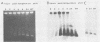Abstract
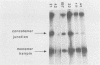
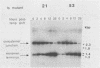
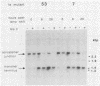
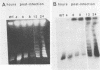
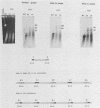
Vaccinia virus replicates in the cytoplasm of infected cells, generating transient replicative intermediates containing the DNA for the terminal sequences as concatemeric junctions. The processing of the terminal sequences for a series of vaccinia virus conditional lethal mutants at the nonpermissive temperature was analyzed by restriction enzyme digestion and Southern blot hybridization of DNA isolated from infected cells. Three phenotypes were observed: DNA replication negative (Rep-), DNA replication positive but concatemer resolution negative (Rep+ Res-), and DNA replication positive and concatemer resolution positive (Rep+ Res+). Interestingly, all six Rep+ Res- mutants from separate complementation groups were defective in late protein synthesis. Isatin beta-thiosemicarbazone, a drug that blocks late protein synthesis, also prevented resolution of concatemers. Orthogonal field gel electrophoresis of the DNA generated by the late defective mutants revealed a distribution of linear genome multimers. The multimers were processed into mature monomers after a shift to the permissive temperature in the presence of cytosine arabinoside for all the Rep+ Res- mutants except ts22, an irreversible mutant which cleaves RNA late in infection (R.F. Pacha and R.C. Condit, J. Virol. 56:395-403, 1985). Genome formation can be divided into two stages: DNA replication, which generates concatemers, and resolution, which processes concatemers into monomers with hairpin termini. Early viral genes are required for the former, and late viral genes are required for the latter.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldick C. J., Jr, Moss B. Resistance of vaccinia virus to rifampicin conferred by a single nucleotide substitution near the predicted NH2 terminus of a gene encoding an Mr 62,000 polypeptide. Virology. 1987 Jan;156(1):138–145. doi: 10.1016/0042-6822(87)90444-2. [DOI] [PubMed] [Google Scholar]
- Ball L. A. High-frequency homologous recombination in vaccinia virus DNA. J Virol. 1987 Jun;61(6):1788–1795. doi: 10.1128/jvi.61.6.1788-1795.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudy B. M., Venkatesan S., Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982 Feb;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- Baroudy B. M., Venkatesan S., Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A. Isolation and preliminary characterization of temperature-sensitive mutants of vaccinia virus. Virology. 1981 Aug;113(1):224–241. doi: 10.1016/0042-6822(81)90150-1. [DOI] [PubMed] [Google Scholar]
- Condit R. C., Motyczka A., Spizz G. Isolation, characterization, and physical mapping of temperature-sensitive mutants of vaccinia virus. Virology. 1983 Jul 30;128(2):429–443. doi: 10.1016/0042-6822(83)90268-4. [DOI] [PubMed] [Google Scholar]
- DeLange A. M., McFadden G. Efficient resolution of replicated poxvirus telomeres to native hairpin structures requires two inverted symmetrical copies of a core target DNA sequence. J Virol. 1987 Jun;61(6):1957–1963. doi: 10.1128/jvi.61.6.1957-1963.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLange A. M., Reddy M., Scraba D., Upton C., McFadden G. Replication and resolution of cloned poxvirus telomeres in vivo generates linear minichromosomes with intact viral hairpin termini. J Virol. 1986 Aug;59(2):249–259. doi: 10.1128/jvi.59.2.249-259.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M., Holowczak J. A. Replication of vaccinia DNA in mouse L cells. I. In vivo DNA synthesis. Virology. 1977 May 1;78(1):57–75. doi: 10.1016/0042-6822(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Esteban M., Holowczak J. A. Replication of vaccinia DNA in mouse L cells. III. Intracellular forms of viral DNA. Virology. 1977 Oct 15;82(2):308–322. doi: 10.1016/0042-6822(77)90006-x. [DOI] [PubMed] [Google Scholar]
- Evans D. H., Stuart D., McFadden G. High levels of genetic recombination among cotransfected plasmid DNAs in poxvirus-infected mammalian cells. J Virol. 1988 Feb;62(2):367–375. doi: 10.1128/jvi.62.2.367-375.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Alberts B. M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986 Dec 5;47(5):793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Blankenship M. L., Ben-Porat T. Replication of herpesvirus DNA. V. Maturation of concatemeric DNA of pseudorabies virus to genome length is related to capsid formation. J Virol. 1980 Mar;33(3):1151–1164. doi: 10.1128/jvi.33.3.1151-1164.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakritz N., Foglesong P. D., Reddy M., Baum S., Hurwitz J., Bauer W. R. A vaccinia virus DNase preparation which cross-links superhelical DNA. J Virol. 1985 Mar;53(3):935–943. doi: 10.1128/jvi.53.3.935-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchlinsky M., Garon C. F., Moss B. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J Mol Biol. 1988 Feb 5;199(3):399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- Merchlinsky M., Moss B. Resolution of linear minichromosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell. 1986 Jun 20;45(6):879–884. doi: 10.1016/0092-8674(86)90562-3. [DOI] [PubMed] [Google Scholar]
- Mosig G. The essential role of recombination in phage T4 growth. Annu Rev Genet. 1987;21:347–371. doi: 10.1146/annurev.ge.21.120187.002023. [DOI] [PubMed] [Google Scholar]
- Moss B., Katz E., Rosenblum E. N. Vaccinia virus directed RNA and protein synthesis in the presence of rifampicin. Biochem Biophys Res Commun. 1969 Aug 22;36(5):858–865. doi: 10.1016/0006-291x(69)90688-3. [DOI] [PubMed] [Google Scholar]
- Moss B., Rosenblum E. N., Katz E., Grimley P. M. Rifampicin: a specific inhibitor of vaccinia virus assembly. Nature. 1969 Dec 27;224(5226):1280–1284. doi: 10.1038/2241280a0. [DOI] [PubMed] [Google Scholar]
- Moyer R. W., Graves R. L. The mechanism of cytoplasmic orthopoxvirus DNA replication. Cell. 1981 Dec;27(2 Pt 1):391–401. doi: 10.1016/0092-8674(81)90422-0. [DOI] [PubMed] [Google Scholar]
- Nagaya A., Pogo B. G., Dales S. Biogenesis of vaccinia: separation of early stages from maturation by means of rifampicin. Virology. 1970 Apr;40(4):1039–1051. doi: 10.1016/0042-6822(70)90150-9. [DOI] [PubMed] [Google Scholar]
- Pacha R. F., Condit R. C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985 Nov;56(2):395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington T. H., Follett E. A., Szilágyi J. F. Events in vaccinia virus-infected cells following the reversal of the antiviral action of rifampicin. J Gen Virol. 1970 Dec;9(3):225–237. doi: 10.1099/0022-1317-9-3-225. [DOI] [PubMed] [Google Scholar]
- Pennington T. H. Isatin-beta-thiosemicarbazone causes premature cessation of vaccinia virus-induced late post-replicative polypeptide synthesis. J Gen Virol. 1977 Jun;35(3):567–571. doi: 10.1099/0022-1317-35-3-567. [DOI] [PubMed] [Google Scholar]
- Pogo B. G. Elimination of naturally occurring crosslinks in vaccinia virus DNA after viral penetration into cells. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1739–1742. doi: 10.1073/pnas.74.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogo B. G., O'Shea M., Freimuth P. Initiation and termination of vaccinia virus DNA replication. Virology. 1981 Jan 15;108(1):241–248. doi: 10.1016/0042-6822(81)90543-2. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Seto J., Celenza L. M., Condit R. C., Niles E. G. Genetic map of the vaccinia virus HindIII D Fragment. Virology. 1987 Sep;160(1):110–119. doi: 10.1016/0042-6822(87)90051-1. [DOI] [PubMed] [Google Scholar]
- Sherman G., Bachenheimer S. L. DNA processing in temperature-sensitive morphogenic mutants of HSV-1. Virology. 1987 Jun;158(2):427–430. doi: 10.1016/0042-6822(87)90214-5. [DOI] [PubMed] [Google Scholar]
- Southern E. M., Anand R., Brown W. R., Fletcher D. S. A model for the separation of large DNA molecules by crossed field gel electrophoresis. Nucleic Acids Res. 1987 Aug 11;15(15):5925–5943. doi: 10.1093/nar/15.15.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulos D. D., Roberts B. E., Panicali D. L., Cohen L. K. Delineation of the viral products of recombination in vaccinia virus-infected cells. J Virol. 1988 Mar;62(3):1046–1054. doi: 10.1128/jvi.62.3.1046-1054.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. L., Condit R. C. Marker rescue mapping of vaccinia virus temperature-sensitive mutants using overlapping cosmid clones representing the entire virus genome. Virology. 1986 Apr 15;150(1):10–20. doi: 10.1016/0042-6822(86)90261-8. [DOI] [PubMed] [Google Scholar]
- Thompson C. L., Hooda-Dhingra U., Condit R. C. Fine structure mapping of five temperature-sensitive mutants in the 22- and 147-kilodalton subunits of vaccinia virus DNA-dependent RNA polymerase. J Virol. 1989 Feb;63(2):705–713. doi: 10.1128/jvi.63.2.705-713.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems T. E., Walliker D., Smith C. L., do Rosario V. E., Maloy W. L., Howard R. J., Carter R., McCutchan T. F. A histidine-rich protein gene marks a linkage group favored strongly in a genetic cross of Plasmodium falciparum. Cell. 1987 Jun 5;49(5):633–642. doi: 10.1016/0092-8674(87)90539-3. [DOI] [PubMed] [Google Scholar]
- White J. H., Richardson C. C. Processing of concatemers of bacteriophage T7 DNA in vitro. J Biol Chem. 1987 Jun 25;262(18):8851–8860. [PubMed] [Google Scholar]
- Wittek R., Menna A., Müller H. K., Schümperli D., Boseley P. G., Wyler R. Inverted terminal repeats in rabbit poxvirus and vaccinia virus DNA. J Virol. 1978 Oct;28(1):171–181. doi: 10.1128/jvi.28.1.171-181.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson B., Joklik W. K. The inhibition of vaccinia virus multiplication by isatin-beta-thiosemicarbazone. Proc Natl Acad Sci U S A. 1965 Sep;54(3):946–953. doi: 10.1073/pnas.54.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]