Abstract
The high-molecular-weight serine proteinase inhibitors (serpins) are restricted, generally, to inhibiting proteinases of the serine mechanistic class. However, the viral serpin, cytokine response modifier A, and the human serpins, antichymotrypsin and squamous cell carcinoma antigen 1 (SCCA1), inhibit different members of the cysteine proteinase class. Although serpins employ a mobile reactive site loop (RSL) to bait and trap their target serine proteinases, the mechanism by which they inactivate cysteine proteinases is unknown. Our previous studies suggest that SCCA1 inhibits papain-like cysteine proteinases in a manner similar to that observed for serpin–serine proteinase interactions. However, we could not preclude the possibility of an inhibitory mechanism that did not require the serpin RSL. To test this possibility, we employed site-directed mutagenesis to alter the different residues within the RSL. Mutations to either the hinge or the variable region of the RSL abolished inhibitory activity. Moreover, RSL swaps between SCCA1 and the nearly identical serpin, SCCA2 (an inhibitor of chymotrypsin-like serine proteinases), reversed their target specificities. Thus, there were no unique motifs within the framework of SCCA1 that independently accounted for cysteine proteinase inhibitory activity. Collectively, these data suggested that the sequence and mobility of the RSL of SCCA1 are essential for cysteine proteinase inhibition and that serpins are likely to utilize a common RSL-dependent mechanism to inhibit both serine and cysteine proteinases.
The high-molecular-weight serine proteinase inhibitors (serpins) comprise a superfamily of structurally well conserved proteins present in plants, animals, fungi, and viruses (1). In higher vertebrates, serpins regulate proteolytic events associated with coagulation, fibrinolysis, apoptosis, and inflammation (reviewed in ref. 2). Unlike small-molecular-weight serine proteinase inhibitors, such as those of the Kazal and Kunitz families, serpins inhibit serine proteinases via a nonstandard, suicide substrate-like mechanism (3–5). Although the sequence of events are not known precisely, this mechanism involves exposure of the reactive site loop (RSL) of the serpin to the active site of the proteinase (3, 5). After the RSL is bound by the active site of the proteinase, the serpin undergoes a major conformational rearrangement characterized by partial or full insertion of the RSL into β-sheet A (5), RSL cleavage, and formation of a covalent serpin–enzyme complex. In addition, this conformational change deforms the active site of the enzyme, thereby impeding deacylation and contributing to the stability of the covalent complex (6). However, if the rate of the loop insertion is retarded, or if stabilizing interactions between the serpin and the proteinase are lost, then the enzyme completes the deacylation step and escapes inhibition (7). In this latter case, the complex dissociates into an inactivated, cleaved serpin and an active proteinase. Thus, a serpin can serve as a typical substrate or an inhibitor depending on the ability of the molecule to undergo a conformational change and trap the proteinase before the deacylation steps.
In general, serpins are restricted to inhibiting proteinases of only the serine mechanistic class. However, at least three serpins are now known to demonstrate cross-class inhibition of several different types of cysteine proteinases: the viral serpin cytokine response modifier A (crmA) inhibits caspase-1 (8); the human serpin antichymotrypsin (ACT) inhibits a putative bovine prohormone thiol processing enzyme (9); and the human serpin squamous cell carcinoma antigen 1 (SCCA1) inhibits the papain-like cysteine proteinases cathepsins (cat) S, K, and L (10, 11). The inhibition of cysteine proteinases by SCCA1 was unexpected, since the nearly identical serpin, SCCA2, inhibits the chymotrypsin-like serine proteinases catG and human mast cell chymase (12).
Recently, we showed that SCCA1 inhibits the papain-like cysteine proteinases with 1:1 stoichiometry, at rates kass > 105 M−1⋅s−1, and a kdiss that was unmeasurable (11). Moreover, we detected stable complexes by PAGE and a single cleavage site that mapped to the RSL. These findings suggest that SCCA1 inhibits cysteine proteinases via an RSL-dependent mechanism. However, since the active site geometry and chemistry of cysteine proteinases differ somewhat from those of serine proteinases, it is conceivable that SCCA1 inhibits cysteine proteinases via a different mechanism, and RSL cleavage simply represents a secondary substrate reaction.
To determine whether the RSL of SCCA1 was required for SCCA1 inhibitory function, we employed site-directed mutagenesis to alter different residues within the loop. These data show that the RSL of SCCA1 is essential for cysteine proteinase inhibition and that serpins are likely to employ a common RSL-dependent mechanism to inhibit both serine and cysteine proteinases.
MATERIALS AND METHODS
Construction of Mutants.
Mutations were generated by using the Quick Change mutagenesis kit from Stratagene and verified by DNA sequencing. Plasmid templates were pGEX/SCCA1 and pGEX/SCCA2, glutathione S-transferase (GST) fusion protein vectors with the complete coding sequence of SCCA1 or SCCA2 (12).
The RSL swap mutants were constructed by restriction digest and ligation. Two StuI/BamHI fragments were isolated from both pGEX/SCCA1 and pGEX/SCCA2: a 5-kbp fragment containing the pGEX vector and the RSL, and a 1-kbp fragment containing the remainder of the SCC coding sequence. The 1-kbp fragments were religated to the 5-kbp fragments of the opposite molecule, forming pGEX/SCCA1 with an SCCA2 RSL [SCCA1(2)] and pGEX/SCCA2 with an SCCA1 RSL [SCCA2(1)]. This domain swap mutated the P14, P4, P3, P2, P1, P1′, P2′, P3′, P10′ residues [Schechter and Berger numbering scheme and relative to the conserved sequences in the RSL (13)]. Correct orientations were verified by DNA sequencing (14).
Enzymes, Inhibitors, and Substrates.
SCCA mutants were purified by the bulk method using the GST fusion protein purification kit (Pharmacia), as described previously (12). Proteins were expressed in BL21 Escherichia coli. Purity of mutant proteins were assayed via SDS/PAGE and visualized by Coomassie blue staining. Concentration of the proteins were determined via Bradford Assay (Bio-Rad).
Recombinant catS was prepared as described (15, 16). Human trypsin and the substrate for cathepsin G (catG) and chymotrypsin, Succinyl-Ala-Ala-Pro-Phe-para-nitroanilide (Succ-AAPF-pNA), were purchased from Sigma. Chymotrypsin, α1-proteinase inhibitor, and catG were purchased from Athens Research & Technology (Athens, GA). The trypsin substrate, Glu-Gly-Arg-pNA (EGR-pNA), was purchased from Bachem. The catS substrate, (Z-Phe-Arg)2-R110 ((Z-FR)2-R110), was purchased from Molecular Probes. PBS reaction buffer (0.01 M phosphate buffer/27 mM KCl/137 mM NaCl, pH 7.4) was used with trypsin, catG, and chymotrypsin. Cathepsin reaction buffer, pH 5.5 (50 mM sodium acetate, pH 5.5/4 mM DTT/1 mM EDTA), was used with catS.
Kinetics.
As an initial test for inhibitor activity, proteinase and molar excess (≈2- to 100-fold) of SCCA or SCCA mutant was incubated for 30 min at 25°C, in the appropriate reaction buffer, as described (12). Residual enzyme activity was measured by the addition of substrate. Activity of enzyme in the presence of inhibitor was compared with an uninhibited control.
The inhibition of catS and trypsin was measured under pseudo-first-order conditions by using the progress-curve method (17), as described (12). The Km for trypsin with EGR-pNA in PBS is 477 μM.
The rate of association of inhibitor with catG was determined under second-order conditions (18), as described (12).
SDS/PAGE.
For the AlaP14Arg gel, SCCA1 (1.25 μg) or SCCA1 AlaP14Arg (1.4 μg) with or without catS (0.3 μg) was incubated (volume 10 μl) at 25°C for 15 min in cathepsin reaction buffer, pH 5.5. For the ThrP3′Ala gel, SCCA1 (5.0 μg), SCCA1 ThrP3′Ala (5.0 μg), or α1-proteinase inhibitor (2.7 μg) with or without chymotrypsin (1.0 μg) was incubated (volume 10 μl) at 25°C for 15 min in PBS reaction buffer. For the GlyP2Arg gel, SCCA1 (5.0 μg) or SCCA1 GlyP2Arg (6.0 μg) with or without trypsin (1.0 μg) was incubated (volume 10 μl) at 25°C for 15 min in PBS reaction buffer. Subsequent to incubation, proteins were mixed with 3× loading buffer (6% SDS/30% glycerol/187.5 mM Tris⋅HCl, pH 6.5/0.03% phenol red/2% 2-mecaptoethanol), heated to 95°C for 5 min, and separated by SDS/PAGE, as described (12). PAGE gels were either 8% (AlaP14Arg gel) or 10% (GlyP2Arg and ThrP3′Ala gels) acrylamide (%T/%C = 19:1). Proteins separated by SDS/PAGE were visualized by staining with Coomassie brilliant blue R-250 (0.25%).
Matrix-Associated Laser Desorption Ionization (MALDI) MS.
Trypsin (5 μM) in PBS reaction buffer (20 μl) was mixed with SCCA1 GlyP2Arg (7 μM) for 5 min at 25°C and then lyophilized. The reaction mixture components were separated by MALDI MS at the Wistar Protein Microchemistry Facility (Philadelphia).
RESULTS
For comparison with other inhibitory-type serpins, including SCCA2, the Schechter–Berger numbering scheme of the RSL was utilized (13). For SCCA1, this scheme designated a conserved Gly of P15 and a consensus cleavage site between P1Ser and P1′Ser (11). By definition, however, the actual P1-P1′ bond must be determined experimentally and, depending on the target proteinases, can differ even within the same molecule (19). Previous studies show that catS cleaves in the RSL of SCCA1 between residues P2Gly and P1Ser (11, 20). If RSL cleavage is a consequence of the inhibitory mechanism rather than a by-product of secondary catalytic events, then, by the Schechter and Berger numbering scheme (13), the P2Gly is actually the functional P1 residue for the SCCA1–catS interaction. However, for comparative purposes, we will continue to designate this Gly residue as P2 (Table 1).
Table 1.
SCCA1 RSL mutants
| Serpin | RSL hinge*
|
RSL variable
|
kass (105 M−1⋅s−1)†
|
|||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P 15 | 14 | 13 | 12 | 11 | 10 | 9 | 8 | 7 | 6 | 5 | 4 | 3 | 2 | 1 | 1′ | 2′ | 3′ | 4′ | 5′ | catS | catG | try | CT | |
| Wild types | ||||||||||||||||||||||||
| SCCA2 | G | V | E | A | A | A | A | T | A | V | V | V | V | E | L | S | S | P | S | T | 0 | 1.0‡ | 0 | 0 |
| SCCA1 | G | A | E | A | A | A | A | T | A | V | V | G | F | G | S | S | P | T | S | T | 1.0§ | 0 | 0 | 0 |
| Mutants | ||||||||||||||||||||||||
| AlaP14Arg | . | R | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 0 | 0 | 0 | |
| AlaP14Thr | . | T | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | 3.9 | 0 | 0 | |
| SCCA1(2) | . | V | . | . | . | . | . | . | . | . | . | V | V | E | L | S | S | P | . | . | 0 | 0.7 | ||
| SCCA2(1)¶ | . | A | . | . | . | . | . | . | . | . | . | G | F | G | S | S | P | T | . | . | 1.2 | 0 | ||
| GlyP2Arg | . | . | . | . | . | . | . | . | . | . | . | . | . | R | . | . | . | . | . | . | 0.2 | 0 | 0.4 | 0 |
| GlyP2Glu | . | . | . | . | . | . | . | . | . | . | . | . | . | E | . | . | . | . | . | . | 1.1 | 0 | ||
| PheP3Ala | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | 0 | 0 | ||
| PheP3Leu | . | . | . | . | . | . | . | . | . | . | . | . | L | . | . | . | . | . | . | . | 3.3 | 0 | 0 | |
| ThrP3′Ala | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | 0.9 | 0 | 0 | |
Schecter and Berger numbering scheme (13), but, for comparative purposes, numbering relative to conserved P15Gly.
catS, cathepsin S; catG, cathepsin G; try, trypsin; CT, chymotrypsin. Second-order rate constants were calculated under second-order conditions (catG) and pseudo-first-order conditions (catS and trypsin). 0, no inhibition seen under conditions used.
Ref. 12.
Ref. 11.
SCCA2 framework with SCCA1 RSL.
Mutation of the RSL Hinge Region.
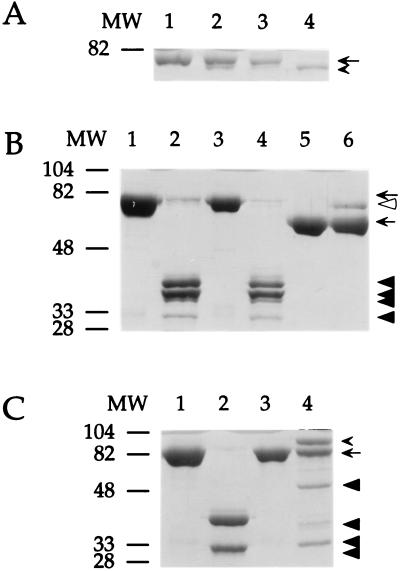
The amino acid sequence of the RSL, in part, determines whether a serpin serves as an inhibitor or a substrate for a serine proteinase. Functionally, the RSL can be divided into two segments: a proximal (P15-P9) hinge region and a distal (P9-P5′) variable region. The hinge region is well conserved among serpins because at least this segment of the loop inserts into β-sheet A during proteinase binding and after RSL cleavage (reviewed in ref. 2). In contrast, the amino acid sequences of the variable portions of serpins resemble substrates in that they are complementary to the subsite specificities of the target proteinases. If SCCA1 employs its RSL to inhibit cysteine proteinases, then amino acid changes within the hinge and variable regions should modify inhibitory activity. The flexibility of the hinge is centered around an uncharged P14 residue (5, 21). For several serpins, mutation of the P14 residue to a charged residue blocks loop insertion and abolishes serine proteinase inhibitory activity (22). Thus, to determine whether a similar mutation of the P14 residue could alter catS inhibitory activity, the P14 Ala of SCCA1 was mutated to an Arg, by site-directed mutagenesis. Inhibitory activity against catS was lost completely (Fig. 1 and Table 1). Moreover, the catS cleavage pattern of the AlaP14Arg SCCA1 mutant, visualized by SDS/PAGE (Fig. 2A, lane 4), was similar to that of catS-cleaved wild-type SCCA1 (11) (Fig. 2A, lane 2) and suggested that the RSL was still accessible to catS cleavage.
Figure 1.
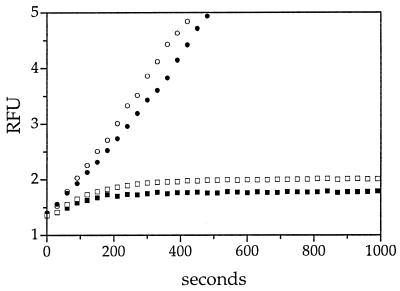
Kinetic analysis of SCCA1 hinge mutants. The effect of hinge mutations on the inhibition of catS was measured under pseudo-first-order conditions by using the progress-curve method. Wild-type SCCA1 (□), SCCA1 AlaP14Thr mutant (■), or SCCA1 AlaP14Arg mutant (•) were incubated with catS and substrate at 25°C in cathepsin buffer, pH 5.5. The progress of catS inactivation was followed by measuring the relative fluorescence (RFU) over time. The rate of inhibition of catS by the AlaP14Thr SCCA1 mutant was kass = 3.9 × 105 M−1⋅s−1. CatS without inhibitor (○) served as a control. A set of curves from a representative experiment is depicted. Concentrations of reagents: inhibitor, 250 nM; enzyme, 10 nM; substrate [(Z-FR)2-R110], 5 μM.
Figure 2.
SDS/PAGE analysis of the interaction of SCCA1 mutants and different proteinases. SCCA1 or SCCA1 mutants were incubated with proteinases at 25°C for 15 min in reaction buffer and then heated at 95°C for 5 min in gel-loading buffer. Protein mixtures were separated by SDS/PAGE and stained with Coomassie blue. Positions of the molecular weight markers are noted to the left of the gel. The molecular mass of SCCA1 and SCCA1 mutants was ≈71 kDa; catS, ≈24 kDa; chymotrypsin, ≈25 kDa; α1-proteinase inhibitor, ≈52 kDa; and trypsin, ≈22 kDa. (A) The interaction of the AlaP14Arg SCCA1 mutant with catS. Lanes: 1, SCCA1 (1.25 μg); 2, SCCA1 with catS (0.3 μg); 3, SCCA1 AlaP14Arg (1.4 μg); 4, SCCA1 AlaP14Arg with catS. The positions of the native (arrow) and cleaved (arrowhead) inhibitors are indicated. (B) The interaction of the ThrP3′Ala SCCA1 mutant with chymotrypsin. Lanes: 1, SCCA1 (5.0 μg); 2, SCCA1 with chymotrypsin (1.0 μg); 3, SCCA1 ThrP3′Ala (5.0 μg); 4, SCCA1 ThrP3′Ala with chymotrypsin; 5, α1-proteinase inhibitor (2.7 μg); 6, α1-proteinase inhibitor with chymotrypsin serve as controls for chymotrypsin activity. The positions of the inhibitors (arrows), α1-proteinase inhibitor–chymotrypsin complex (open arrowhead), and the wild-type and mutant SCCA1 degradation products (closed arrowheads) are indicated. (C) Formation of SDS–stable complexes of trypsin and SCCA1 GlyP2Arg. Lanes: 1, SCCA1 (5.0 μg); 2, SCCA1 with trypsin (1.0 μg); 3, SCCA1 GlyP2Arg (6.0 μg); 4, SCCA1 GlyP2Arg and trypsin. The positions of the inhibitors (arrow), SCCA1 GlyP2Arg-trypsin complex (arrowhead), and the wild-type and mutant degradation products (closed arrowheads) are indicated.
Interestingly, the putative P14 of SCCA1 is an Ala and not the consensus Thr. To determine whether P14Ala is itself critical for catS inhibition, an AlaP14Thr mutant was derived. This mutation enhanced the inhibitory interaction of SCCA1 with catS as manifested by an almost 4-fold increase in the second-order rate constant (kass = 3.9 × 105 M−1⋅s−1) (Fig. 1). Collectively, these data suggested that the P14 residue, which is permissive to neutral but not charged residues, was critical for the inhibition of catS by SCCA1.
Variable Region Swap Between SCCA1 and SCCA2.
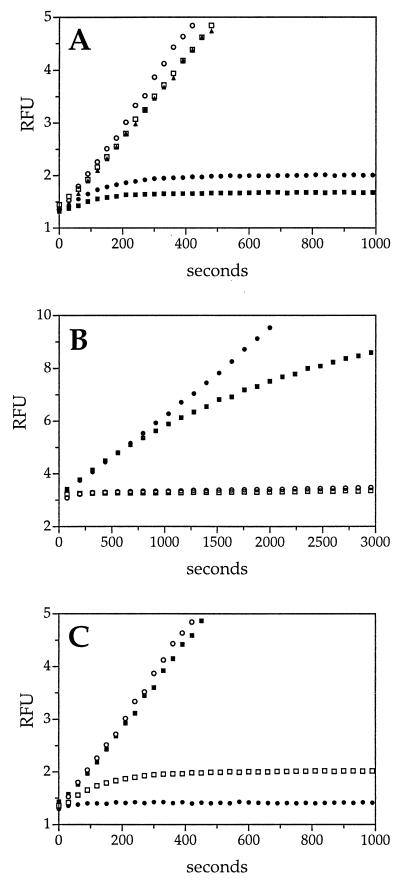
Next, we sought to determine whether the variable portion of the RSL was necessary for SCCA1 inhibitory activity. Since SCCA1 and SCCA2 are nearly identical, a swap of the C termini would alter the variable residues located at P4-P3′. If the variable region of the RSL is critical for catS inhibition, then a recombinant SCCA1 containing the C terminus of SCCA2 [SCCA1(2)] should fail to inhibit catS, but may inhibit catG (SCCA2 inhibits the serine proteinase cathepsin G) (12). As predicted, SCCA1(2) no longer inhibited catS (Fig. 3A), but inhibited catG (kass = 0.7 × 105 M−1⋅s−1) at a rate similar to that of wild-type SCCA2 (kass = 1.0 × 105 M−1⋅s−1) (12). Conversely, a recombinant SCCA2 containing the C terminus of SCCA1 [SCCA2(1)] inhibited catS (kass = 1.2 × 105 M−1⋅s−1) at a rate similar to that of wild-type SCCA1 (kass = 1.0 × 105 M−1⋅s−1) (11). These results suggested that the variable segment of the SCCA1 (and SCCA2) RSL were critical for inhibition and defining target specificity. Moreover, the reciprocal gain and loss of cysteine proteinase activity by SCCA2(1) and SCCA1(2), respectively, suggested that there are no unique motifs within the framework of the SCCA1 molecule that independently account for cysteine proteinase inhibitory activity.
Figure 3.
Kinetic analysis of RSL variable-region mutants. The effect of mutations in the variable region of the RSL on catS inhibition was measured under pseudo-first-order conditions by using the progress-curve method, as described in Fig. 1. Sets of curves from representative experiments are depicted. (A) Loop swap mutants. The rates of product formation by catS in the presence of wild-type SCCA2 (□), the SCCA1(2) mutant (▴) (containing the SCCA2 RSL), wild-type SCCA1 (•), and SCCA2(1) mutant (■) (containing the SCCA1 RSL). CatS without inhibitor (○) served as a control. The rate of catS inhibition by SCCA2(1) was kass = 1.2 × 105 M−1⋅s−1. Concentrations of reagents: inhibitor, 250 nM; enzyme, 10 nM; substrate [(Z-FR)2-R110], 5 μM. (B) P1 mutants. The rate of inhibition of catS by the GlyP2Arg SCCA1 mutant (■) (kass = 1.8 × 104 M−1⋅s−1) and the GlyP2Glu SCCA1 mutant (□) (kass = 1.1 × 105 M−1⋅s−1) was compared with that of wild-type SCCA1 (○). The curve of the GlyP2Glu SCCA1 mutant is obscured by that of the wild-type SCCA1. CatS without inhibitor (•) served as a control. Concentrations of reagents: inhibitor, 360 nM; enzyme, 13.5 nM; substrate [(Z-FR)2-R110], 10 μM. (C) P2 mutants. The rate of inhibition of catS by the PheP3Ala SCCA1 mutant (■) and the PheP3Leu SCCA1 mu tant (•) (kass = 3.3 × 105 M−1⋅s−1) was compared with that of wild-type SCCA1 (□). CatS without inhibitor (○) served as a control. Concentrations of reagents: inhibitor, 250 nM; enzyme, 10 nM; substrate [(Z-FR)2-R110], 5 μM.
Mutation of the Functional P1 and P2 Residues.
The previous result suggested that critical residues responsible for cysteine proteinase inhibition lie within the RSL variable segment of SCCA1. To determine whether any single amino acid might be required for the inhibitory interaction, different residues within the RSL of SCCA1 were mutated. For most serpin–serine proteinase interactions, a mutation in the P1 residue (i.e., the residue that occupies the S1 subsite of the serine proteinase and forms an ester bond with the active-site serine) alters the inhibitory activity of the molecule (reviewed in ref. 2). Yet, mutation of the P2Gly of SCCA1 (i.e., the functional P1 relative to catS cleavage site) to charged residues such as Arg (GlyP2Arg kass = 1.8 × 104 M−1⋅s−1) or Glu (GlyP2Glu kass = 1.1 × 105 M−1⋅s−1) had little effect on the inhibition of catS (Fig. 3B). This result was not surprising as, in contrast to serine proteinases, it is the residue occupying the S2 subsite (i.e., the functional P2 residue of the substrate) of catS that determines substrate binding (15, 16). Thus, if the RSL of SCCA1 acts as a substrate in the context of an inhibitory reaction, then mutation of the P3Phe (i.e., the functional P2) should modify inhibitory activity.
As shown by progress-curve analysis (Fig. 3C), a PheP3Ala mutant of SCCA1 lost catS inhibitory activity. In contrast, a mutation with a more conservative change of PheP3Leu retained catS inhibitory activity (kass = 3.3 × 105 M−1⋅s−1). This finding agreed with studies using small peptide substrates which show that catS has similar preferences for Phe and Leu in the functional P2 position (15, 16). These data suggested that the P3Phe behaves as a functional P2 residue when interacting with catS and that this residue was critical for SCCA1 inhibitory activity.
SCCA1 as a Serine Proteinase Inhibitor.
In addition to inhibiting cysteine proteinases, crmA and ACT also inhibit the serine proteinases granzyme B and catG, respectively (18, 23). If SCCA1 inhibits cysteine proteinases via an RSL-dependent mechanism, then this serpin might be capable of inhibiting serine proteinases, as well. Although we were unable to identify a target serine proteinase for SCCA1 (11), Nawata et al. (20, 24) cloned a tumor-derived ThrP3′Ala mutant SCCA1 molecule that inhibits chymotrypsin activity in a protein degradation assay. To determine whether this single amino acid difference could alter the specificity of wild-type SCCA1, we generated a ThrP3′Ala mutant. Using sensitive chromogenic peptide substrates and high concentrations of pure recombinant proteins, no inhibition of chymotrypsin was detected. Furthermore, both wild-type and mutant SCCA1 were susceptible to extensive degradation by chymotrypsin (Fig. 2B, lanes 2 and 4). The ThrP3′Ala mutant was still a potent inhibitor of catS (kass = 0.9 × 105 M−1⋅s−1).
Although the ThrP3′Ala SCCA1 mutant failed to inhibit chymotrypsin under our assay conditions, we sought to determine whether any other single amino acid mutation could change SCCA1 to an inhibitor of serine proteinases. Analysis of substrate preference of various other serine proteinases revealed that trypsin prefers to cleave after an Arg residue. Since we had generated a GlyP2Arg mutant SCCA1, we tested the inhibitory activity of this mutant versus trypsin. This mutant SCCA1 inhibited trypsin at a near 1:1 stoichiometry with a second-order rate constant of kass = 3.9 × 104 M−1⋅s−1 (Fig. 4 A and B). This rate is similar to that of catS inhibition by the same mutant (kass = 1.8 × 104 M−1⋅s−1) (Fig. 3B). Furthermore, characteristic of a serpin–serine proteinase inhibitory reaction, the GlyP2Arg SCCA1 mutant formed an SDS–stable complex with trypsin when resolved by SDS/PAGE (Fig. 2C, lane 4). Minor degradation products of the GlyP2Arg mutant upon incubation with trypsin (Fig. 2C, lane 4) may be a result of inefficient cleavage of the thrombin site located between the GST moiety (Mr ≈ 26,000) and the GlyP2Arg mutant (Mr ≈ 45,000). Furthermore, this cleavage may account for the modest increase in stoichiometry of inhibition (1.25) measured for the interaction between the GlyP2Arg mutant and trypsin.
Figure 4.
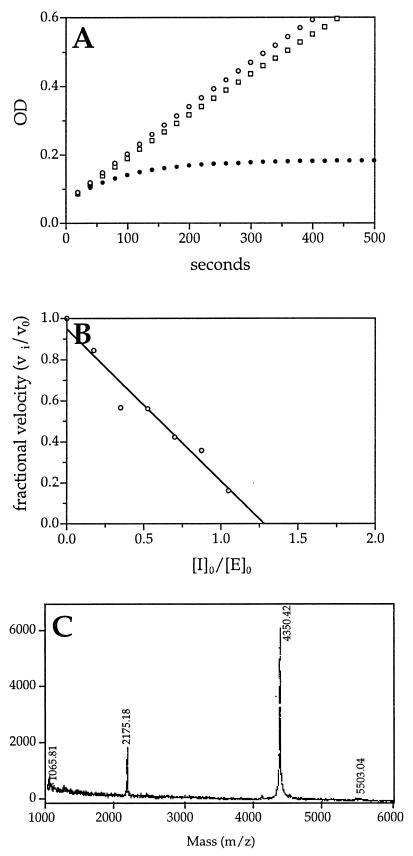
Kinetic and biochemical analysis of the interaction of trypsin and SCCA1 GlyP2Arg mutant. (A) Kinetic analysis of trypsin inhibition by SCCA1 GlyP2Arg using the progress-curve method. Trypsin (7.7 nM) alone (○) or with SCCA1 (□, 509 nM) or SCCA1 GlyP2Arg (•, 517 nM) was incubated with substrate (EGR-pNA, 0.8 mM) at 25°C in PBS reaction buffer under pseudo-first-order conditions. The progress of trypsin inactivation was followed by measuring the ΔA405 for the reaction over time. The SCCA1 GlyP2Arg mutant inhibited trypsin with a second-order rate constant of kass = 3.9 × 104 M−1⋅s−1. A set of curves from a representative experiment is depicted. (B) Stoichiometry of inhibition of trypsin by SCCA1 GlyP2Arg. Trypsin (4 μM) was incubated with different concentrations of SCCA1 GlyP2Arg (0–4 μM) at 25°C for 30 min in PBS. Residual trypsin activity was measured by adding the substrate (EGR-pNA) and measuring the ΔA405. Fractional activity was the ratio of the velocity of inhibited enzyme (vi) to the velocity of uninhibited control (v0). The stoichiometry of inhibition was determined by using linear regression to extrapolate the inhibitor and enzyme ratio, resulting in complete inhibition. (C) MALDI analysis of trypsin and SCCA1 GlyP2Arg cleavage products. Trypsin (2.2 μg) was incubated with SCCA1 GlyP2Arg (10 μg) for 5 min at 25°C. The reaction mixture then was separated by MALDI MS.
If the GlyP2Arg SCCA1 mutant was inhibiting trypsin in an RSL-dependent fashion, then the scissile bond should be located between the P2Arg and P1Ser (i.e., the P2 Arg becomes the functional P1). Moreover, cleavage of this bond should yield a novel C-terminal fragment with Mr = 4,351.6. To determine whether this was the case, trypsin and GlyP2Arg SCCA1 were incubated and the resulting cleavage fragments were resolved by using MALDI MS. A major peak of 4,350.4 Da was detected (Fig. 4C). This result suggested that trypsin cleaved the mutant SCCA1 molecule between the P2Arg and the P1Ser, and that a single amino acid change within the RSL was capable of converting SCCA1 into an inhibitor of either a cysteine or a serine proteinase.
DISCUSSION
There are now at least three examples of serpins inhibiting proteinases belonging to the cysteine mechanistic class. These include the viral serpin crmA and the human serpins ACT and SCCA1 (8, 10, 11, 25). However, the mechanism of this type of inhibition is unclear. Serpins neutralize their target serine proteinases by undergoing a conformational change that traps the enzyme in a covalent intermediate (reviewed in ref. 2). The inhibitory complexes are stabilized by enzymatic cleavage of the RSL (3–6). The ability to detect a cleavage site within the RSL of SCCA1 after its interaction with catS suggested that serpins employ a similar RSL-dependent mechanism to inhibit cysteine proteinases. However, cleavage of the RSL per se does not indicate the presence of an inhibitory reaction. Because of its exposure on the surface of the molecule, the RSL is very susceptible to proteolysis. Indeed, proteinases from different mechanistic classes are known to inactivate serpins by simple RSL cleavage (26–29). For example, catL inactivates α1-proteinase inhibitor by cleavage at MetP1-P1′Ser and at GluP5-P4Ala (29). Thus, the RSL cleavage associated with the SCCA1-catS interaction could represent an ancillary cleavage event unrelated to the true inhibitory mechanism. However, the results of this study indicate that the RSL of SCCA1 indeed does play an essential role in the inhibition of cysteine proteinases and that serpins are likely to employ a common RSL-dependent mechanism to inhibit both cysteine and serine proteinases.
Previous studies of inhibition of serine proteinases by serpins demonstrate that alterations to the hinge region (P15-P9) affect serpin activity by altering the RSL mobility and the rate at which the RSL inserts into the serpin. Mutation of the P14 residue to charged residues with large side chains blocks RSL insertion and abrogates inhibitory activity while still allowing for an RSL substrate (noninhibitory) reaction. In contrast, mutation of P14 to uncharged residues has little effect (5, 21, 22). Similar findings were observed with the SCCA1 mutants. The AlaP14Arg mutant lost catS inhibitory activity, whereas the AlaP14Thr SCCA1 mutant had a modest 4-fold increase in the kass. Similarly, mutation of the P14Thr of crmA to an Arg residue abolishes inhibitory activity (30). These loop-insertion mutants suggested that, as with serpin–serine proteinase interactions, flexibility of the RSL was important for the inhibition of papain- and caspase-like cysteine proteinases.
The results of the RSL swap mutants SCCA1(2) and SCCA2(1) underscored the importance of the RSL in the inhibition of cysteine proteinases by SCCA1. The loss of catS inhibitory activity by SCCA1(2) again suggested that there were no motifs within the body of the SCCA1 molecule that independently regulated cysteine proteinase activity. Moreover, the gain of catS inhibitory activity of SCCA2(1) suggested that, as seen with serpin–proteinase interactions, the variable portion of the RSL dictated the specificity of the target proteinase.
Intrinsic to the mechanism of serpin–serine proteinase inhibition is that the variable region of the RSL should appear, in part, as a substrate for the target proteinase. This notion is supported by the RSL swap mutants and confirmed by single mutants of the functional P1 and P2 residues. Unlike serine proteinases, catS displays binding specificity for the residue occupying the S2, and not the S1, subsite (i.e., the substrate P2, and not P1, residue) (15, 16). We showed that mutations to the P2Gly of SCCA1 (i.e., the functional P1) to charged residues, Arg or Glu, had little effect on the inhibitory activity of SCCA1. However, mutation of the P3Phe (i.e., the functional P2 residue) to an Ala, a residue not preferred by catS, abolished SCCA1 inhibitory activity. In contrast, mutation of the P3Phe to Leu (a residue preferred by catS) had little effect on catS inhibition. These results demonstrated that single amino acid changes in the proteinase recognition sequence of the RSL were sufficient to disrupt serpin–cysteine proteinase interactions.
Although we have yet to determine whether SCCA1 inhibits any serine proteinase, the serpins crmA and ACT can target certain serine proteinases. Previous studies suggested that a tumor-derived SCCA1, containing a single ThrP3′Ala mutation, inhibits the serine proteinase chymotrypsin. However, when we used sensitive, small chromogenic peptides, neither wild-type SCCA1 nor our ThrP3′Ala SCCA1 mutant inhibited chymotrypsin. Although this discrepancy could be a result of differences in protein preparations, the assay conditions employed by Nawata et al. (20) do not preclude inhibition by SCCA1 via a simple substrate reaction. Indeed, SDS/PAGE analysis of the interaction of both wild-type and the ThrP3′Ala mutant SCCA1 with chymotrypsin revealed extensive degradation of both serpins by this enzyme. Thus, the mutant SCCA1 “inhibition” of chymotrypsin may be due to competition between substrates rather than a true inhibitory reaction.
Although a serine proteinase target for wild-type SCCA1 has yet to be identified, SCCA1 may prove to be a dual, cross-class inhibitor, such as crmA and ACT. A GlyP2Arg SCCA1 mutant, in addition to inhibiting catS, inhibited the serine proteinase trypsin. This demonstrated that via a single RSL point mutation, SCCA1 could inhibit either a serine or a cysteine proteinase. Thus, there appeared to be no structural motif within the SCCA1 molecule that precluded its functioning as a serine proteinase inhibitor.
Acknowledgments
We thank Dr. Sule Çataltepe and Philip Pemberton for useful discussions and Ms. Kelly Ames for assistance with the manuscript. This work was supported by a Breast Cancer Research Grant from the Massachusetts Department of Public Health and grants from the National Institutes of Health (HD28475, CA69331, and CA73031) and the Smokeless Tobacco Research Council.
ABBREVIATIONS
- serpin
high-molecular-weight serine proteinase inhibitor
- RSL
reactive site loop
- crmA
cytokine response modifier A
- ACT
antichymotrypsin
- SCCA
squamous cell carcinoma antigen
- cat
cathepsin
- GST
glutathione S-transferase
- MALDI
matrix-assisted laser desorption ionization
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Hunt L T, Dayhoff M O. Biochem Biophys Res Commun. 1980;95:864–871. doi: 10.1016/0006-291x(80)90867-0. [DOI] [PubMed] [Google Scholar]
- 2.Gettins P G W, Patston P A, Olson S T. Serpins: Structure, Function and Biology. Austin, TX: R. G. Landes and Chapman & Hall; 1996. [Google Scholar]
- 3.Travis J, Guzdek A, Potempa J, Watorek W. Biol Chem Hoppe-Seyler. 1990;71:3–11. [PubMed] [Google Scholar]
- 4.Patston P A, Gettins P, Beechem J, Schapira M. Biochemistry. 1991;30:8876–8882. doi: 10.1021/bi00100a022. [DOI] [PubMed] [Google Scholar]
- 5.Carrell R W, Evans D L I. Curr Opin Struct Biol. 1992;2:438–446. [Google Scholar]
- 6.Matheson N R, van Halbeek H, Travis J. J Biol Chem. 1991;266:13489–13491. [PubMed] [Google Scholar]
- 7.Patston P A, Gettins P G, Schapira M. Ann N Y Acad Sci. 1994;714:13–20. doi: 10.1111/j.1749-6632.1994.tb12026.x. [DOI] [PubMed] [Google Scholar]
- 8.Komiyama T, Ray C A, Pickup D J, Howard A D, Thornberry N A, Peterson E P, Salvesen G. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 9.Hook V Y, Purviance R T, Azaryan A V, Hubbard G, Krieger T J. J Biol Chem. 1993;268:20570–20577. [PubMed] [Google Scholar]
- 10.Takeda A, Yamamoto T, Nakamura Y, Takahashi T, Hibino T. FEBS Lett. 1995;359:78–80. doi: 10.1016/0014-5793(94)01456-b. [DOI] [PubMed] [Google Scholar]
- 11.Schick C, Pemberton P A, Shi G-P, Kamachi Y, Cataltepe S, Bartuski A J, Gornstein E R, Bromme D, Chapman H A, Silverman G A. Biochemistry. 1998;37:5258–5266. doi: 10.1021/bi972521d. [DOI] [PubMed] [Google Scholar]
- 12.Schick C, Kamachi Y, Bartuski A J, Cataltepe S, Schechter N M, Pemberton P A, Silverman G A. J Biol Chem. 1997;272:1849–1855. doi: 10.1074/jbc.272.3.1849. [DOI] [PubMed] [Google Scholar]
- 13.Schechter I, Berger A. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 14.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brömme D, Bonneau P R, Lachance P, Storer A C. J Biol Chem. 1994;269:30238–30242. [PubMed] [Google Scholar]
- 16.Brömme D, Bonneau P R, Lachance P, Wiederanders B, Kirschke H, Peters C, Thomas D Y, Storer A C, Vernet T. J Biol Chem. 1993;268:4832–4838. [PubMed] [Google Scholar]
- 17.Morrison J F, Walsh C T. Adv Enzymol Relat Areas Mol Biol. 1988;61:201–301. doi: 10.1002/9780470123072.ch5. [DOI] [PubMed] [Google Scholar]
- 18.Beatty K, Bieth J, Travis J. J Biol Chem. 1980;255:3931–3934. [PubMed] [Google Scholar]
- 19.Potempa J, Shieh B-H, Travis J. Science. 1988;241:699–700. doi: 10.1126/science.2456616. [DOI] [PubMed] [Google Scholar]
- 20.Nawata S, Tsunaga N, Numa F, Tanaka T, Nakamura K, Kato H. Electrophoresis. 1995;16:1027–1030. doi: 10.1002/elps.11501601173. [DOI] [PubMed] [Google Scholar]
- 21.Huntington J A, Fan B, Karlsson K E, Deinum J, Lawrence D A, Gettins P G W. Biochemistry. 1997;36:5432–5440. doi: 10.1021/bi9702142. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence D A, Olson S T, Palaniappan S, Ginsburg D. J Biol Chem. 1994;269:27657–27662. [PubMed] [Google Scholar]
- 23.Quan L T, Caputo A, Bleackley R C, Pickup D J, Salvesen G S. J Biol Chem. 1995;270:10377–10379. doi: 10.1074/jbc.270.18.10377. [DOI] [PubMed] [Google Scholar]
- 24.Suminami Y, Kishi F, Sekiguchi K, Kato H. Biochem Biophys Res Commun. 1991;181:51–58. doi: 10.1016/s0006-291x(05)81380-4. [DOI] [PubMed] [Google Scholar]
- 25.Hwang S-R, Kohn A B, Hook V Y H. Proc Natl Acad Sci USA. 1994;91:9579–9583. doi: 10.1073/pnas.91.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei D, Majmudar G, Weiss S J. J Biol Chem. 1994;269:25849–25855. [PubMed] [Google Scholar]
- 27.Morihara K, Tsuzuki H, Harada M, Iwata T. J Biochem (Tokyo) 1984;95:795–804. doi: 10.1093/oxfordjournals.jbchem.a134671. [DOI] [PubMed] [Google Scholar]
- 28.Kress L F. J Cell Biochem. 1986;32:51–58. doi: 10.1002/jcb.240320106. [DOI] [PubMed] [Google Scholar]
- 29.Johnson D A, Barrett A J, Mason R W. J Biol Chem. 1986;261:14748–14751. [PubMed] [Google Scholar]
- 30.Tewari M, Quan L T, O’Rourke K, Desnoyers S, Zeng Z, Beidler D R, Poirier G G, Salvesen G S, Dixit V M. Cell. 1995;81:801–809. doi: 10.1016/0092-8674(95)90541-3. [DOI] [PubMed] [Google Scholar]