Abstract
Succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti, is composed of polymerized octasaccharide subunits, each of which consists of one galactose and seven glucoses with succinyl, acetyl, and pyruvyl modifications. Production of specific low molecular weight forms of R. meliloti exported and surface polysaccharides, including succinoglycan, appears to be important for nodule invasion. In a previous study of the roles of the various exo gene products in succinoglycan biosynthesis, exoP, exoQ, and exoT mutants were found to synthesize undecaprenol-linked fully modified succinoglycan octasaccharide subunits, suggesting possible roles for their gene products in polymerization or transport. Using improved techniques for analyzing succinoglycan biosynthesis by these mutants, we have obtained evidence indicating that R. meliloti has genetically separable systems for the synthesis of high molecular weight succinoglycan and the synthesis of a specific class of low molecular weight oligosaccharides consisting of dimers and trimers of the octasaccharide subunit. Models to account for our unexpected findings are discussed. Possible roles for the ExoP, ExoQ, and ExoT proteins are compared and contrasted with roles that have been suggested on the basis of homologies to key proteins involved in the biosynthesis of O-antigens and of certain exported or capsular cell surface polysaccharides.
To invade the nodules it elicits on alfalfa and establish a nitrogen-fixing symbiosis, Rhizobium meliloti strain Rm1021 needs to be able to synthesize either of two exopolysaccharides, succinoglycan or EPS II (1–5). Succinoglycan is a complex acidic exopolysaccharide (6) composed of polymerized octasaccharide subunits, each of which consists of one galactose (Gal) and seven glucoses (Glcs), and carries succinyl, pyruvyl, and acetyl modifications (7–9). A failure of R. meliloti to synthesize either exopolysaccharide leads to the formation of immature nodules that are devoid of bacteria or bacteroids (1–3, 5, 10, 11). The defect in nodule invasion is apparently caused by the infection thread failing to initiate or aborting at an early stage (12, 13). Several pieces of evidence suggest that for both succinoglycan (14, 15) and EPS II (16, 17), the generation of specific low molecular weight forms of the exopolysaccharide is required for successful nodule invasion to occur and that these may serve as signals to the plant.
The mechanism of succinoglycan biosynthesis is of interest, not only because of the importance of succinoglycan for symbiosis, but also because this exopolysaccharide is representative of a major class of bacterial exopolysaccharides, such as xanthan gum (18), that are synthesized on undecaprenol carriers (19–22). In a previous study (23), our lab had assigned biosynthetic functions to most of the exo genes (24, 25) that are required for succinoglycan biosynthesis. Interestingly, exoP, exoQ, and exoT mutants, all of which fail to fluoresce under UV on Calcofluor plates and do not synthesize succinoglycan, were capable of synthesizing fully modified lipid-linked succinoglycan octasaccharides (23). This observation suggested that the ExoP, ExoQ, and ExoT proteins had roles in the polymerization and/or export of succinoglycan. However, in this earlier study, the radiolabeled nucleotide sugars UDP-Gal and UDP-Glc were introduced into R. meliloti cells that had been permeabilized by a Tris-EDTA/freeze-thaw protocol that evidently inactivates the machinery for polymerizing the succinoglycan octasaccharide subunits (23).
Using an improved method for introducing radiolabeled nucleotide sugars that preserves succinoglycan polymerizing activity (26, 27), we observed things that led us to the unexpected conclusion that R. meliloti can polymerize succinoglycan subunits by two alternative mechanisms. One is an exoP/exoQ-dependent mechanism that yields high molecular weight succinoglycan whereas the other is an exoP/exoT-dependent mechanism that yields a class of specific low molecular weight succinoglycan oligosaccharides consisting of dimers and trimers of the octasaccharide subunit that may be important for the interaction of R. meliloti with its plant host.
MATERIALS AND METHODS
Bacterial Strains and Media.
Strains used were all derivatives of R. meliloti Rm1021: exoB294 (galE)∷Tn5 exoR95::Tn5–233; exoB294∷Tn-Tp exoR95∷Tn5-233 exoY210∷Tn5; exoB294::Tn5-Tp exoR95∷Tn5-233 exoQ332∷Tn5; exoB294∷Tn5-Tp exoR95::Tn5-233 exoP468∷Tn5; exoB294∷Tn5-Tp exoR95::Tn5-233 exoT274∷TnphoA (23). R. meliloti was grown in Luria–Bertani broth with 2.5 mM MgSO4 and 2.5 mM CaCl2 added. Antibiotic concentrations (28) and media (29) have been described.
Electroporation of Nucleotide Sugars.
Using a modification of the method of Semino et al. (26), a mid-log culture of a R. meliloti exoR galE (exoB) strain or an exoP, exoQ, or exoT derivative grown in the supplemented Luria–Bertani described above with streptomycin, was centrifuged and the cell pellet washed repeatedly with 5% (vol/vol) glycerol. The cells were resuspended in 5% (vol/vol) glycerol to a final concentration of 8–10 mg of protein/ml. In an 0.4-ml electroporation cell, freshly prepared R. meliloti cells (≈1 mg of protein) were mixed with 24 nmol of UDP-Glc, 0.41 pmol (0.6 × 106 cpm) of UDP-[14C]Gal (≈300 μCi/mmol; 1 Ci = 37 GBq) and the final concentration was brought to 150 μl with water and kept at 0°C. Electroporation was performed at 1,500 V, 400 Ω, and 25 μF with a Bio-Rad Gene Pulser Transfection Apparatus. The resulting time constant was between 7.0–8.2. The electroporated solution was transferred to an incubation vial containing Tris⋅HCl (pH 8.2) and MgCl2 to a final concentration of 70 and 10 mM, respectively, in a total volume of 200 μl. The preparation was shaken at 30°C for 60 min. The reaction was stopped by adding 5 μl of 250 mM EDTA adjusted to pH 8.0 and boiling for 5 min.
HPLC Anion-Exchange Chromatography Coupled with Pulsed Amperometric Detection (HPAEC-PAD).
A Dionex DX 500 chromatography system (Dionex) equipped with a pulsed amperometric detector (ED 40, Dionex) was used. Monosaccharides and their alditols were separated on a CarboPac MA-1 column (4 × 250 mm, Dionex) with an isocratic NaOH concentration of 0.5 M at a rate of 0.4 ml/min. The pulsed amperometric detector was operated at 0.1 mC sensitivity by using the following waveforms (potentials and durations) for carbohydrate detection: E1 = +0.05 V (T1 = 0–0.4 s), E2 = +0.75 V (T2 = 0.41–0.6 s), E3 = −0.l5 V (T3 = 0.61–1 s). The resulting chromatographic data were integrated and plotted by using the Dionex PeakNet system.
Analysis of the Low Molecular Weight Succinoglycan.
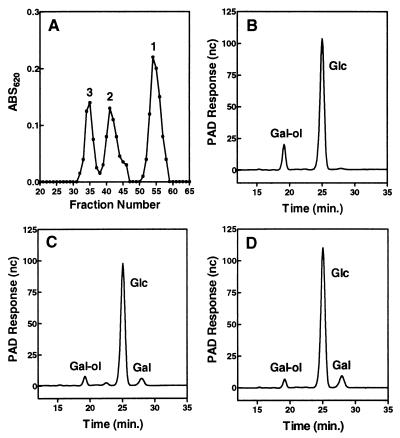
Low molecular weight succinoglycan was prepared as described (14, 15) and fractionated on a column (1 × 110 cm) of Bio-Gel P-6 (fine mesh, Bio-Rad), which was pre-equilibrated and eluted with pyridinium acetate buffer (0.1 M, pH 5.0). The fractions for peaks 1, 2 and 3 (see Fig. 1A) were individually pooled and lyophilized, which were further purified by repeating the chromatography on the same Bio-Gel P-6 column. For monosaccharide composition analysis, 9 μg (Glc equivalent) of the individual samples was diluted to 200 μl with water, mixed with 200 μl of 4 M trifluoroacetic acid in a 2.0 ml screw cap tube (Sarstedt), and hydrolyzed in a heating block at 100°C for 3.5 h. The hydrolyzates were evaporated by SpeedVac (Savant) and used for HPAEC-PAD analysis. For the determination of degree of polymerization (d.p.) of each low molecular weight succinoglycan, 9 μg (Glc equivalent) of the individual samples was first reduced with 0.1 M NaBH4 in 5 mM NaOH in a total volume of 200 μl at 37°C for 7 h in a 2-ml screw cap microtube. Then the reaction mixture was neutralized with 1 M acetic acid. The mixture was passed through a column (0.5 × 2 cm) of Dowex 50W-X4 (H+ form) to remove sodium ion and washed with water, and the eluents were evaporated to dryness and coevaporated with methanol (3 × 0.5 ml) to remove traces of boric acid. The NaBH4-treated samples thus obtained were subjected to acidic hydrolysis and subsequent HPAEC-PAD analysis as described above.
Figure 1.
Analysis of low molecular weight succinoglycan. (A) Low molecular weight succinoglycan was fractionated on a Bio-Gel P-6 (1 ml/fraction) column. The individual samples (peaks 1, 2, and 3) were subjected to NaBH4 reduction and acidic hydrolysis and then subjected to HPAEC analysis (B, C, and D, respectively) (see Materials and Methods).
Fractionation of the Incorporation Products.
The cells were centrifuged and washed three times with 70 mM Tris⋅HCl buffer, pH 8.2 (200 μl each wash). The washings were combined with the incubation supernatant. This fraction contains the excess of sugar nucleotides and the water-soluble products liberated during the incubation period. Lipid-linked intermediates were isolated by a method derived from procedures described for R. meliloti (19, 20, 23) and fractionated by TLC (23). Gel filtration chromatography was carried out on a Bio-Gel P-4 (400 mesh; Bio-Rad) column (1 × 133 cm), Bio-Gel P-6 (400 mesh; Bio-Rad) column (1 × 133 cm) or Bio-Gel A5 m (200–400 mesh) column (1 × 30) in 0.1 M pyridinium acetate (pH 5.0). Ferritin and CoCl2 were used as indicators of total exclusion and inclusion, respectively. Fractions of 1.0 ml were collected. Radioactivity was detected by liquid scintillation counting in Hydrofluor (National Diagnostics).
RESULTS
Analysis of the Low Molecular Weight Succinoglycan.
Interpretations of the data presented in this paper critically depend on knowing the identity of the low molecular weight forms of succinoglycan produced by R. meliloti. A previous study (14) stated that the succinoglycan found in the culture supernatant consists of tetramers, trimers, and monomers of the succinoglycan repeating subunit in addition to the high molecular weight polymer. This limited range along with the curious fact that dimers of the succinoglycan subunit were not reported, led us to re-examine the identity of these low molecular weight forms by using HPAEC-PAD as a sensitive analytical tool. When the low molecular weight succinoglycan was fractionated on a Bio-Gel P-6 column by using pyridinium acetate buffer (0.1 M, pH 5.0) as the eluent, three major carbohydrate peaks were detected (anthrone-sulfuric acid assay) and separated (Fig. 1A). Each peak represents a succinoglycan species because none of them are produced by R. meliloti exo mutants such as exoY that are completely deficient in succinoglycan production. In addition, compositional analysis by HPAEC-PAD revealed that the three peaks all consisted of Glc and Gal in a ratio of 7:1, regardless of their molecular size, indicating that they are monomers or oligomers of succinoglycan. To determine the d.p., each peak fraction was subjected to NaBH4 reduction. This treatment would convert the reducing end Gal to a galactitol (Gal-ol) moiety and, after complete acidic hydrolysis, the samples would give Gal-ol, Gal, and Glc in a ratio of 1: (n-1): 7n (where n represents the d.p.). Under our running conditions of HPAEC-PAD (see Materials and Methods), the Gal-ol, Glc, and Gal were well separated and appeared at 19.2, 25.1, and 28.0 min., respectively (Fig. 1 B–D). Quantitative determination of the ratios (Table 1) clearly showed that peak 1 consisted of monomers of succinoglycan octasaccharide subunit (with d.p. of 1); peak 2 consisted of dimers of the octasaccharide subunit (with d.p. of 2); and peak 3 consisted of trimers of the octasaccharide subunit (with d.p. of 3). This assignment of low molecular weight succinoglycan was in agreement with the data yielded by mass spectrometry of the reduced and deacylated samples of each peak fraction (data not shown).
Table 1.
Degree of polymerization (d.p) of LMW succinoglycan oligosaccharides
| Samples | Gal-ol/Gal | Gal-ol/Glc | Average d.p. (n) |
|---|---|---|---|
| Peak-1 | 1:0 (n = 1.0) | 1:6.7 (n = 1.0) | 1.0 |
| Peak-2 | 1:1.0 (n = 2.0) | 1:17* (n = 2.4) | 2.2 |
| Peak-3 | 1:2.0 (n = 3.0) | 1:21 (n = 3.0) | 3.0 |
*Small amounts of β-1,2 cyclic glucan found in peak 2 may have accounted for the slightly smaller than expected ratio of Gal-ol/Glc.
Synthesis of Succinoglycan After Electroporation of Sugar Nucleotides into an exoR galE Strain.
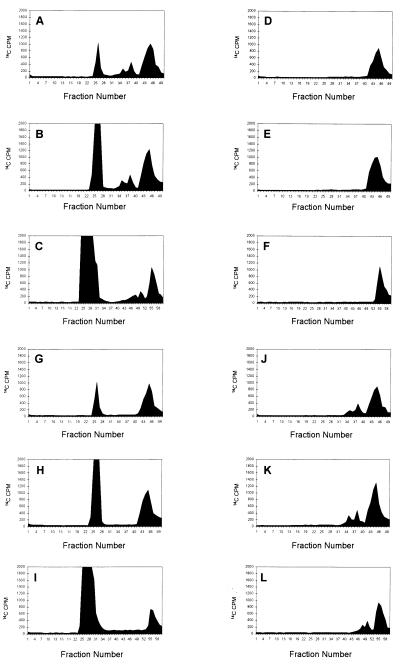
Our analyses of succinoglycan biosynthesis were carried out in an exoR galE background (23). The exoR mutation results in increased expression of the exo genes (30) whereas the galE (formerly exoB) gene encodes the UDP-Glc-4-epimerase (31, 32) that normally provides the UDP-Gal needed for succinoglycan biosynthesis. After electroporation of UDP-[14C]Gal and unlabeled UDP-Glc into an exoR galE strain and a period of incubation, the lipid-linked intermediates were extracted and the oligosaccharides they carried were removed by mild acid hydrolysis. Examination of the resulting oligosaccharides revealed not only the fully modified octasaccharide subunits observed in our previous study (23) but also additional higher molecular weight material at the origin of the TLC. P4 column chromatography of the oligosaccharides that had been cleaved from the lipid carrier revealed that, in addition to the monomeric octasaccharide subunit, radiolabeled material was present that eluted at the positions for the dimers and trimers of the octasaccharide described above as well as material large enough to be excluded from the column (Fig. 2A).
Figure 2.
Gel filtration chromatography of the [14C]Gal-labeled oligosaccharides from exoR galE derivatives. All strains carried the exoR95∷Tn5 mutation so that the exo genes are expressed constitutively and a galE mutation, which blocks UDP-Gal synthesis, so that the strains cannot synthesize succinoglycan or succinoglycan intermediates until UDP-[14C]Gal is introduced into the cell by electroporation. (A) The lipid linked oligosaccharides extracted from electroporated exoR galE cells with solvent 1203 (23), subjected to mild acid hydrolysis, and chromatographed using a Bio-Gel P4 column. (B) The Bio-Gel P4 chromatography of oligosaccharides isolated from the culture supernatants of electroporated exoR galE cells. (C) The same material as in B but chromatographed through a Bio-Gel P6 column. (D–F) The same analyses as A, B, and C, respectively except that exoR galE exoP cells were used. (G–I) The same analyses as A, B, and C, respectively, except that exoR galE exoT cells were used. (J–L) The same analyses as A, B, and C, respectively but exoR galE exoQ cells were used.
After the electroporation and incubation, the medium was also found to contain radiolabeled material that eluted from a P4 column at the positions expected for the monomeric succinoglycan octasaccharide and for dimers and trimers of this octasaccharide subunit. In addition, higher molecular weight material was present that was excluded from the P4 column (Fig. 2B) as well as from a P6 column (Fig. 2C). None of these species were present if the cells also carried an exoY mutation, which specifically blocks the first step of succinoglycan biosynthesis. In addition, this high molecular weight material was treated with a preparation of Cytophaga arvensicola succinoglycan depolymerase, an enzyme that specifically cleaves succinoglycan at its Gal-β(1, 4)-Glc bond to yield the same fully modified octasaccharide subunit that is produced biosynthetically (33). Before treatment, this high molecular weight material was excluded from a Bio-Gel A5 m column (5,000,000 Da exclusion limit) but after it was almost completely degraded to a lower molecular form, which migrated at the same position as the fully modified succinoglycan octasaccharide upon TLC (data not shown). Taken together, these observations indicated that the use of the electroporation protocol allowed succinoglycan biosynthesis to proceed considerably farther that in our previous study by using the Tris-EDTA/freeze-thaw protocol (23) in which we had only observed lipid-linked monomeric octasaccharide subunits and only monomeric octasaccharides were present in the medium.
An exoP Mutation Blocks the Polymerization of Succinoglycan Octasaccharide Subunits.
We similarly electroporated UDP-[14C]Gal and unlabeled UDP-Glc into an R. meliloti exoR galE exoP strain and examined both the lipid-linked oligosaccharides and the oligosaccharides present in the medium. In contrast to the parental exoR galE strain, the only lipid-linked intermediate extracted from the exoP468∷Tn5 derivative, after the mild acid hydrolysis, eluted from a P4 column at the same position as the monomeric succinoglycan octasaccharide subunit (Fig. 2D). Furthermore, the only radiolabeled species found in the medium eluted from P4 and P6 columns at the same position as the succinoglycan octasaccharide subunit (see Fig. 2 E and F). Because the Tn5 insertion in the exoP468 allele disrupts the exoP ORF within the N-terminal periplasmic domain, it most likely results in complete loss of exoP function (25). Thus, these observations indicate that loss of exoP function blocks any polymerization of the succinoglycan octasaccharide subunits.
An exoT Mutant Produces a High Molecular Weight Form of Succinoglycan But Not Certain Low Molecular Oligosaccharides of Succinoglycan.
A similar analysis of the radiolabeled material synthesized by a R. meliloti exoR galE exoT mutant revealed marked differences from the exoR galE parent and the exoP derivative. In addition to the monomeric succinoglycan octasaccharide subunit, the lipid-linked fraction that had been subjected to mild acid hydrolysis also included radiolabeled material that was of high enough molecular weight to be excluded from a P4 column (Fig. 2G). Strikingly, no material eluted from the P4 column at the positions expected from dimers and trimers of the octasaccharide subunit. Furthermore, analyses of the radiolabeled material in the medium showed that it contained both a species that eluted from the P4 column at the position expected for the monomeric succinoglycan octasaccharide subunit and a form that was excluded from both P4 and P6 columns (Fig. 2 H and I). As with the lipid-linked intermediates, no material was present that eluted at the positions expected for dimers and trimers of the octasaccharide subunit. These observations indicated an exoT mutant synthesizes high molecular weight forms of succinoglycan without accumulating dimers and trimers of the octasaccharide either on lipid carriers or in the medium.
An exoQ Mutant Produces Certain Low Molecular Weight Oligosaccharides of Succinoglycan But Not Higher Molecular Weight Succinoglycan.
Electroporation of radiolabeled UDP-[14C]Gal and UDP-Glc into an R. meliloti exoR galE exoQ mutant revealed that its ability to synthesize succinoglycan was different from the exoR galE parent, the exoP derivative, and the exoT derivative. In addition to the monomeric succinoglycan octasaccharide subunit, the lipid-linked fraction that had been subjected to mild acid hydrolysis treatment also included radiolabeled material that eluted from the P4 column at the positions expected for dimers and trimers of the octasaccharide subunit (Fig. 2J). However, in contrast to the exoR galE parent and its exoT derivative, no radiolabeled material of high enough molecular weight to be excluded from a P4 column was present in the lipid-linked fraction. Furthermore, analyses of the radiolabeled material in the medium showed that it contained, not only a species that eluted from the P4 column at the position expected for the monomeric succinoglycan octasaccharide subunit, but also species eluting at the positions expected for oligosaccharides consisting of dimers and trimers of the octasaccharide subunit. The material did not contain high molecular weight polysaccharide (Fig. 2 K and L). As with the lipid-linked intermediates, no material was present that was excluded from both P4 and P6 columns. These observations indicated an exoQ mutant synthesizes dimers and trimers of the octasaccharide without accumulating high molecular weight forms of succinoglycan either on lipid carriers or in the medium.
DISCUSSION
We have analyzed the roles of the R. meliloti ExoP, ExoQ, and ExoT proteins in the latter stages of the biosynthesis of succinoglycan, a symbiotically important exopolysaccharide. A R. meliloti strain that is wild type with respect to these three functions synthesizes: (i) fully modified octasaccharide subunits, (ii) dimers and trimers of the octasaccharide, and (iii) high molecular weight succinoglycan. Analysis of exoP, exoQ, and exoT derivatives by using an electroporation protocol to introduce radiolabeled nucleotide sugars revealed that each mutant had a different defect in succinoglycan biosynthesis. The exoP derivative produced only the octasaccharide subunit. The exoT derivative produced octasaccharide subunit and also high molecular weight succinoglycan. The exoQ derivative produced octasaccharide subunit and also dimers and trimers of the octasaccharide. In each case, the same species were seen both free in the medium and in lipid-linked forms. The analytical method we used is extremely sensitive, thus enabling the detection of the small amounts of polymerized radiolabeled succinoglycan found in the medium of exoQ and exoT mutants (0.002–0.003 pmol of succinoglycan hexose/108 cells after the 1-hr time course of the experiment). This may account for the fact that we were able to detect the synthesis of succinoglycan material in exoQ and exoT mutants despite the fact that exoP, exoQ, and exoT derivatives of R. meliloti form nonfluorescing colonies under UV when grown on Calcofluor-containing medium. It is also possible that the electroporation procedure may have slightly damaged the cells and facilitated release of polymerized succinoglycan material into the medium despite the lack of ExoQ and ExoT function. In any case, the differences between the exoP, exoQ, and exoT mutants are so striking it seems likely that they reflect physiologically relevant roles of these three gene products.
Our unexpected observation that we can genetically separate the synthesis of oligosaccharides consisting of dimers and trimers of the octasaccharide subunit from the synthesis of high molecular weight succinoglycan is intriguing because of the apparent role of low molecular weight forms of exopolysaccharides in nodule invasion by R. meliloti (14–16) and because of the suggestion that the tetramer of the octasaccharide might be the specific species required in the case of succinoglycan (14). The evidence we have provided here indicates that the species previously identified as a tetramer of succinoglycan octasaccharide subunits (14) is, in fact, a trimer. It is additionally interesting because R. meliloti is known to modulate the molecular weight distribution of the succinoglycan it produces in response to environmental conditions (34). It is possible that the direct synthesis of a specific size class of succinoglycan oligosaccharides, perhaps in response to some environmental conditions encountered during the nodulation process, might play an important role in the organism’s ability to enter into a productive symbiosis with its plant host. Furthermore, it appears that R. meliloti has a substantial genetic investment in controlling the molecular weight of the succinoglycan it synthesizes because it also encodes at least two secreted proteins, ExoK (25, 35) and also ExsH (36), that can depolymerize succinoglycan to yield low molecular weight material (36). Our observation that exoK exsH double mutants are symbiotically proficient (36) raises the possibility that low molecular oligosaccharides necessary for nodule invasion are produced by the direct biosynthetic mechanism, which our investigations have revealed, rather than by degradation of high molecular weight succinoglycan polymer.
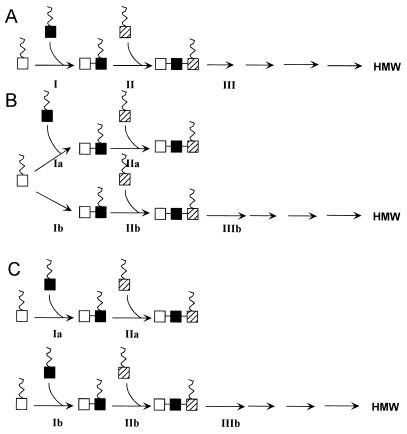
Our analyses of the properties of the exoP, exoQ, and exoT mutants have interesting implications for the latter stages of succinoglycan biosynthesis, in particular for how the octasaccharide subunits are polymerized to yield the specific oligosaccharide size class consisting of dimers and trimers of the octasaccharide subunit as well as high molecular weight succinoglycan. Formally, there are three classes of models for how the polymerization of succinoglycan subunits could occur. In the linear pathway model (Fig. 3A), all low molecular weight lipid-linked oligosaccharides can potentially be elongated to high molecular weight succinoglycan, after a mechanism similar to the one previously proposed for xanthan gum, where the new repeating subunits are transferred from the lipid carrier to the reducing end of the growing polysaccharide (22). A pool of dimers and trimers of the octasaccharide subunit would accumulate if step III was slow compared with all other polymerization steps. In the branched pathway model (Fig. 3B), the pool of lipid-linked dimers of the octasaccharide subunit is partitioned between two separate pathways, one of which specifically produces dimers and trimers of the octasaccharide subunit that are not further elongated and while the other specifically produces high molecular weight succinoglycan. In the parallel pathway model (Fig. 3C), the dimer and trimer class of oligosaccharides is synthesized by a pathway that is completely separate from the one which produces high molecular weight succinoglycan. The latter two models differ from the first in that they involve the synthesis of a specific class of trimers of the octasaccharide subunit that are not substrates for further elongation.
Figure 3.
Models for succinoglycan polymerization of octasaccharide subunits. (A) Linear pathway model for polymerization of lipid-linked succinoglycan subunits (see Discussion). Chain growth is shown occurring from the reducing end as proposed for xanthan gum (22). (B) Branched pathway model for polymerization of lipid-linked succinoglycan subunits. (C) Parallel pathway model for polymerization of lipid-linked succinoglycan subunits.
For all of the models, the very high molecular weight succinoglycan can reach without accumulating tetramers and higher oligomers of the octasaccharide subunits can be explained if the polymerization steps after step II proceed with very high processivity. Regardless of which model is being considered, our observation that the same succinoglycan species were found in both lipid-linked forms and in the medium supports our previous speculation (23) that polymerization of the octasaccharide subunits occurs on a lipid carrier.
For all three models, the failure of the exoP null mutant to accumulate any polymerized octasaccharide subunits whatsoever could be explained economically by postulating that ExoP is required for the polymerization event that generates the dimer of the octasaccharide subunit, perhaps even being the polymerase that carries out that specific polymerization step. Such a hypothesis does not exclude the possibility that ExoP could be required for some or all of the subsequent polymerization steps in any of the models. An alternative possibility is that ExoP has no polymerization function of its own but instead regulates the degree of polymerization of succinoglycan by controlling polymerization activities of one or more proteins, ExoT and ExoQ being the prime candidates. If that were the case, to account for our results, it would be necessary to postulate that both ExoT and ExoQ can synthesize the dimer of the octasaccharide subunit because polymerized product greater than a dimer is seen in both exoT and exoQ mutants. Regardless of how ExoP acts, its C-terminal cytoplasmic domain is not obligatorily required because cells containing only the membrane-anchored N-terminal periplasmic domain of ExoP (37) can synthesize high molecular weight succinoglycan under high osmotic conditions (38).
The possible roles that the ExoQ and ExoT proteins, each of which appear to span the membrane several times, might play in succinoglycan biosynthesis depend on which of the three models is being considered. In both the branched and parallel pathway models, ExoQ could serve as a highly processive polymerase that efficiently elongates dimers of the octasaccharide to yield high molecular weight material whereas ExoT could serve as a decidedly nonprocessive polymerase that efficiently elongates dimers of the octasaccharide to yield only trimers. Whether ExoQ and ExoT also would synthesize the dimer of the octasaccharide itself would depend on the considerations regarding ExoP function that are discussed above. Using the linear pathway model (Fig. 3A), the results we observed could be explained by postulating that ExoT promotes termination of polymerization after Step III whereas ExoQ suppresses termination of polymerization after step III. The balance of their opposing action then would give rise to the distinctive distribution of dimers and trimers and high molecular weight material that is observed in a wild-type cell. It is difficult to accommodate the possibility of ExoQ or ExoT being a polymerase within a linear pathway model.
The mechanism of succinoglycan biosynthesis shares certain key features with the biosynthetic mechanisms of various other exopolysaccharides (e.g., xanthan gum, acetan, colanic acid), certain O-antigens, and certain capsular polysaccharides (39). For all of these, the carbohydrate subunit is sequentially assembled on an undecaprenol carrier on the cytoplasmic face of the plasma membrane and then subsequently polymerized in a blockwise fashion. The synthesis of this class of O-antigen has been the object of intense investigation and three gene products—Wzz (Rol/Cld), Wzx (RfbX), and Wzy (Rfc)—have been identified as being crucial for the steps subsequent to the assembly of the undecaprenol-linked subunit.
Becker et al. (37) suggested that ExoP, as well as proteins involved in polysaccharide synthesis in other bacteria, might be a member of the Wzz (Rol/Cld) family of proteins involved in chain length determination (40–42) and subsequently additional proteins have been similarly classified (43). A careful analysis of the proteins associated with the biosynthesis of surface polymers reveals two major groups of “Wzz (Rol/Cld)-like” proteins. The first group, which is mostly involved in capsule biosynthesis, is ≈42,000 Da in molecular mass and shares homology between themselves [e.g., CtrB (44), BexC (45)]. The second group, which is mostly involved in exopolysaccharide biosynthesis, contains proteins that are larger in size (≈80,000 Da) and has a large C-terminal cytoplasmic domain that Wzz lacks. Most of the proteins in this group have similarities among themselves [e.g., ExoP, AmsA (46), Wzc (47), and EpsB (48) share very similar hydrophilicity plots]. Our results are not compatible with ExoP solely carrying out a dispensable regulatory role in succinoglycan polymerization and indicate that ExoP must play some critical role in succinoglycan biosynthesis that is either enzymatic (e.g., it could catalyze the formation of dimers of the octasaccharide as discussed above) or structural (e.g., it could form a complex with ExoQ and ExoT that is necessary for their function).
Wzx (RfbX) is widely referred to as being a “flippase” that is thought to translocate the undecaprenol-linked O-antigen subunits across the membrane so that the carbohydrate moiety is on the periplasmic side (40, 49). The ExoT protein is a member of a group of proteins that share homology and predicted topological similarities with the Wzx (RfbX) protein (47). Our observation that exoT mutants can still synthesize high molecular weight succinoglycan, a process that presumably takes place in the periplasm, cannot be accounted for by simply postulating that ExoT is the flippase for succinoglycan biosynthesis and raise the possibility that the Wzx (RfbX) protein may have an additional role besides being a flippase or may even have a different role entirely. Considerable evidence indicates that Wzy (Rfc) is the polymerase that responsible for polymerizing the O-antigen subunits (39, 40), an event that occurs on the periplasmic face of the cytoplasmic membrane (50). The ExoQ protein shows only limited homology with one polysaccharide synthesis gene in searches of the current databases. The size and predicted topology of ExoQ are reminiscent of Wzy (Rfc) proteins and our branched and parallel pathway models are compatible with ExoQ serving as a highly processive polymerase that yields high molecular weight succinoglycan. Our results suggest that some caution should be exercised in efforts to assign possible functions to polysaccharide synthesis genes on the basis of sequence homology alone without concomitant biochemical studies (51).
It will be interesting to see whether other bacteria besides R. meliloti similarly have a biosynthetic mechanism for producing a specific size class of oligosaccharide related to exported or cell surface polysaccharides and, if so, whether these play as-yet-undiscovered roles in interactions with other organisms.
Acknowledgments
This work was supported by U.S. Public Health Service Grant GM31030 (to G.C.W.). J.E.G. was supported by a Post-Doctoral Fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. L.E.C.-T. was partly supported by National Science Foundation Grant MCB-9733532 to J.E.G., and C.E.S. was supported by U.S. Public Health Service Grant GM31318 to P. W. Robbins.
ABBREVIATIONS
- HPAEC-PAD
HPLC anion-exchange chromatography coupled with pulsed amperometric detection
- d.p.
degree of polymerization
- Glc
glucose
- Gal
galactose
- Gal-ol
galactitol
References
- 1.Leigh J A, Signer E R, Walker G C. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finan T M, Hirsch A M, Leigh J A, Johansen E, Kuldau G A, Deegan S, Walker G C, Signer E R. Cell. 1985;40:869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- 3.Glazebrook J, Walker G C. Cell. 1989;56:661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 4.Zhan H J, Levery S B, Lee C C, Leigh J A. Proc Natl Acad Sci USA. 1989;86:3055–3059. doi: 10.1073/pnas.86.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller M, Arnold W, Kapp D, Müller P, Niehaus K, Shmidt M, Quandt J, Weng W M, Pühler A. In: Pseudomonas: Biotransformations, Pathogenesis, and Evolving Biotechnology. Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Washington, DC: Am. Soc. Microbiol.; 1990. pp. 91–97. [Google Scholar]
- 6.Harada T, Harada A. In: Polysaccharides in Medicinal Applications. Dumitriu S, editor. New York: Dekker; 1996. pp. 21–58. [Google Scholar]
- 7.Aman P, McNeil M, Franzen L-E, Darvill A G, Albersheim P. Carbohydr Res. 1981;95:263–282. [Google Scholar]
- 8.Jansson P-E, Kenne L, Lindberg B, Ljunggren H, Ruden U, Svensson S. J Am Chem Soc. 1977;99:3812–3815. doi: 10.1021/ja00453a049. [DOI] [PubMed] [Google Scholar]
- 9.Reinhold B B, Chan S Y, Reuber T L, Marra A, Walker G C, Reinhold V N. J Bacteriol. 1994;176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keller M, Müller P, Simon R, Pühler A. Mol Plant-Microbe Int. 1988;1:267–274. [Google Scholar]
- 11.Leigh J A, Walker G C. Trends Genet. 1994;10:63–67. doi: 10.1016/0168-9525(94)90151-1. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch A M. New Phytol. 1992;122:211–237. doi: 10.1111/j.1469-8137.1992.tb04227.x. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H P, Walker G C. J Bacteriol. 1998;80:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Battisti L, Lara J C, Leigh J A. Proc Natl Acad Sci USA. 1992;89:5625–5629. doi: 10.1073/pnas.89.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urzainqui A, Walker G C. J Bacteriol. 1992;174:3403–3406. doi: 10.1128/jb.174.10.3403-3406.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.González J E, Reuhs B L, Walker G C. Proc Natl Acad Sci USA. 1996;93:8636–8641. doi: 10.1073/pnas.93.16.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González J E, York G M, Walker G C. Gene. 1996;179:141–146. doi: 10.1016/s0378-1119(96)00322-8. [DOI] [PubMed] [Google Scholar]
- 18.Vanderslice R W, Doherty D H, Capage M A, Betlach M R, Hassler R A, Henderson N M, Ryan-Graniero J, Tecklenburg M. In: Biomedical and Biotechnological Advances in Industrial Polysaccharides. Crescenzi V, Dea I C M, Paoletti S, Stivala S S, Sutherland I W, editors. New York: Gordon and Breach; 1988. pp. 145–156. [Google Scholar]
- 19.Tolmasky M E, Staneloni R J, Ugalde R A, Leloir L F. Arch Biochem Biophys. 1980;203:358–364. doi: 10.1016/0003-9861(80)90187-3. [DOI] [PubMed] [Google Scholar]
- 20.Tolmasky M E, Staneloni R J, Leloir L F. J Biol Chem. 1982;257:6751–6757. [PubMed] [Google Scholar]
- 21.Ielpi L, Couso R O, Dankert M A. Biochem Biophys Res Comm. 1981;102:1400–1408. doi: 10.1016/s0006-291x(81)80167-2. [DOI] [PubMed] [Google Scholar]
- 22.Ielpi L, Couso R O, Dankert M A. J Bacteriol. 1993;175:2490–2500. doi: 10.1128/jb.175.9.2490-2500.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reuber T L, Walker G C. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- 24.Glucksmann M A, Reuber T L, Walker G C. J Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glucksmann M A, Reuber T L, Walker G C. J Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semino C E, Dankert M A. J Gen Microbiol. 1993;139:2745–2756. doi: 10.1099/00221287-139-11-2745. [DOI] [PubMed] [Google Scholar]
- 27.Semino C E. Ph.D thesis. Argentina.: University of Buenos Aires; 1994. [Google Scholar]
- 28.Glazebrook J, Walker G C. Methods Enzymol. 1991;204:398–418. doi: 10.1016/0076-6879(91)04021-f. [DOI] [PubMed] [Google Scholar]
- 29.Leigh J A, Reed J W, Hanks J F, Hirsch A M, Walker G C. Cell. 1987;51:579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- 30.Reuber T L, Long S, Walker G C. J Bacteriol. 1991;173:426–434. doi: 10.1128/jb.173.2.426-434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buendia A M, Enenkel B, Köplin R, Niehaus K, Arnold W, Pühler A. Mol Microbiol. 1991;5:1519–1530. doi: 10.1111/j.1365-2958.1991.tb00799.x. [DOI] [PubMed] [Google Scholar]
- 32.Canter-Cremers H C J, Batley M, Redmond J W, Eydems L, Breedveld M W, Zevenhuizen L P T M, Pees E, Wijffelman C J, Lugtenberg B J J. J Biol Chem. 1990;265:21122–21127. [PubMed] [Google Scholar]
- 33.Harada T. In: Methods in Carbohydrate Chemistry. BeMiller J N, Manners D J, Sturgeon R J, editors. New York: Wiley; 1994. pp. 155–163. [Google Scholar]
- 34.Breedveld M W, Zevenhuizen L P T M, Zehnder A J B. J Gen Microbiol. 1990;136:2511–2519. [Google Scholar]
- 35.Becker A, Kleickmann A, Keller M, Arnold W, Pühler A. Mol Gen Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- 36.York G M, Walker G C. Mol Microbiol. 1997;25:117–134. doi: 10.1046/j.1365-2958.1997.4481804.x. [DOI] [PubMed] [Google Scholar]
- 37.Becker A, Niehaus K, Pühler A. Mol Microbiol. 1995;16:191–203. doi: 10.1111/j.1365-2958.1995.tb02292.x. [DOI] [PubMed] [Google Scholar]
- 38.Becker A, Pühler A. J Bacteriol. 1998;180:395–399. doi: 10.1128/jb.180.2.395-399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitfield C, Valvano M A. Adv Microb Physiol. 1993;35:135–246. doi: 10.1016/s0065-2911(08)60099-5. [DOI] [PubMed] [Google Scholar]
- 40.Whitfield C, Amor P A, Koplin R. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 41.Bastin D A, Stevenson G, Brown P K, Haase A, Reeves P R. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 42.Morona R, van den Bosch L, Manning P A. J Bacteriol. 1995;177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitfield C. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 44.Frosch M, Edwards U, Kraube K, Weisgerber C. Mol Microbiol. 1991;5:1251–1263. doi: 10.1111/j.1365-2958.1991.tb01899.x. [DOI] [PubMed] [Google Scholar]
- 45.Kroll J S, Loynds B, Brophy L M, Moxon E R. Mol Microbiol. 1990;4:1853–1862. doi: 10.1111/j.1365-2958.1990.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 46.Bugert P, Geider K. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 47.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Schell M. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 49.Liu D, Cole R A, Reeves P R. J Bacteriol. 1996;178:2102–7210. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGrath B C, Osborn M J. J Bacteriol. 1991;173:649–654. doi: 10.1128/jb.173.2.649-654.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R, et al. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]