.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol. Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 8.Ramos CH. Mapping subdomains in the C-terminal region of troponin I involved in its binding to troponin C and to thin filament. J. Biol. Chem. 1999;274:18189–18195. doi: 10.1074/jbc.274.26.18189. [DOI] [PubMed] [Google Scholar]
- 9.Narita A, Yasunaga T, Ishikawa T, Mayanagi K, Wakabayashi T. Ca2+-induced switching of troponin and tropomyosin on actin filaments as revealed by electron cryo-microscopy. J. Mol. Biol. 2001;308:241–261. doi: 10.1006/jmbi.2001.4598. [DOI] [PubMed] [Google Scholar]
- 10.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca2+-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 11.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, et al. Ca2+-regulated structural changes in troponin. Proc. Natl. Acad. Sci. USA. 2005;102:5038–5043. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murakami K, Yumoto F, Ohki SY, Yasunaga T, Tanokura M, Wakabayashi T. Structural basis for Ca2+-regulated muscle relaxation at interaction sites of troponin with actin and tropomyosin. J. Mol. Biol. 2005;352:178–201. doi: 10.1016/j.jmb.2005.06.067. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman RM, Blumenschein TM, Sykes BD. An interplay between protein disorder and structure confers the Ca2+ regulation of striated muscle. J. Mol. Biol. 2006;361:625–633. doi: 10.1016/j.jmb.2006.06.031. [DOI] [PubMed] [Google Scholar]
- 14.Zot HJ, Potter JD. Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annu. Rev. Biophys. Biophys. Chem. 1987;16:535–559. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]
- 15.Farah CS, Reinach FC. The troponin complex and regulation of muscle contraction. FASEB J. 1995;9:755–767. doi: 10.1096/fasebj.9.9.7601340. [DOI] [PubMed] [Google Scholar]
- 16.Pirani A, Vinogradova MV, Curmi PMG, King WA, Fletterick RJ, Craig R, et al. An atomic model of the thin filament in the relaxed and Ca2+-activated states. J. Mol. Biol. 2006;357:707–717. doi: 10.1016/j.jmb.2005.12.050. [DOI] [PubMed] [Google Scholar]
- 17.Foster DB, Huang R, Hatch V, Craig R, Graceffa P, Lehman W, et al. Modes of caldesmon binding to actin: sites of caldesmon contact and modulation of interactions by phosphorylation. J. Biol. Chem. 2004;279:53387–53394. doi: 10.1074/jbc.M410109200. [DOI] [PubMed] [Google Scholar]
- 18.Isambert H, Venier P, Maggs AC, Fattoum A, Kassab R, Pantaloni D, et al. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg MJ, Wang C-L, Lehman W, Moore JR. Modulation of actin mechanics by caldesmon and tropomyosin. Cell Motil. Cytoskeleton. 2008;65:156–164. doi: 10.1002/cm.20251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landis CA, Bobkova A, Homsher E, Tobacman LS. The active state of the thin filament is destabilized by an internal deletion in tropomyosin. J. Biol. Chem. 1997;272:14051–14056. doi: 10.1074/jbc.272.22.14051. [DOI] [PubMed] [Google Scholar]
- 21.Pirani A, Xu C, Hatch V, Craig R, Tobacman LS, Lehman W. Single particle analysis of relaxed and activated muscle thin filaments. J. Mol. Biol. 2005;346:761–772. doi: 10.1016/j.jmb.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 22.Owen C, DeRosier DJ. A 13-Å map of the actin-scruin filament from the Limulus acrosomal process. J. Cell Biol. 1993;123:337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 24.Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J. Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trachtenberg S, DeRosier DJ. Three-dimensional structure of frozen hydrated flagellar filament of Salmonella typhimurium. J. Mol. Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- 26.Holmes KC, Angert I, Kull FJ, Jahn W, Schröder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 27.Lorenz M, Poole KJV, Popp D, Rosenbaum G, Holmes KC. An atomic model of the unregulated thin filament obtained by X-ray fiber diffraction on oriented actin-tropomyosin gels. J. Mol. Biol. 1995;246:108–119. doi: 10.1006/jmbi.1994.0070. [DOI] [PubMed] [Google Scholar]
- 28.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Tobacman LS, et al. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J. Struct. Biol. 2006;155:273–284. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson RE, Sun Y-B, Mercier P, Brack AS, Sykes BD, Corrie JET, et al. In situ orientations of protein domains: Troponin C in skeletal muscle fibers. Mol. Cell. 2003;11:865–874. doi: 10.1016/s1097-2765(03)00096-0. [DOI] [PubMed] [Google Scholar]
- 30.Sun YB, Brandmeier B, Irving M. Structural changes in troponin in response to Ca2+ and myosin binding to thin filaments during activation of skeletal muscle. Proc. Natl. Acad. Sci. 2006;103:17771–17776. doi: 10.1073/pnas.0605430103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler H. 3D reconstruction and processing of volumetric data in cryo-electron tomography. J.Struct. Biol. 2007;157:126–137. doi: 10.1016/j.jsb.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data: To help in understanding the experimental data presented, we provide statistical evidence to indicate the boundaries of cTerm-TnI densities in our reconstructions (Item 1), images of single thin filaments used to generate averaged tomograms (Item 2), and a graph showing the inhibition of actin-activated S1 ATPase by cTerm-TnI (Item 3).
Supplementary data - Item 1. Statistical significance of densities contributing to cTerm-TnI in reconstructions shown in figure 2 and figure 3. Right: Five consecutive cross-sections covering an axial distance of 28 Å were made through the reconstruction of cTerm-TnI decorated actin-tropomyosin. (Since azimuthally related actin subunits are staggered by 27.5 Å, the density profiles of actin monomers (55 Å long) and actin-associated proteins are all represented over this distance). Left: The region of the reconstruction (displayed at 2 sigma over the mean density) that was sectioned is marked. Difference maps (not shown) comparing corresponding sections were calculated by subtracting densities in F-actin-tropomyosin controls from those in cTerm-TnI decorated F-actin-tropomyosin. The significance of the densities contributing to the difference, representing cTerm-TnI, was evaluated by using a Student’s t-test22,25. Significant differences at >99% confidence levels that are accounted for by cTerm-TnI are shown (in cyan) superimposed on maps of corresponding actin (gold) and tropomyosin (magenta). Note that the entire cTerm-TnI extension, including the parts of TnI that extend between azimuthally related actin subunits (arrowhead), and those that abut and drape over tropomyosin (arrow, asterisk) are significant. Similar analysis of differences between cTerm-TnI decorated F-actin (no tropomyosin) also show significant cTerm-TnI densities at these confidence levels (not shown).
Supplementary data - Item 2. Longitudinal sections through tomograms of negatively stained filaments reconstituted from actin, tropomyosin and intact troponin. Top: (a, b) Sections through tomograms of two individual thin filaments (prior to cutting filaments into ~40 nm segments for alignment and averaging, see Fig. 4). Bottom (c, d) An enlargement of the central sections in the top panel. Arrows indicate positions of troponin core domain bulges and arrowhead, actin subunits in the longitudinal slices.
Supplementary data - Item 3. Inhibitory effect of cTerm-TnI on actin-activated S1 MgATPase rate. Increasing concentrations of cTerm-TnI were added to samples containing myosin S1 (1 µM) and either 3 µM actin (solid line), or 3 µM actin with saturating concentrations of tropomyosin (2 µM, dashed line). The ATPase inhibition (in the absence of tropomyosin) is consistent with a cTerm-TnI - actin affinity of 3 × 106 M−1, assuming inhibition is proportional to fractional saturation of actin with the fragment. ATPase rates were normalized to those in the absence of the TnI construct, which under these conditions are also higher in the absence of tropomyosin.