Abstract
We describe a novel biphasic regulation of Il2 transcription in naïve CD4+ T cells. Few (~5%) CD4+ T cells transcribe Il2 within 6 h of α-TCR-β + α-CD28 stimulation (early phase). Most naïve CD4+ T cells do not initiate Il2 transcription until after an additional ~12 h of T cell stimulation (late phase). In comparison, essentially all previously activated (Pre-Ac) CD4+ T cells that transcribe Il2 do so with an early phase response. Late phase Il2 expression mostly requires c-Rel, CD28, and TNFR signaling. In contrast, early phase transcription is only partly c-Rel and CD28 dependent and TNFR independent. There was also increased stable DNA accessibility at the Il2 locus and elevated c-Rel expression in resting Pre-Ac CD4+ cells. Upon T cell activation, a faster and greater increase in DNA accessibility as well as c-Rel nuclear expression were observed in Pre-Ac CD4+ cells relative to naïve CD4+ T cells. In addition, both acetylated histone H3 (Ac-H3) and total H3 decreased at the Il2 locus upon re-challenge of Pre-Ac CD4+ T cells, while increased Ac-H3 with no change in total H3 was observed following activation of naïve CD4+ T cells. We propose a model in which nucleosome disassembly facilitates rapid initiation of Il2 transcription in CD4+ T cells, and suggest that a threshold level of c-Rel must be reached for Il2 promoter activity in both naïve and Pre-Ac CD4+ T cells. This is provided, at least partially, by TNFR signaling during priming, but not during recall.
Keywords: T lymphocyte, nucleosome, chromatin, cytokines, gene regulation, transcription factors
Introduction
IL-2 plays a critical role in maintaining T cell homeostasis. Signaling through the high affinity IL-2 receptor (IL-2R) regulates a plethora of immunological parameters, including the proliferation, differentiation, and survival of effector T cells and the production and function of regulatory T cells (1). Dysregulation of IL-2 bioavailability has profound consequences including lethal autoimmunity (2–7). IL-2 is mainly produced by activated peripheral CD4+ T cells, but regulation of its production is complex. For example, memory CD4+ T cells make IL-2 faster and more abundantly than do naïve CD4+ T cells (8); CD4+CD25+ T regulatory CD4+ T cells don’t make IL-2 at all (9); and naïve CD4+ T cells made anergic through deprivation of co-stimulation or converted to regulatory T cells in the periphery lose their ability to make IL-2 upon restimulation (10, 11).
In quiescent naïve CD4+ T cells, the Il2 locus is maintained in a transcriptionally silent state and is transiently activated following Ag presentation to the TCR in the context of an appropriate MHC molecule and costimulation (6, 12–15). Induction of Il2 expression is controlled at the level of transcription through the assembly of transcription factor complexes, coregulators, chromatin-remodeling complexes, and complexes responsible for signal-specific histone modifications at the promoter and enhancer regions (16). A minimal essential regulatory region is located in the first 300 bp region upstream of the transcription start site of the Il2 gene. Numerous well characterized cis-acting elements for multiple inducible and constitutive transcription factors, including members of the AP-1, NF-κB, NFAT, and Oct families are assembled at this site (17, 18) as well as multiple binding sites for the architectural protein HMGI(Y) (19). Maximal induction of Il2 gene expression requires all of these elements (20). The CD28 response element (RE) located between −164 bp and −152 bp upstream of the transcription start site is particularly critical for Il2 gene transcription (18, 21–23). A number of studies demonstrate that changes in chromatin structure accompany induction of Il2 transcription (20, 24–27). Acetylation, methylation, and phosphorylation of histones as well as demethylation of individual CpGs occupying the −300 to +1 proximal promoter accompany transcriptional activation (20, 28–31), and it is likely that a functional cooperativity among transcription factors and epigenetic mechanisms governs the transient nature of Il2 expression.
The objective of this study was to investigate whether rapid IL-2 production was associated with changes at the level of chromatin structure. At the cellular level, rapid IL-2 production in CD4+ T cells during re-challenge has been attributed to a greater frequency of Ag-specific memory cells, a lower threshold for activation, less dependence on accessory costimulation, and an increased ability to respond to a wider range of APC. However, the underlying molecular mechanisms remain elusive. To determine whether chromatin structure contributes to the kinetics of Il2 transcription, we studied MNase hypersensitivity and histone modifications during antigen priming and re-challenge of a population of mono-TCR-specific CD4+ T cells. We identify a previously unrecognized biphasic regulation for the induction of Il2 transcription. The late phase is mostly cRel-, CD28-, and TNFR-dependent, while the early phase is much less so. The Shannon laboratory has previously associated nucleosome occupancy within the regulatory region [from −60 bp to −210 bp] of the human Il2 gene with impaired transcription factor binding and promoter silencing (32). Our data now suggest that Il2 promoter nucleosome loss only occurs during secondary re-activation, not during primary antigen stimulation. This loss contributes to a more rapid onset of Il2 production, but does not act as the ON/OFF switch for transcriptional initiation. We propose a model in which stable chromatin modifications in the proximal promoter of the Il2 gene during priming of CD4+ T cells allow for a T cell intrinsic mechanism of rapid Il2 transcription in a secondary immune re-challenge.
Materials and Methods
Mice
All the mice used in these experiments were obtained from the National Institute of Allergy and Infectious Diseases contract facility at Taconic Farms (Germantown, NY), an American Association for the Accreditation of Laboratory Animal Care accredited specific pathogen-free barrier and housed in sterile caging at the National Institutes of Health. B10.A (H-2a) TCR-Cyt 5C.C7, Rag2Δ/Δ mice (33) carry the Vα11/Vβ3 CD4+ T cell Tg receptor specific for the moth cytochrome c (MCC) 88–103 and pigeon cytochrome c 81–104 peptides and are referred to as wild-type (WT). The IL-2-GFP knock-in mouse (34) was received on a C57BL/6 (N11) Rag1Δ/Δ (N2) background and crossed twice onto the B10.A 5C.C7 Rag2Δ/Δ background before selecting for homozygosity of the 5C.C7 TCR transgene, the GFP allele, and the B10.A MHC. B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ IL-2-GFP heterozygous (IL-2-GFPWT/KI) mice carrying one WT IL-2 allele and one ‘knocked in’ IL-2 GFP allele and B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ IL-2-GFP homozygous (IL-2-GFPKI/KI) mice carrying two ‘knocked in’ GFP alleles were used in these experiments. IL-2-GFPWT/KI mice were obtained as F1 offspring from B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ IL-2-GFPKI/KI X B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ WT matings. The TNFR P55Δ/Δ TNFR P75Δ/Δ mouse generated by Peschon et al (35) was received on a C57BL/6 (N4) background. These mice were crossed five times onto the B10.PL background and then crossed onto the B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ background for 4–6 times before intracrossing and selecting for the 5C.C7 TCR transgene, the p55Δ/Δ, p75Δ/Δ, and Rag2Δ/Δ alleles, and the B10.A MHC.
Cell preparations
Naïve CD4+ T cells were obtained from cervical, axillary, inguinal, and popliteal lymph nodes. To generate Pre-Ac CD4+ T cells, single-cell lymph node preparations were stimulated with moth cytochrome c peptide in the presence of irradiated (3,000 rad) B10.A splenocytes for 72 h, then expanded 20 fold in 10 U/ml of rIL-2 (Biosource International) for 14 days before isolation on a Ficoll gradient and use as Pre-Ac CD4+ T cells. The NIH3T3 fibroblast-transformed cell line (NIH/Swiss strain, CRL-1658) (ATTC) was used as a non-T cell control. FACS was generally used to purify populations of Vβ3 TCR+ CD4+ T cells. Briefly, cells from B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ mice were labeled with Abs to B220, CD11b, CD11c, CD16/32, and NK1.1 prior to sorting. At times, Miltenyi magnetic beads rather than flow cytometry were used for negative selection. Purity was typically >95% Vβ3+CD4+ T cells in either case. At other times, FACS was used to positively purify either Vα11+TCR+CD4+ or Vβ3+CD4+ T cells; purity achieved was typically >99%. CD4+ T cells were stimulated with plate-bound anti-TCR-β (clone H57; 1–10 μg/ml). Some cultures were supplemented with ascites fluid containing the 37.51 mAb to CD28 (a gift from J. Allison, Memorial Sloan-Kettering Cancer Center, New York, NY) at a final dilution of 1/1000. Pentoxifylline (PF) and cyclosporin A (CsA) were purchased from Sigma; treatments (300 μg/ml and 200 ng/ml, respectively) were selected on titrations for maximal suppression of GFP expression with minimal cell death as measured by Annexin V and 7-AAD staining at 48 h (data not shown) (36, 37). PE-conjugated CD120a and CD120b, purchased from BD Pharmingen, were used for detecting surface TNFRI and TNFRII expression.
RNA purification and real-time quantitative PCR
Total RNA was extracted using the Qiagen RNeasy kit and treated with RNase-free DNAse I. Taqman reverse transcription reagents, probes, and primers for murine Il2 and c-rel were purchased from ABI. For analyses, Il2 and c-rel samples were normalized to GAPDH; the normalized threshold cycle (Ct) values were subtracted from the target Ct values of each sample (ΔCt). Relative levels of target mRNA were calculated as 2−ΔΔCt.
IL-2 secretion
The cytokine secretion assay-detection kit (Miltenyi Biotec) was used to quantify IL-2 secretion as previously described (38).
MNase sensitivity
MNase digestion was performed essentially as described previously (39). Briefly, 5 million purified CD4+ T cells/digestion were lysed in a 5% sucrose homogenation buffer, nuclei were separated using sucrose gradient centrifugation, and aliquots of 1 million nuclei were digested with 25, 50, or 100 U MNase (Roche) in 1 mM CaCl2 with the middle concentration optimized to generate predominantly tri-, di-, and mononucleosomes. Proteins were extracted via overnight proteinase K digestion. DNA was extracted using phenol:chloroform, precipitated with ethanol and 3M sodium acetate, and then dissolved in TE buffer to 0.4 ng/ml. Genomic DNA (5 ng) was used to perform SYBR® green quantitative PCR on an ABI PRISM 7700 or 7900 sequence detector (ABI) using SYBR® green PCR master mix (ABI) and primers spanning the proximal promoter of the murine Il2 gene sequence (GenBank accession no. M39728) and as described previously (26, 27); the primer set and the corresponding amplified region are: G = −1982 to −1890; F = −652 to −460; E = −459 to −342; D = −309 to −225; C = −201 to −111; B = −110 to −16; A = +38 to +107. Data are expressed as described previously (40).
Chromatin immunoprecipitation (ChIP)
The ChIP protocol described here is a variation of a previously published protocol (40). Briefly, nuclei were either prepared from 5 × 107 naïve or Pre-Ac CD4+ T cells crosslinked with 1% formaldehyde and DNA was sheared by sonication to lengths of 200 to 1000 bp using a Bioruptor (Diagenode). Alternatively, since the c-Rel RE overlaps a nucleosome binding site, 5 × 107 naïve or Pre-Ac CD4+ T cells were digested with 200 U MNase at RT for 15 min to obtain a primarily mononucleosome preparation. One-tenth of the sonicated or digested material was kept as an input sample; the remainder was divided into five aliquots and precleared with salmon sperm DNA/protein A agarose. Rabbit polyclonals to histone H3 (H3; Abcam), diacetylated (K9, K14) histone H3 (AcH3; Abcam), c-Rel (Santa Cruz), and control rabbit IgG (Santa Cruz) were added and incubated overnight. Protein–DNA crosslinking was reversed by incubation at 65°C for 4 h with 0.2 M NaCl. The protein was digested at 45°C for 1h with proteinase K. DNA was purified by phenol–chloroform extraction followed by ethanol precipitation. Each immunoprecipitation was done at least three times. Chromatin immune precipitates were quantified using SYBR® green quantitative PCR and primers amplifying −201 bp to −111 bp (region containing c-Rel κB site) or −1982 bp to −1890 bp (non c-Rel RE control) of the Il2 promoter. Unbound chromatin in the no-Ab-added sample was used as an input control. Data are reported as fold induction of each crosslinked sample as determined by comparing the Ct value of the target to the Ct value of the unstimulated naïve T cell immune precipitates.
Immunofluorescent detection and analysis of c-Rel nuclear translocation
Freshly isolated lymphocytes or rested Pre-Ac CD4+ T cells from 5C.C7 transgenic mice were stimulated with 10 μg/ml α-TCR-β + soluble α-CD28 (as described above) for the times indicated. Cells were stained with α-c-Rel (Santa Cruz), Alexa Fluor 555 (Invitrogen), PE-Texas Red-conjugated α-CD4 (Caltag), and DRAQ5 (BioStatus, Ltd., Leicestershire, UK), then analyzed for fluorescence image-based intracellular and nuclear c-Rel translocation using the ImageStream® 100 multispectral imaging flow cytometer (Amnis Corp.) as described previously (41). Briefly, a minimum of 7400 CD4+ T cells were collected and analyzed. Cells were excited with a 488 nm laser light and imaged on a time delay integration (TDI) CCD camera. Light was then passed through a spectral decomposition element to direct different spectral bands to spatially distinct channels on the TDI CCD detector. The images of individual cells were then optically decomposed into a series of sub-images with each corresponding to a different color component and each having an identical spatial registry of pixels from channel to channel. C-Rel translocation was assessed by the similarity of pixel intensities between the c-Rel image in channel 4 (Alexa Fluor 555) and the nuclear image in channel 6 based on the image mask of DRAQ5 on a pixel-by-pixel basis for each cell. The similarity score for each cell was calculated using a log transformation of Pearson’s correlation coefficient. Positive translocation events were assessed by comparison with the similarity score for the negative control (correlation of darkfield scatter image with the nuclear image) followed by visual inspection of progressive bins on the similarity score histogram using the composite image of c-Rel stain and nuclear stain. Data were analyzed using the IDEAS software package (Amnis Corp.).
Results
Onset of transcription of the Il2 gene in the majority of naïve CD4+ T cells is delayed until approximately 12–18 h after TCR ligation
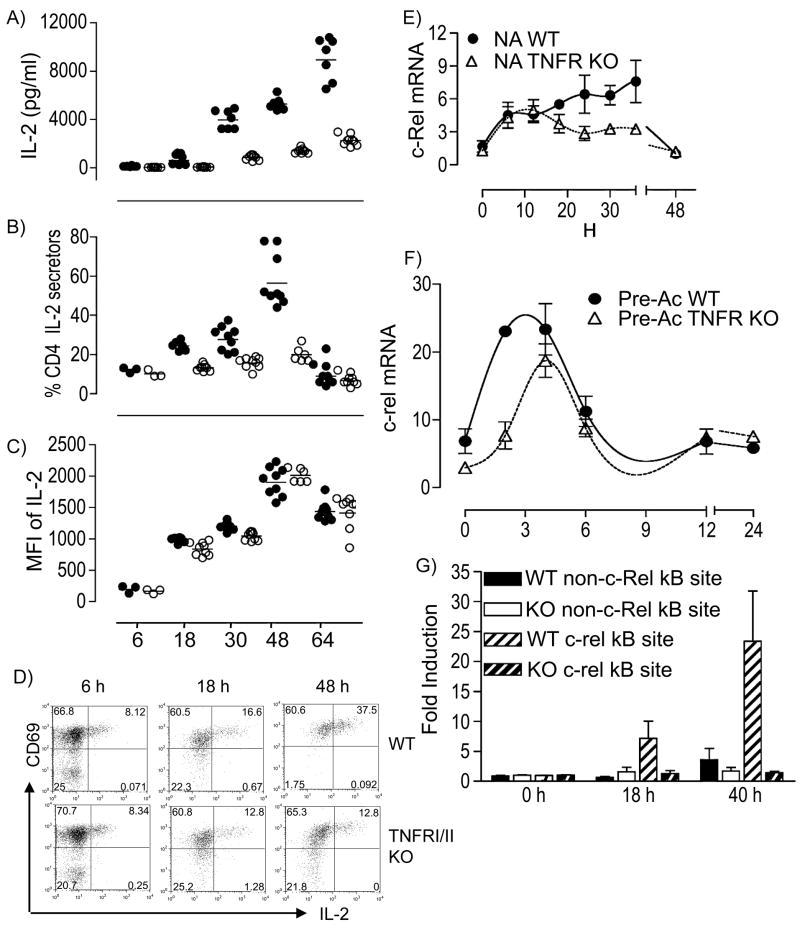
Vβ3+Vα11+CD4+ T cells from B10.A Rag2Δ/Δ 5C.C7 TCR mice were used to compare rates of induction and duration of steady-state Il2 mRNA expression during priming relative to rechallenge of CD4+ cells. Following restimulation of resting effector Pre-Ac CD4+ T cells with α-TCR-β + α-CD28, steady-state Il2 mRNA expression was rapid and transient, with peak production by 6 h (Fig. 1A). Induction of Il2 mRNA in naïve CD4+ T cells was minimal (although detectable) during this same period but was followed by a prolonged burst of expression beginning ~ 10–12 h after TCR ligation, peaking at 18 h and then persisting for up to 42 h (Fig. 1A). The small amount of Il2 mRNA produced by naïve T cells during the early phase correlated with 5% of cells actively secreting IL-2 protein (Fig. 1B); peak IL-2 protein production was at 48 h. In contrast, > 60% of Pre-Ac CD4+ T cells actively secrete IL-2 at 6 h (Fig. 1B). Surface expression of CD44 and CD62L on naïve CD4+/Vβ3/Vα11 cells did not differ between early and late IL-2 producers (data not shown). In addition, no demonstrable difference in constitutive CD4, Vβ3 or Vα11 TCR, CD2, CD27, CD28, CD103, CD127, CD122, CD132, CD134, CD137, CD154, CD223, CD278, CTLA-4, or PD-1 surface expression or the induction (rate and magnitude) of CD69 or CD25 surface expression between these two populations (data not shown), thus arguing against the possibility that early IL-2 producers represent a subpopulation of cells with effector or memory status. Collectively, these results establish that the limited Il2 mRNA produced by naïve CD4+ T cells early after T cell stimulation reflects a very low frequency of IL-2 producers at that time. Pre-Ac CD4+ T cells were also heterogenous for IL-2 production; but no differences in Vβ3, CD4, CD69, or CD25 expression were noted between IL-2 producers and non-producers.
FIGURE 1.
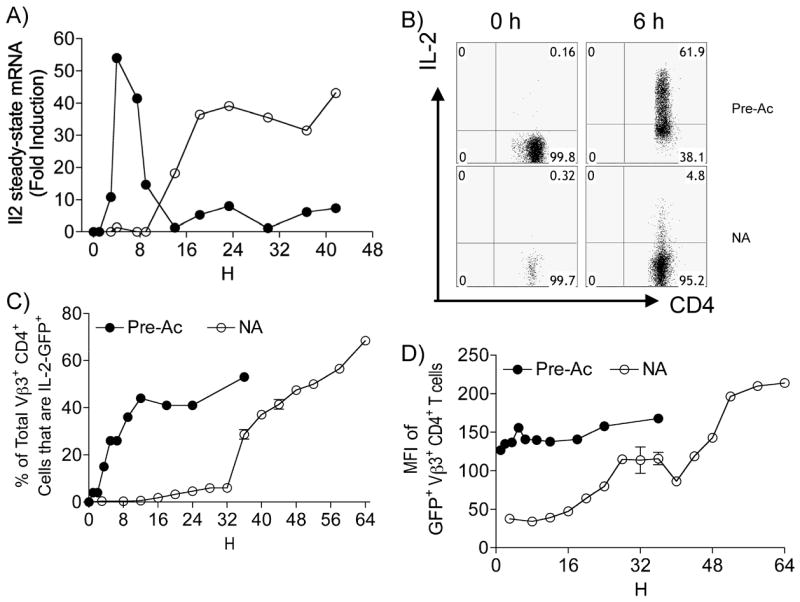
Rapid Il2 transcription in previously activated (Pre-Ac) CD4+ T cells. Freshly isolated naive (○) or resting Pre-Ac (●) WT CD4+ T cells were stimulated with α-TCR-β + α-CD28 for the times indicated. A, Cells were lysed and steady-state Il2 mRNA was quantified using qPCR; data are representative of one of two experiments; values are expressed relative to GAPDH mRNA. B, The frequency of IL-2 producers was determined by in vitro cytokine capture assay; 7-AAD−Vβ3+ cells were gated for analyses; one of four experiments is shown. C, D, Freshly isolated naïve (○) or resting Pre-Ac (●) CD4+ T cells from IL-2-GFPKI//KI mice were stimulated with α-TCR-β + α-CD28 for the times indicated. The frequency (C) and MFI (D) of 7-AAD− Vβ3+ IL-2 GFP+ CD4+ T cells were analyzed by flow cytometry.
IL-2 GFP knock-in mice were then used to determine whether the different rates of Il2 mRNA expression were regulated at the level of promoter activity. The GFP transcript in these transgenic mice lacks the sequence instability elements normally present in the Il2 promoter, and GFP protein stability (T1/2 ~18h) is greater than that of Il2 mRNA (T1/2 < 1h), thus enabling the opportunity to track individual cells as they accumulate over time. As illustrated in Fig. 1C, kinetic analyses of GFP expression reinforced our conclusion of a bimodal Il2 expression pattern in naïve CD4+ T cells and demonstrated regulation at the level of Il2 promoter activity. Pre-Ac T cells showed instead a rapid, unimodal onset that was maximal by 12 h. The magnitude of Il2 expressed within a given naïve cell (mean fluorescence intensity [MFI]) also increased in a time-dependent manner (Fig. 1D). In contrast, maximal IL-2 per cell was evident from the onset in Pre-Ac cells. Overall, these data establish a previously unappreciated time-dependent bimodal regulation of Il2 transcription in naïve T cells and identify a small cohort of naïve CD4+ T cells that upregulate Il2 at a rate similar to that in Pre-Ac T cells.
Accessibility of the Il2 proximal promoter differs between naïve and Pre-Ac CD4+ T cells
Given that competence of a cell to transcribe the Il2 gene is associated with chromatin remodeling at the Il2 promoter, we investigated if accessibility changes might contribute to the rate of onset for Il2 transcription. CD4+ T cells, MNase digestion, RT-PCR, and a series of seven primers spanning the Il2 promoter (−1980 bp to +107 bp) were used to compare chromatin accessibility, as previously described (27). This region of the Il2 locus contains cis acting elements (RE) for transcription factors including NF-κB, AP-1, NFAT, and Oct family members, a CD28RE located at −164 bp to −152 bp, at least one nucleosome positioned at −200 bp to − 60 bp, and at least one and perhaps two regions that have been shown to be DNase I hypersensitive prior to activation, located from −313 to −361 and at the TATA cis element (Fig. 2A) (16, 25, 27, 42). In our studies, we found naïve CD4+ T cells to be slightly but reproducibly more sensitive to MNase digestion relative to fibroblasts, a population of cells that never transcribes the Il2 gene. More importantly, they were significantly less sensitive to digestion than resting Pre-Ac T cells (Fig. 2B). We further show a different time-dependent increase in MNase sensitivity for naïve versus Pre-Ac T cells (Figs. 2C and 2D) that closely parallels the temporal increase in the frequency of IL-2 producers (Fig. 1C). This suggests that the progressively increasing hypersensitivity of nucleosomal DNA might be a consequence of an increased frequency of responding cells rather than a time-dependent change in cell-intrinsic chromatin modification. Maximal MNase sensitivity in naïve T cells (12 fold) occurred at ~40 h and was considerably smaller (50-fold) than the rapid 600-fold increase observed in Pre-Ac cells at 6 h (compare Figs. 2C and 2D).
FIGURE 2.
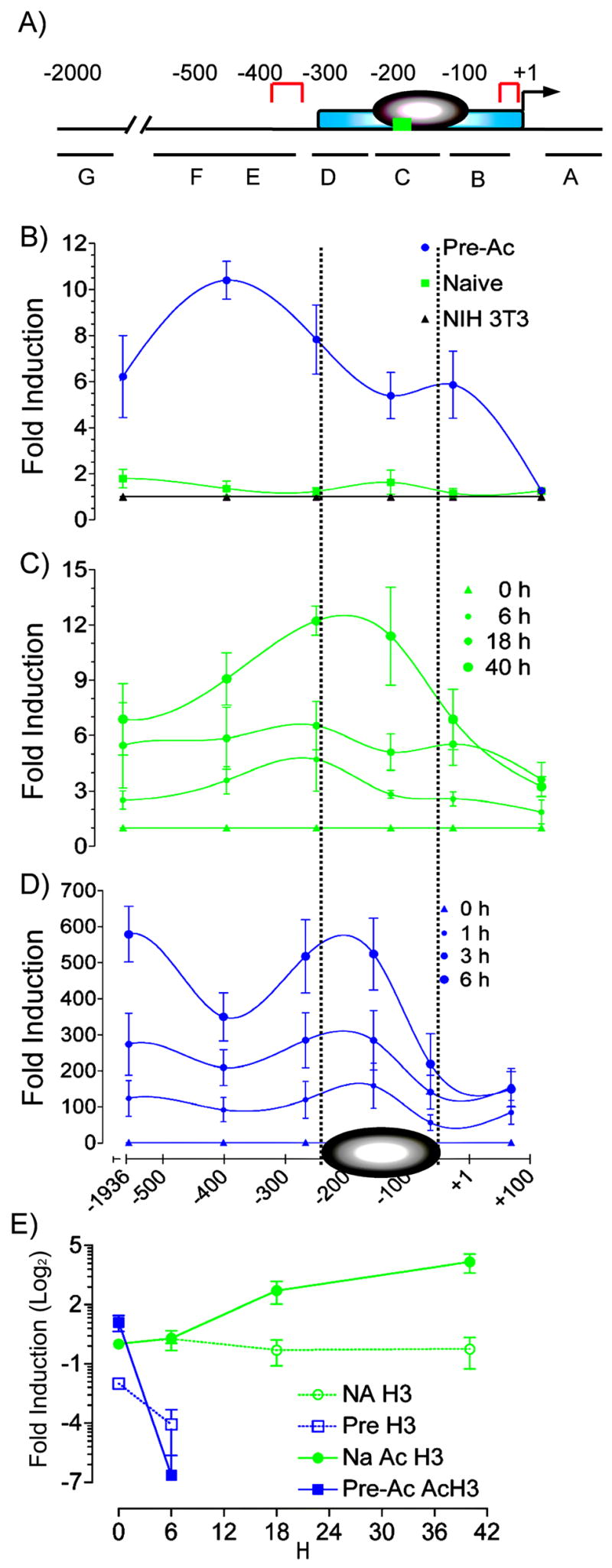
MNase sensitivity at the −300 bp Il2 promoter correlates with the kinetics of Il2 gene transcription. A, The proximal promoter of the mouse Il2 promoter. The minimal essential regulatory region [−300 to +1] (blue), nucleosome [−200 to −60] (gray), hypersensitivity sites in resting cells [−361 to −313 and TATA box] (red) c-Rel κB site [−164 to −152] (green), and the primer sets in the corresponding amplified regions (G = −1982 to −1890; F = −652 to −460; E = −459 to −342; D = −309 to −225; C = −201 to −111; B = −110 to −16; A = +38 to +107) are shown. B, Nuclei were prepared from unstimulated naive (▪) or Pre-Ac (●) WT CD4+ T cells or NIH3T3 fibroblasts (▲) digested with MNase and analyzed using qPCR with seven amplicons across the Il2 gene promoter. The generated Ct values were converted to fold increase relative to undigested genomic DNA using the 2^Ct method. C, Nuclei were isolated from naive CD4+ T cells that were stimulated with α-TCR-β + α-CD28 for 0, 6, 18, or 40 h and analyzed for MNase sensitivity. D, Nuclei were isolated from Pre-Ac CD4+ T cells that were stimulated with α-TCR-β + α-CD28 for 0, 1, 3, or 6 h and analyzed for MNase sensitivity. In both C and D, the generated Ct values for stimulated cells were converted to fold increase relative to respective digested unstimulated controls using the 2Ct method. Means ± SEM are shown. E, ChIP. SYBR® green qPCR and primer set C [−201 to −111] were used to determine total and acetylated histone H3 from sonicated nuclei obtained from naïve and Pre-Ac WT CD4+ T cells following α-TCR-β + α-CD28 for the times indicated. Data are expressed as fold induction relative to unstimulated naïve cells.
To test whether the MNase hypersensitivity associated with activation of Pre-Ac T cells was a consequence of nucleosome loss or altered nucleosome structure and/or composition, we measured total and acetylated levels of histone H3 at the Il2 locus using primers that span the −201 bp to −111 bp region. Total and acetylated H3 were reduced in Pre-Ac cells compared with naïve T cells at 6 h (Fig. 2E). These results correlate an increase in MNase sensitivity with nucleosome loss in re-challenged Pre-Ac T cells in a manner similar to that reported by Shannon and colleagues (30). In contrast to Pre-Ac cells, however, we observed an increase in acetylated histone H3 and no change in total histone H3 in naïve T cells activated for up to 48 h (Fig. 2E). These results suggest that nucleosome loss is not essential for Il2 transcription.
Enhanced basal expression of c-Rel in resting, Pre-Ac CD4+ T cells
c-Rel is essential for IL-2 production in naïve CD4+ T cells, presumably—at least in part—because of its requirement for induction of localized chromatin accessibility at the proximal Il2 promoter following T cell activation (27). A similar role for c-Rel in effector cells is unclear (43). To determine whether c-Rel expression correlates with the rate of induction of Il2 transcription and/or magnitude of MNase accessibility, we used multispectral imaging flow cytometry (41) to quantify the frequency of CD4+ T cells expressing c-Rel. Prior to re challenge, the frequency of Pre-Ac CD4+ T cells producing detectable amounts of c-Rel was 26% (trial 1) to 41% (trial 2) (Fig. 3A). After activation, essentially all (98%) of the Pre-Ac cells rapidly upregulated c-Rel with nuclear accumulation at any given time in 50% to 83% of these. The majority of naïve T cells (up to 89%) also received signaling sufficient to produce detectable c-Rel, albeit quite slowly (Fig. 3A). This slow increase in c-Rel expression paralleled the delayed onset of Il2 transcription described in Fig. 1A. Finally, using ChIP (Fig. 3B), we found that c-rel promoter occupancy was rapidly upregulated in Pre-Ac cells, i.e., within 6 h of T cell stimulation, while c-Rel binding in naïve cells was delayed, i.e., 18 to 40 h following activation.
FIGURE 3.
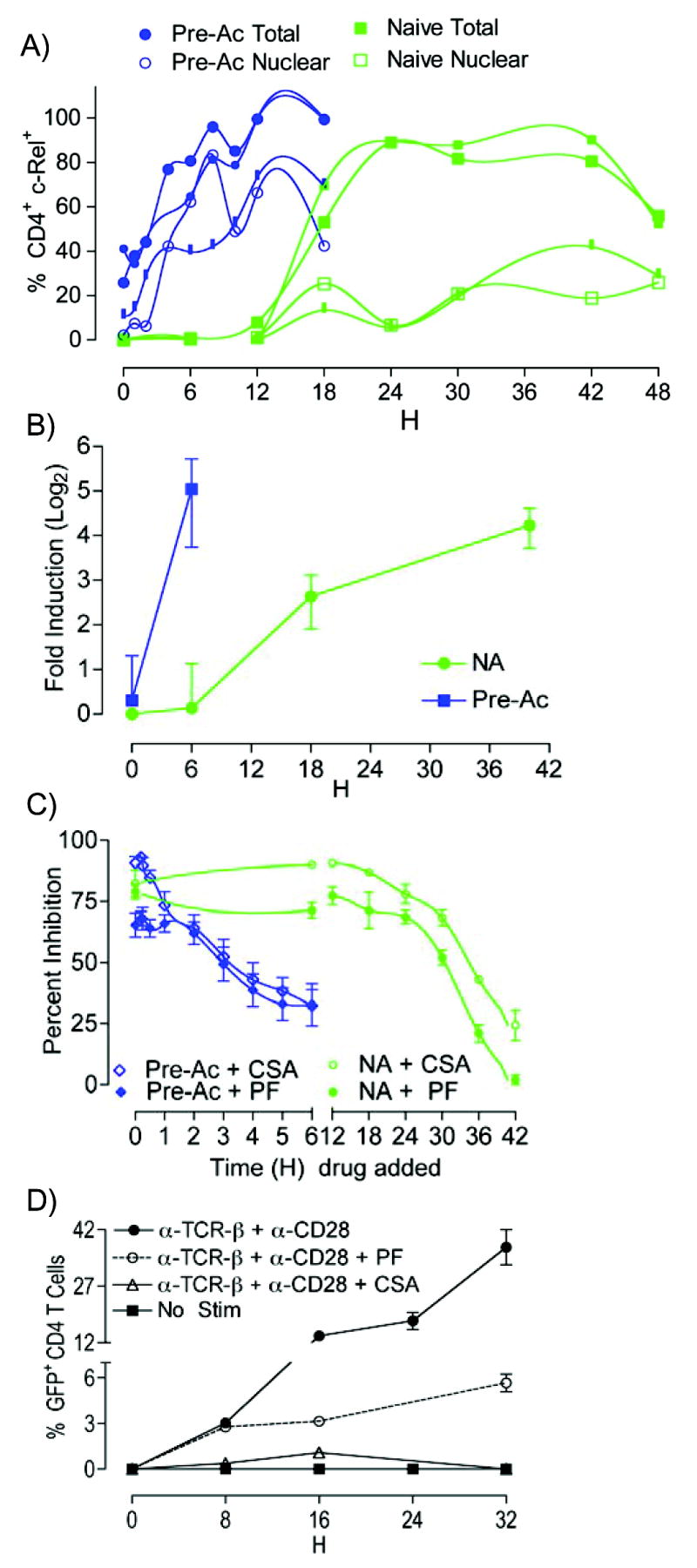
Enhanced basal c-Rel expression in Pre-Ac CD4+ T cells and inhibition by PF. A, Pre-Ac (blue) or naïve (green) WT CD4+ T cells were stimulated with α-TCR-β + α-CD28 for the times indicated, paraformaldehyde fixed, stained with α-c-Rel, α-CD4, and DRAQ5, and then analyzed for intracellular c-Rel expression and c-Rel nuclear translocation using the ImageStream® 100 multispectral imaging flow cytometer. B, ChIP. SYBR® green qPCR, and primer set C [−201 to −111] were used to determine c-Rel promoter occupancy in naïve and Pre-Ac WT CD4+ T cells following α-TCR-β + α-CD28 for the times indicated; data are expressed as fold induction relative to unstimulated naïve cells. C, Pre-Ac (blue) or naïve (green) IL-2-GFPKI/WT CD4+ T cells were stimulated with α-TCR-β + α-CD28; PF or CsA was added at various times after T cell activation as indicated. Cells were harvested at 54 h (naïve) or 24 h (Pre-Ac) and gated on 7-AAD−CD4+ T cells for GFP analyses. D, PF or CsA were added to IL-2-GFPKI/WT CD4+at the time of α-TCR-β + α-CD28 stimulation. Cells were harvested for GFP analyses at the times indicated.
Given the overlapping induction profiles of c-Rel and Il2 following T cell activation in naive and Pre-Ac cells (compare Figs. 1A and 3A), we used a pharmacologic inhibitor of c-Rel transcription, pentoxifylline (PF) to compare the causal role of c-Rel for transcription of Il2 in naïve and Pre-Ac CD4+ T cells. For these experiments, IL-2-GFPWT/KI CD4+ T cells were stimulated with α-TCR-β + α-CD28 in the presence of PF for either 54 h (naïve) or 24 h (Pre-Ac). Addition of PF prior to or at the time of T cell activation reduced the frequency of IL-2 producers by 85% ± 4% (naïve) or 56% ± 11% (Pre-Ac) (Fig. 3C). The subpopulation of naive CD4+ T cells that resisted suppression by PF appears to represent those cells that rapidly transcribe Il2 (early-phase IL-2 producers) (Fig. 3D). Adding cyclosporine A (CsA), an inhibitor of NFAT nuclear translocation, mostly overcame the PF resistance (Figs. 3C and 3D). This suggests that this small cohort of naïve cells that seemingly does not require c-Rel, does in fact need NFAT to transcribe Il2. Nevertheless, induction of Il2 transcription in the majority of naïve CD4+ T cells appears to also require newly synthesized c-Rel - a form that can be readily translocated into the nucleus (39). In contrast, substantially more Pre-Ac cells (44%) transcribed Il2 in the presence of PF. However, because PF inhibits c-rel transcription, the reduced efficacy of this drug may be a consequence of the constitutive c-Rel expression in a large cohort (~ 30% to 40%) of Pre-Ac T cells (Fig. 3A).
TNF-α regulates late phase c-Rel transcription in naïve CD4+ T cells
TNF-α has been shown to regulate c-Rel activity in CD4+ T cells and IL-2 production in CD8+ T cells (44). We hypothesized that TNFR signaling might provide the costimulation necessary for the late-phase Il2 transcription in CD4+ T cells. To test this, CD4+ T cells from B10.A TCR-Cyt 5C.C7 Rag2Δ/Δ TNFR P55Δ/Δ TNFR P75Δ/Δ mice were stimulated with α-TCR-β + α-CD28 in vitro. Supernatants were analyzed for IL-2 by ELISA (Fig. 4A). Relative to WT, IL-2 secretion from the double-deficient TNFR CD4+ T cells was greatly impaired at times after 18 h. This reduction corresponded to a decreased frequency of IL-2 producers (Fig. 4B) but not MFI (Fig. 4C). This suggests that TNFR signaling is required for late-phase IL-2 production. We then stimulated FACS-sorted, purified naïve CD4+ T cells with α-TCR-β + α-CD28 to show that this TNFR requirement was T cell autonomous (Fig. 4D). Unaltered CD69 surface expression (Fig. 4D) indicated unperturbed proximal TCR signaling in TNFRΔ/Δ CD4+ T cells (45). Overall, we interpret these results to mean that T cell autonomous TNFR P55 and/or TNFR P75 costimulation overcomes a threshold for activation of the Il2 promoter during late- but not early-phase IL-2 production.
FIGURE 4.
Defective IL-2 production in TNFRΔ/Δ CD4+ T cells. IL-2 production in naïve WT (●) or TNFRΔ/Δ (○) CD4+ T cells was measured by ELISA (A) or cytokine capture (B, C) at different times after α-TCR-β + α-CD28 stimulation. Data shown are from three separate experiments in triplicate; bar indicates geometric mean. D, CD4+ FACS-sorted WT or TNFRΔ/Δ T cells were stimulated with α-TCR-β + α-CD28 for 0, 6, 18, or 48 h; IL-2 production was quantified using cytokine capture. Data are representative of three separate experiments. Naïve (E) or Pre-Ac (F) WT or TNFRΔ/Δ CD4+ T cells were FACS purified, stimulated with α-TCR-β + α-CD28 for the times indicated, lysed, and analyzed for steady-state c-rel mRNA using qPCR. The data are normalized to GAPDH and expressed as fold induction (mean ± SEM) relative to unstimulated naïve WT and are from three separate experiments. G, ChIP analyses to determine c-Rel association with the Il2 promoter. MNase digested nuclei were obtained from WT or TNFRΔ/Δ CD4+ T cells that had been stimulated with α-TCR-β + α-CD28 for the times indicated. Data are expressed as fold induction relative to unstimulated naïve cells.
Given that new c-rel transcription is required for IL-2 production (43), we hypothesized that reduced c-Rel expression may contribute to impaired Il2 expression in naïve CD4+ T cells from TNFRΔ/Δ -deficient mice. To test this, we quantified c-rel mRNA following activation of CD4+ T cells from WT and TNFRΔ/Δ mice. Constitutive c-rel mRNA was detectable in both naïve and Pre-Ac cells but was 5-fold greater in Pre-Ac T cells (Fig. 4E vs. Fig. 4F). Following activation, naïve CD4+ T cells rapidly upregulated c-rel mRNA (plateau at 2 h; data not shown), and this was followed by a persistent, slow and steady increase during the 18 to 36 h window after T cell activation. This second, prolonged wave was not seen in the TNFRΔ/Δ cells, and suggests that TNFR signaling is required to maintain maximal expression beyond the initial early burst of Il2 transcription (Fig. 4E). The effect of TNFR deficiency in Pre-Ac CD4+ T cells was different. Specifically, the basal level was lower and the time it took to reach peak c-rel mRNA production was slower; however the magnitude of c-rel mRNA achieved by 4 h was not significantly different from that of WT CD4+ T cells (Fig. 4F). These data establish that regulation of c-Rel expression differs between naïve and Pre-Ac CD4+ T cells and suggest that TNFR signaling may function to maintain a threshold of bioavailable c-Rel necessary for late-phase Il2 promoter activity in naïve cells. In agreement with this hypothesis, Fig. 4G shows that c-Rel binding to the proximal Il2 promoter in a ChIP assay was also impaired in naïve CD4+ cells from TNFRΔ/Δ mice following T cell activation.
CD28 but not TNFR costimulation is required for optimal induction of Il2 transcription in resting, Pre-Ac CD4+ T cells
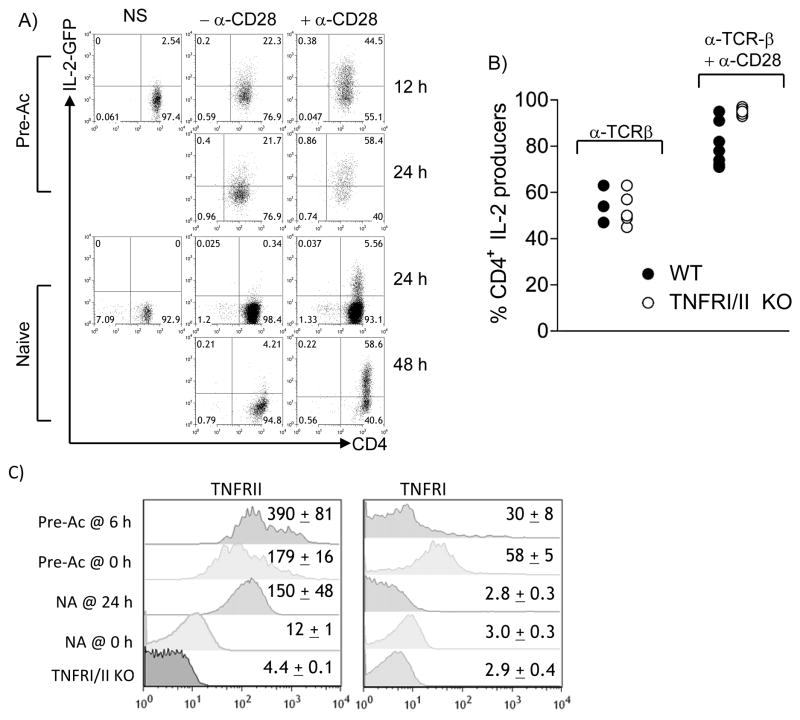
We next compared the relative roles of TNFR and CD28 costimulation in IL-2 production in naïve and Pre-Ac CD4+ T cells. CD28/B7 costimulation is critical for IL-2 production in naïve CD4+ T cells (10); however, CD28-independent IL-2 production has been suggested for effector CD4+ T cells (46). Moreover, augmentation of IL-2 production by CD28 has been suggested to be entirely c-Rel dependent (43). This is consistent with CD28-mediated degradation of the inhibitory IκBα (43) and a c-Rel binding κB motif in the Il2 promoter (22). Using FACS-purified IL-2-GFPKI/KI CD4+ T cells, we observed that CD28 costimulation is required for Il2 transcription in both naïve and Pre-Ac CD4+ T cells, although the latter are only partially dependent (Fig. 5A). Early (24 h) as well as late (48 h) Il2 transcription in naïve IL-2-GFPKI/KI CD4+ T cells stimulated with α-TCR alone was barely detectable (Fig. 5A). In contrast, by 24 h, 22% of the total Pre-Ac CD4+ T cells responded to α-TCR in the absence of CD28 (Fig. 5A). Nonetheless, costimulation provided by α-CD28 increased the IL-2-GFP+ frequency at 24 h to 58% and the production of GFP by individual cells (MFI) by 50-fold. This partial requirement for CD28 costimulation in Pre-Ac cells is consistent with the partial c-Rel-dependent IL-2 production in most Pre-Ac T cells (see Fig. 3C).
FIGURE 5.
CD28 but not TNFR costimulation is required for optimal induction of Il2 transcription in resting, Pre-Ac CD4+ T cells. (A) IL-2-GFP expression in naïve or Pre-Ac IL-2-GFPKI/WT CD4+ T cells following stimulation with α-TCR-β alone (-α -CD28) or α-TCR-β + α-CD28 for the times indicated; cells are gated on 7-AAD−Vβ3+ for analyses. (B) Pre-Ac WT (●) or TNFRΔ/Δ (○) CD4+ T cells were re-challenged with α-TCR-β or α-TCR-β + α-CD28; IL-2 production is reported as the frequency of CD4+ T cells producing IL-2 at 6 h Data from (A) and (B) are representative of two separate experiments. (C) Naïve or Pre-Ac WT CD4+ T cells were stimulated with α-TCR-β + α-CD28 for 24 h or 6 h, respectively and gated on CD4 for analyses. Unstimulated lymph node CD4+ T cells from TNFRΔ/Δ mice are also gated for negative controls. The MFI ± SEM for three separate experiments of TNFRI and TNFRII are shown. Data shown is from one of three representative experiments
Upregulation of TNFR II following activation of naïve T cells
Unlike CD28 costimulation, TNFR signaling was not required for IL-2 production during re-challenge (Fig. 5B), in contrast to its major role in the late phase of IL-2 production by naïve T cells (Fig. 4A). To look more closely at this pathway, we examined TNF receptor expression on naïve and Pre-Ac CD4+ T cells. TNFRII expression is low, but detectable, in resting naïve cells and was upregulated 10 fold upon activation (Fig. 5C). TNFRII expression remained elevated in resting Pre-Ac cells and further increased during re-challenge. In contrast, the small amount of TNFRI in resting naïve cells was down-regulated upon activation. TNFRI expression was highest in resting Pre-Ac cells, and this was also rapidly down-regulated upon activation. Thus, the lack of an apparent role for TNFR in IL-2 production in Pre-Ac cells does not stem from an absence of TNF receptors. Instead, CD28 costimulation alone may be adequate for c-Rel induction in Pre-Ac cells. Alternatively, other inducible TNFR/TNF ligand superfamily members—including CD134 (OX40)/OX40L, CD137 (4-1BB)/4-1BBL, HVEM/LIGHT, CD30/CD30L, GITR/GITRL, or CD27/CD70 may compensate for the loss of TNFR/TNF signaling. Finally, the upregulation of TNFR II following activation of naïve T cells may be the rate limiting step required for late IL-2 production.
Discussion
The kinetics of IL-2 production differs between naïve and memory CD4+ T cells. This change in IL-2 bioavailability can significantly alter effector function and the outcome of an adaptive immune response. Rapid IL-2 production in memory CD4+ T cells is attributed to a host of factors including recognition of a wider range of antigen presenting cells, a lower threshold for activation, and less stringency for costimulation (8); yet the molecular mechanism(s) underlying the kinetics of Il2 transcription remain enigmatic. Here, we contribute to the unraveling of this important question by presenting new mechanistic information regarding the regulation of Il2 promoter activity. First, we identify a novel biphasic regulation of Il2 transcription in naïve CD4+ T cells. Second, we establish that chromatin structure and histone modifications at the proximal Il2 promoter differ markedly between naïve and Pre-Ac CD4+ T cells. Third, we report enhanced c-Rel expression in resting Pre-Ac CD4+ T cells, and suggest that this transcription factor may contribute to more extensive chromatin remodeling. Fourth, we suggest that the presence of a nucleosome in the proximal Il2 promoter may play a role in delaying the time of onset for Il2 transcription, but that it does not serve as an all-or-nothing ON/OFF-switch for Il2 transcription initiation. Lastly, we identify a role for TNFR signaling as a critical mediator of c-Rel activity, and presumably chromatin remodeling and IL-2 production in naïve, but not Pre-Ac CD4+ T cells.
Our present results expand upon earlier experiments suggesting that naïve CD4+ T cells transcribe only a small amount of Il2 mRNA in comparison to Pre-Ac T cells. Using single cell analysis of Il2 promoter activity, we show that most naïve CD4+ T cells, in fact, do transcribe Il2–but with delayed kinetics (Fig. 1). The time-dependent increase in the MFI pattern of GFP expression in naïve T cells (Fig. 1D) is consistent with independent transcriptional competence at each of the two Il2 alleles. These new findings further support our previously described two-step quantitative biallelic model for expression of the Il2 locus (47, 48). In contrast to naïve T cells, GFP expression in Pre-Ac cells was rapid and MFI was maximal from the onset. This is consistent with rapid promoter activity at both alleles. This kinetic contrast in Il2 transcription between naïve and Pre-Ac T cells led us to speculate that there was differential regulation at the level of chromatin structure.
In agreement with this hypothesis, our MNase hypersensitivity assays showed a striking difference in accessibility at the regulatory region of the Il2 promoter between naïve and Pre-Ac CD4+ T cells (Fig. 2B). DNA from resting Pre-Ac cells was more readily digested in comparison to unstimulated naïve cells. This observation establishes, for the first time, a stable increase in DNA accessibility at the Il2 locus following antigen priming. We speculate that a poised chromatin structure in resting Pre-Ac cells permits rapid binding of readily available c-Rel (Fig. 3A), NFAT (49), high mobility group proteins (32) and perhaps other trans-acting transcription factors (50, 51) to their respective cognate response elements within the Il2 proximal promoter to potentiate rapid chromatin remodeling and biallelic transcription (48). Following activation, both naïve and Pre-Ac T cells showed an increase in MNase hypersensitivity; however, the effect in Pre-Ac cells was 50 fold greater (Fig. 2C and D). We attribute this difference to nucleosome loss seen only in Pre-Ac T cells (Fig 2E). The ChIP assays indicate differences at the level of histone modification between naïve and Pre-Ac cells. Specifically, we observed a decrease in both Ac-H3 and total H3 following re-challenge of Pre-Ac cells; presumably the observed H3 hypoacetylation reflects nucleosome eviction. As yet, it is unclear whether the nucleosome is lost from the DNA in trans or rather removed by sliding along the DNA in cis (52, 53). Nonetheless, we think that this nucleosome is removed from both of the Il2 alleles upon re-challenge, and that this removal accounts for the rapid and maximal GFP expression from the onset of transcription in Pre-Ac cells. In contrast, there was no change in the total amount of H3 associated with the Il2 promoter in naïve T cells. Instead, the amount of Ac-H3 increased. This suggests that the nucleosome remains at each of the two Il2 alleles during antigen priming of naïve cells, although transcription is eventually initiated despite its presence. These findings are supported by previous observations from other labs reporting a loss of total H3 and Ac-H3 following activation of EL-4 cells, a “Pre-Ac” tumor population (30, 54). In contrast, Wells et. al., (55) recently showed an increased Ac-H3 with no change in total H3 following activation of a population of around 85% naïve CD4+ cells. Our naïve versus Pre-Ac results now provide a plausible biological explanation for the accuracy of both of these sets of experiments.
Collectively, our results show that removal of a pre-existing nucleosome in the proximal Il2 promoter is not essential for transcription. RNA polymerase II (Pol II) can in fact elongate through DNA containing a nucleosome, but the process is cumbersome and slow (56). We think sustained transcription through a nucleosome accounts for the ‘late phase’ in our biphasic model for Il2 transcription and for the eventual ability to produce as much IL-2 as seen with Pre-Ac cells. After initial activation, however, we propose that a stable chromatin remodeling takes place that facilitates nucleosome loss during re-challenge and removal of this rate-limiting step for a rapid onset of Il2 transcription initiation, during restimulation. Despite this more rapid onset, however, the final total IL-2 mRNA and protein production by Pre-Ac T cells is not much greater than that of naïve T cells (Fig 1A and unpublished data). This is because IL-2 mRNA levels also decrease quickly in Pre-Ac T cells following peak activation (Fig. 1A). The mechanism for this might be a decreased mRNA stability in Pre-Ac T cells (57, 58) and/or a greater autocrine negative feedback loop by the burst of IL-2 protein on Il2 transcription -mediated by STAT5 signaling through the IL-2R and induction of the transcriptional repressor Blimp-1 (59–62).
What about the small percentage of naïve T cells that respond early? These cells are from Rag2Δ/Δ TCR transgenic mice and show no surface-marker or size evidence for pre-activation. Nonetheless, their early IL-2 production following α-TCR stimulation behaves in several ways like that of Pre-Ac T cells. Their Il2 transcription (GFP+) is resistant to PF (Fig. 3D) and does not require TNFR signaling (Fig. 4A). It is, however, totally dependent on CD28 costimulation (Fig, 5A) and the MFI of its response (Fig. 1D) suggests that only one allele is being actively transcribed. It is tempting to speculate that this minor population, where there is evidence of rapid, monoallelic and perhaps stochastic Il2 expression, arises from Pol II elongation through a nucleosome-freed promoter.
The question of what determines nucleosome loss at the proximal Il2 promoter is now raised. As alluded to earlier, cell signaling, chromatin remodeling, and histone modifications have all been proposed to destabilize nucleosome interactions with the DNA. Although the underlying mechanism(s) remain unknown, c-Rel convincingly plays a role in chromatin remodeling of several pro-inflammatory genes, including Il2 (27, 63). Naïve CD4+ T cells from c-Rel-deficient mice do not transcribe Il2 and chromatin at the proximal Il2 promoter does not remodel upon T cell activation (27, 43). We now show that c-Rel is at least partially required for Il2 transcription during re-challenge. In addition, c-Rel is over-expressed and more rapidly up-regulated in Pre-Ac CD4+ T cells (Fig. 3A). It is also more readily chromatin bound (Fig. 3B). We correlate c-Rel abundance with a poised Il2 locus in resting Pre-Ac CD4+ T cells and propose that overexpression of c-Rel may facilitate nucleosome displacement by rapidly recruiting other transcription factors (64, 65) and/or TFIID-containing transcription complexes (66). Those cells that do not require c-Rel for Il2 transcription (Fig. 3C) may in fact already have their nucleosomes removed (Fig. 2E). These cells may represent a residual cohort of non-quiescent effector cells that harbor an open chromatin configuration reflective of S-phase DNA generated through “TCR tickling” or peripheral antigen cross-reactivity (67). This hypothesis is consistent with earlier studies showing unperturbed production of IL-2 in antigen-induced effector CD4+ T cells from c-Rel deficient mice (43). Alternatively, the c-Rel-independent cells reflect those cells that have undergone stochastic epigenetic remodelling at the transcriptionally permissive Il2 locus (68).
Finally, TNFR signaling appears to cooperate with TCR signaling and CD28 costimulation to provide a previously undescribed regulation of late, but not early, phase Il2 transcription during antigen priming of naïve CD4+ T cells. Specifically, in naïve T cells, c-Rel is sequestered in the cytoplasm, predominantly as a complex with the inhibitory IκBβ; in contrast, p65/RelA is complexed with both IκBα and IκBβ (37). TCR and CD28 signaling selectively degrade IκBα (69). As a consequence, the majority of NF-κB proteins translocated into the nucleus early after TCR ligation are p65/RelA. These κB proteins are insufficient in most of the T cells to induce the necessary chromatin remodeling of the Il2 locus essential for promoter activity (27). A third signal is required to activate c-Rel/IκBβ. Inducible TNFR can help provide this signal (Fig. 5C). TNFR signaling not only leads to the ubiquitination of IκBβ, but also induces synthesis of c-Rel and IκBα. This ultimately shifts c-Rel mostly into an IκBα complex (43) -a form degradable by TCR signaling. It is likely that different concentrations of c-Rel inhibitory partners, including IκBα, IκBβ, and IκBε, which control nuclear to cytoplasmic oscillation of NF-κB function over time, contribute to the difference in c-Rel nuclear expression between naïve and Pre-Ac cells (Fig. 3A). Alternatively, new c-Rel synthesis in Pre-Ac cells, exceeding the amount of newly synthesized IκBα, will result in enhanced nuclear localization. In our studies and others, abundant constitutive c-Rel, and NFAT (70) in resting, Pre-Ac cells is insufficient for IL-2 production in the absence of TCR signaling (compare Figs. 3A and 1B) and reinforces the concept that cooperative interaction of multiple transcription factors is required to drive transcription at this promoter (20). Using other genetic mutants of Rag2Δ/Δ TCR Tg mouse models, we have determined that neither IL-6 nor IL-1R deficiency impairs IL-2 production in naïve CD4+ T cells (data not shown). This potentially makes TNFR the critical signaling component necessary to achieve the threshold level of c-Rel activity required for Il2 promoter activity. We propose that the small amount of TCR/CD28-inducible, preformed c-Rel/IkBα in naïve and a relatively greater amount in Pre-Ac T cells accounts for the early-phase chromatin remodeling of the Il2 locus, while late-phase IL-2 production in the majority of the naïve T cells is delayed until induction of TNFR II and sufficient concentrations of TNF-α. Addition of soluble TNF-α (up to 10 ng/ml) to the culture medium shortly after T cell activation did not shorten the time to onset of Il2 transcription in naïve T cells (data not shown). Thus, the increase in TNFR II would appear to be the rate limiting step. We are also exploring the possibility of a role for membrane-bound TNF-α in the signaling (71). Overall, we propose a model in which nucleosome loss during re-challenge, but not during antigen priming, facilitates rapid Il2 induction in CD4+ T cells. We suggest that a threshold level of c-Rel must be reached for Il2 promoter activity in both naïve and Pre-Ac CD4+ T cells, and that this is provided, at least partially, by TNFR signaling during priming, but not during recall.
Acknowledgments
We thank Dr. Phil Morrissey for acquiring and analyzing all of the c-Rel multispectral images; Drs. Ann Dean, Gordon Hager, Christine Keifer, Sam John, and Ainhoa Perez-Diez for valuable discussions and technical assistance; Betsy Majane and Dr. Charles Mainhart for maintenance of mouse models; Dr. Kevin Holmes, Carol Henry, Calvin Eigsti, and Tom Moyer for FACS sorting; Scott Greathouse for technical assistance with the graphics; and Drs. Mike Pazin and Howard Young for valuable comments on the manuscript.
Footnotes
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Disclosures
The authors have no financial conflict of interest.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; CsA, cyclosporin A; Ct, threshold cycle; MFI, mean fluorescence intensity; MNase, micrococcal nuclease; PF, pentoxifylline; Pre-Ac, previously activated.
References
- 1.Bachmann MF, Oxenius A. Interleukin 2: from immunostimulation to immunoregulation and back again. EMBO Rep. 2007;8:1142–1148. doi: 10.1038/sj.embor.7401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 3.Schorle H, Holtschke T, Hunig T, Schimpl A, Horak I. Development and function of T cells in mice rendered interleukin-2 deficient by gene targeting. Nature. 1991;352:621–624. doi: 10.1038/352621a0. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H, Kundig TM, Furlonger C, Wakeham A, Timms E, Matsuyama T, Schmits R, Simard JJ, Ohashi PS, Griesser H, et al. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 1995;268:1472–1476. doi: 10.1126/science.7770771. [DOI] [PubMed] [Google Scholar]
- 5.Willerford DM, Chen J, Ferry JA, Davidson L, Ma A, Alt FW. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 1995;3:521–530. doi: 10.1016/1074-7613(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 6.Schimpl A, Berberich I, Kneitz B, Kramer S, Santner-Nanan B, Wagner S, Wolf M, Hunig T. IL-2 and autoimmune disease. Cytokine Growth Factor Rev. 2002;13:369–378. doi: 10.1016/s1359-6101(02)00022-9. [DOI] [PubMed] [Google Scholar]
- 7.Horak I. Immunodeficiency in IL-2-knockout mice. Clin Immunol Immunopathol. 1995;76:S172–173. doi: 10.1016/s0090-1229(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 8.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–223. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi T, Kuniyasu Y, Toda M, Sakaguchi N, Itoh M, Iwata M, Shimizu J, Sakaguchi S. Immunologic self-tolerance maintained by CD25+CD4+ naturally anergic and suppressive T cells: induction of autoimmune disease by breaking their anergic/suppressive state. Int Immunol. 1998;10:1969–1980. doi: 10.1093/intimm/10.12.1969. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz RH. T cell anergy. Annu Rev Immunol. 2003;21:305–334. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fehervari Z, Yamaguchi T, Sakaguchi S. The dichotomous role of IL-2: tolerance versus immunity. Trends Immunol. 2006;27:109–111. doi: 10.1016/j.it.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Malek TR, Bayer AL. Tolerance, not immunity, crucially depends on IL-2. Nat Rev Immunol. 2004;4:665–674. doi: 10.1038/nri1435. [DOI] [PubMed] [Google Scholar]
- 14.Nelson BH. IL-2, regulatory T cells, and tolerance. J Immunol. 2004;172:3983–3988. doi: 10.4049/jimmunol.172.7.3983. [DOI] [PubMed] [Google Scholar]
- 15.Acuto O, Mise-Omata S, Mangino G, Michel F. Molecular modifiers of T cell antigen receptor triggering threshold: the mechanism of CD28 costimulatory receptor. Immunol Rev. 2003;192:21–31. doi: 10.1034/j.1600-065x.2003.00034.x. [DOI] [PubMed] [Google Scholar]
- 16.Bunting K, Wang J, Shannon MF. Control of interleukin-2 gene transcription: a paradigm for inducible, tissue-specific gene expression. Vitam Horm. 2006;74:105–145. doi: 10.1016/S0083-6729(06)74005-5. [DOI] [PubMed] [Google Scholar]
- 17.Jain J, Loh C, Rao A. Transcriptional regulation of the IL-2 gene. Curr Opin Immunol. 1995;7:333–342. doi: 10.1016/0952-7915(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 18.Serfling E, Avots A, Neumann M. The architecture of the interleukin-2 promoter: a reflection of T lymphocyte activation. Biochim Biophys Acta. 1995;1263:181–200. doi: 10.1016/0167-4781(95)00112-t. [DOI] [PubMed] [Google Scholar]
- 19.Himes SR, Reeves R, Attema J, Nissen M, Li Y, Shannon MF. The role of high-mobility group I(Y) proteins in expression of IL-2 and T cell proliferation. J Immunol. 2000;164:3157–3168. doi: 10.4049/jimmunol.164.6.3157. [DOI] [PubMed] [Google Scholar]
- 20.Rothenberg EV, Ward SB. A dynamic assembly of diverse transcription factors integrates activation and cell-type information for interleukin 2 gene regulation. Proc Natl Acad Sci U S A. 1996;93:9358–9365. doi: 10.1073/pnas.93.18.9358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crabtree GR, Clipstone NA. Signal transmission between the plasma membrane and nucleus of T lymphocytes. Annu Rev Biochem. 1994;63:1045–1083. doi: 10.1146/annurev.bi.63.070194.005145. [DOI] [PubMed] [Google Scholar]
- 22.Shapiro VS, Truitt KE, Imboden JB, Weiss A. CD28 mediates transcriptional upregulation of the interleukin-2 (IL-2) promoter through a composite element containing the CD28RE and NF-IL-2B AP-1 sites. Mol Cell Biol. 1997;17:4051–4058. doi: 10.1128/mcb.17.7.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verweij CL, Geerts M, Aarden LA. Activation of interleukin-2 gene transcription via the T-cell surface molecule CD28 is mediated through an NF-kB-like response element. J Biol Chem. 1991;266:14179–14182. [PubMed] [Google Scholar]
- 24.Siebenlist U, Durand DB, Bressler P, Holbrook NJ, Norris CA, Kamoun M, Kant JA, Crabtree GR. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol Cell Biol. 1986;6:3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward SB, Hernandez-Hoyos G, Chen F, Waterman M, Reeves R, Rothenberg EV. Chromatin remodeling of the interleukin-2 gene: distinct alterations in the proximal versus distal enhancer regions. Nucleic Acids Res. 1998;26:2923–2934. doi: 10.1093/nar/26.12.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao S, Procko E, Shannon MF. Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol. 2001;167:4494–4503. doi: 10.4049/jimmunol.167.8.4494. [DOI] [PubMed] [Google Scholar]
- 27.Rao S, Gerondakis S, Woltring D, Shannon MF. c-Rel is required for chromatin remodeling across the IL-2 gene promoter. J Immunol. 2003;170:3724–3731. doi: 10.4049/jimmunol.170.7.3724. [DOI] [PubMed] [Google Scholar]
- 28.Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, Shibuya A, Kawabe Y, Yanagisawa J. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas RM, Gao L, Wells AD. Signals from CD28 induce stable epigenetic modification of the IL-2 promoter. J Immunol. 2005;174:4639–4646. doi: 10.4049/jimmunol.174.8.4639. [DOI] [PubMed] [Google Scholar]
- 30.Chen X, Wang J, Woltring D, Gerondakis S, Shannon MF. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol Cell Biol. 2005;25:3209–3219. doi: 10.1128/MCB.25.8.3209-3219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- 32.Attema JL, Reeves R, Murray V, Levichkin I, Temple MD, Tremethick DJ, Shannon MF. The human IL-2 gene promoter can assemble a positioned nucleosome that becomes remodeled upon T cell activation. J Immunol. 2002;169:2466–2476. doi: 10.4049/jimmunol.169.5.2466. [DOI] [PubMed] [Google Scholar]
- 33.Tanchot C, Barber DL, Chiodetti L, Schwartz RH. Adaptive tolerance of CD4+ T cells in vivo: multiple thresholds in response to a constant level of antigen presentation. J Immunol. 2001;167:2030–2039. doi: 10.4049/jimmunol.167.4.2030. [DOI] [PubMed] [Google Scholar]
- 34.Naramura M, Hu RJ, Gu H. Mice with a fluorescent marker for interleukin 2 gene activation. Immunity. 1998;9:209–216. doi: 10.1016/s1074-7613(00)80603-2. [DOI] [PubMed] [Google Scholar]
- 35.Peschon JJ, Torrance DS, Stocking KL, Glaccum MB, Otten C, Willis CR, Charrier K, Morrissey PJ, Ware CB, Mohler KM. TNF receptor-deficient mice reveal divergent roles for p55 and p75 in several models of inflammation. J Immunol. 1998;160:943–952. [PubMed] [Google Scholar]
- 36.Wang W, Tam WF, Hughes CC, Rath S, Sen R. c-Rel is a target of pentoxifylline-mediated inhibition of T lymphocyte activation. Immunity. 1997;6:165–174. doi: 10.1016/s1074-7613(00)80423-9. [DOI] [PubMed] [Google Scholar]
- 37.Randak C, Brabletz T, Hergenrother M, Sobotta I, Serfling E. Cyclosporin A suppresses the expression of the interleukin 2 gene by inhibiting the binding of lymphocyte-specific factors to the IL-2 enhancer. EMBO J. 1990;9:2529–2536. doi: 10.1002/j.1460-2075.1990.tb07433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKarns SC, Schwartz RH. Distinct effects of TGF-beta 1 on CD4+ and CD8+ T cell survival, division, and IL-2 production: a role for T cell intrinsic Smad3. J Immunol. 2005;174:2071–2083. doi: 10.4049/jimmunol.174.4.2071. [DOI] [PubMed] [Google Scholar]
- 39.Gong QH, McDowell JC, Dean A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the epsilon-globin gene in vivo by 5′ hypersensitive site 2 of the beta-globin locus control region. Mol Cell Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.George TC, Fanning SL, Fitzgeral-Bocarsly P, Medeiros RB, Highfill S, Shimizu Y, Hall BE, Frost K, Basiji D, Ortyn WE, Morrissey PJ, Lynch DH. Quantitative measurement of nuclear translocation events using similarity analysis of multispectral cellular images obtained in flow. J Immunol Methods. 2006;311:117–129. doi: 10.1016/j.jim.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 42.Brunvand MW, Krumm A, Groudine M. In vivo footprinting of the human IL-2 gene reveals a nuclear factor bound to the transcription start site in T cells. Nucleic Acids Res. 1993;21:4824–4829. doi: 10.1093/nar/21.20.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee D, Liou HC, Sen R. c-Rel-dependent priming of naive T cells by inflammatory cytokines. Immunity. 2005;23:445–458. doi: 10.1016/j.immuni.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Pimentel-Muinos FX, Mazana J, Fresno M. Biphasic control of nuclear factor-kappa B activation by the T cell receptor complex: role of tumor necrosis factor alpha. Eur J Immunol. 1995;25:179–186. doi: 10.1002/eji.1830250130. [DOI] [PubMed] [Google Scholar]
- 45.Kim EY, Teh HS. Critical role of TNF receptor type-2 (p75) as a costimulator for IL-2 induction and T cell survival: a functional link to CD28. J Immunol. 2004;173:4500–4509. doi: 10.4049/jimmunol.173.7.4500. [DOI] [PubMed] [Google Scholar]
- 46.Dubey C, Croft M. Accessory molecule regulation of naive CD4 T cell activation. Immunol Res. 1996;15:114–125. doi: 10.1007/BF02918501. [DOI] [PubMed] [Google Scholar]
- 47.Umlauf SW, Beverly B, Kang SM, Brorson K, Tran AC, Schwartz RH. Molecular regulation of the IL-2 gene: rheostatic control of the immune system. Immunol Rev. 1993;133:177–197. doi: 10.1111/j.1600-065x.1993.tb01516.x. [DOI] [PubMed] [Google Scholar]
- 48.Chiodetti L, Barber DL, Schwartz RH. Biallelic expression of the IL-2 locus under optimal stimulation conditions. Eur J Immunol. 2000;30:2157–2163. doi: 10.1002/1521-4141(2000)30:8<2157::AID-IMMU2157>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 49.Dienz O, Eaton SM, Krahl TJ, Diehl S, Charland C, Dodge J, Swain SL, Budd RC, Haynes L, Rincon M. Accumulation of NFAT mediates IL-2 expression in memory, but not naive, CD4+ T cells. Proc Natl Acad Sci U S A. 2007;104:7175–7180. doi: 10.1073/pnas.0610442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204:971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi L, Qiu D, Zhao G, Corthesy B, Lees-Miller S, Reeves WH, Kao PN. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35:2302–2310. doi: 10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14:667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 53.Korber P, Luckenbach T, Blaschke D, Horz W. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol Cell Biol. 2004;24:10965–10974. doi: 10.1128/MCB.24.24.10965-10974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adachi S, Rothenberg EV. Cell-type-specific epigenetic marking of the IL2 gene at a distal cis-regulatory region in competent, nontranscribing T-cells. Nucleic Acids Res. 2005;33:3200–3210. doi: 10.1093/nar/gki637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas RM, Chunder N, Chen C, Umetsu SE, Winandy S, Wells AD. Ikaros enforces the costimulatory requirement for IL2 gene expression and is required for anergy induction in CD4+ T lymphocytes. J Immunol. 2007;179:7305–7315. doi: 10.4049/jimmunol.179.11.7305. [DOI] [PubMed] [Google Scholar]
- 56.Ercan S, Carrozza MJ, Workman JL. Global nucleosome distribution and the regulation of transcription in yeast. Genome Biol. 2004;5:243. doi: 10.1186/gb-2004-5-10-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindstein T, June CH, Ledbetter JA, Stella G, Thompson CB. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 58.Umlauf SW, Beverly B, Lantz O, Schwartz RH. Regulation of interleukin 2 gene expression by CD28 costimulation in mouse T-cell clones: both nuclear and cytoplasmic RNAs are regulated with complex kinetics. Mol Cell Biol. 1995;15:3197–3205. doi: 10.1128/mcb.15.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 60.Martins GA, Cimmino L, Shapiro-Shelef M, Szabolcs M, Herron A, Magnusdottir E, Calame K. Transcriptional repressor Blimp-1 regulates T cell homeostasis and function. Nat Immunol. 2006;7:457–465. doi: 10.1038/ni1320. [DOI] [PubMed] [Google Scholar]
- 61.Villarino AV, Tato CM, Stumhofer JS, Yao Z, Cui YK, Hennighausen L, O’Shea JJ, Hunter CA. Helper T cell IL-2 production is limited by negative feedback and STAT-dependent cytokine signals. J Exp Med. 2007;204:65–71. doi: 10.1084/jem.20061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jameson SC. T cells climb on board Blimp-1. Trends Immunol. 2006;27:349–351. doi: 10.1016/j.it.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Brettingham-Moore KH, Rao S, Juelich T, Shannon MF, Holloway AF. GM-CSF promoter chromatin remodelling and gene transcription display distinct signal and transcription factor requirements. Nucleic Acids Res. 2005;33:225–234. doi: 10.1093/nar/gki161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu X, Prorock C, Ishikawa H, Maldonado E, Ito Y, Gelinas C. Functional interaction of the v-Rel and c-Rel oncoproteins with the TATA-binding protein and association with transcription factor IIB. Mol Cell Biol. 1993;13:6733–6741. doi: 10.1128/mcb.13.11.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cha-Molstad H, Young DP, Kushner I, Samols D. The interaction of C-Rel with C/EBPbeta enhances C/EBPbeta binding to the C-reactive protein gene promoter. Mol Immunol. 2007;44:2933–2942. doi: 10.1016/j.molimm.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 66.Kerr LD, Ransone LJ, Wamsley P, Schmitt MJ, Boyer TG, Zhou Q, Berk AJ, Verma IM. Association between proto-oncoprotein Rel and TATA-binding protein mediates transcriptional activation by NF-kappa B. Nature. 1993;365:412–419. doi: 10.1038/365412a0. [DOI] [PubMed] [Google Scholar]
- 67.Geginat J, Campagnaro S, Sallusto F, Lanzavecchia A. TCR-independent proliferation and differentiation of human CD4+ T cell subsets induced by cytokines. Adv Exp Med Biol. 2002;512:107–112. doi: 10.1007/978-1-4615-0757-4_14. [DOI] [PubMed] [Google Scholar]
- 68.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- 69.Zhou XY, Yashiro-Ohtani Y, Nakahira M, Park WR, Abe R, Hamaoka T, Naramura M, Gu H, Fujiwara H. Molecular mechanisms underlying differential contribution of CD28 versus non-CD28 costimulatory molecules to IL-2 promoter activation. J Immunol. 2002;168:3847–3854. doi: 10.4049/jimmunol.168.8.3847. [DOI] [PubMed] [Google Scholar]
- 70.Serfling E, Berberich-Siebelt F, Avots A. NFAT in lymphocytes: a factor for all events? Sci STKE. 2007:pe42. doi: 10.1126/stke.3982007pe42. [DOI] [PubMed] [Google Scholar]
- 71.Gerspach J, Gotz A, Zimmermann G, Kolle C, Bottinger H, Grell M. Detection of membrane-bound tumor necrosis factor (TNF): an analysis of TNF-specific reagents. Microsc Res Tech. 2000;50:243–250. doi: 10.1002/1097-0029(20000801)50:3<243::AID-JEMT8>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]