Abstract
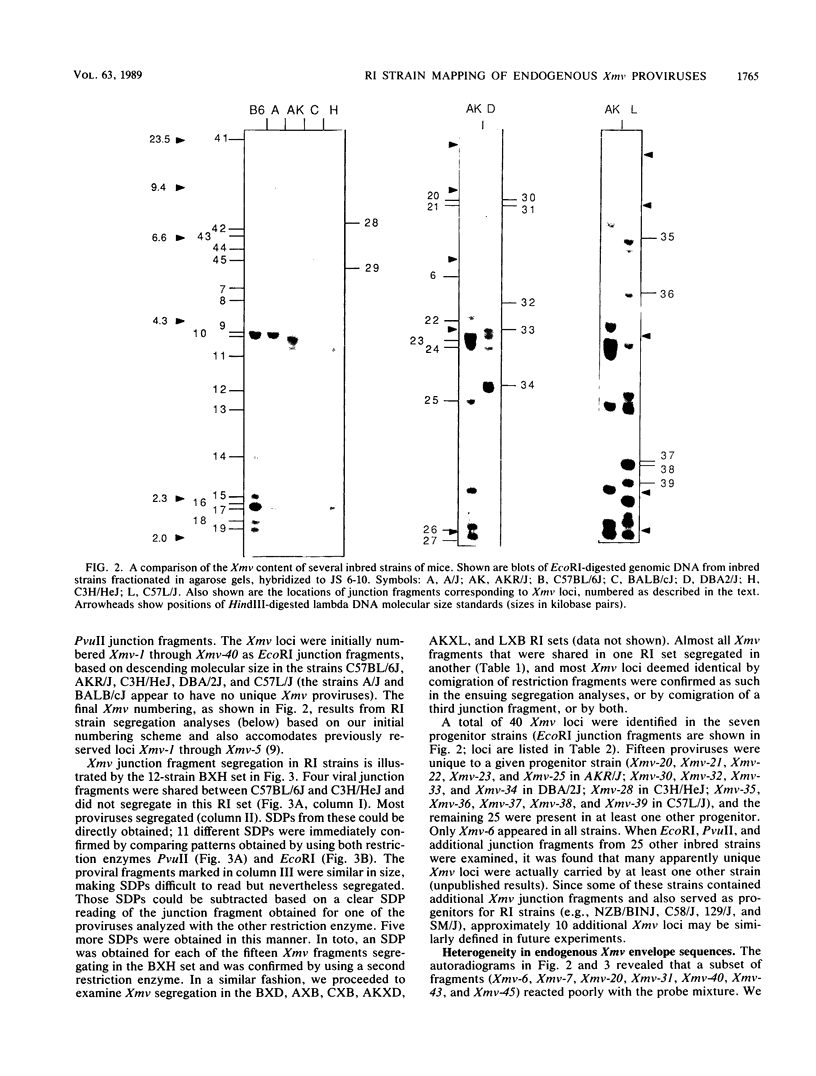
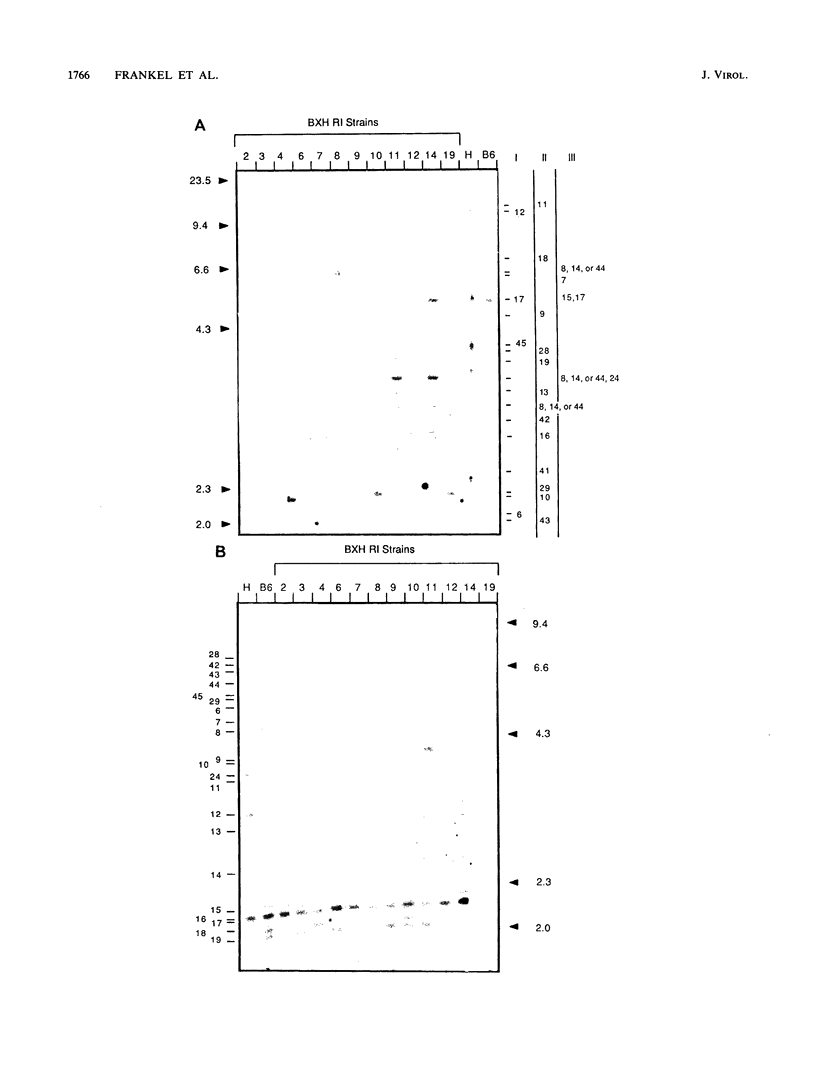
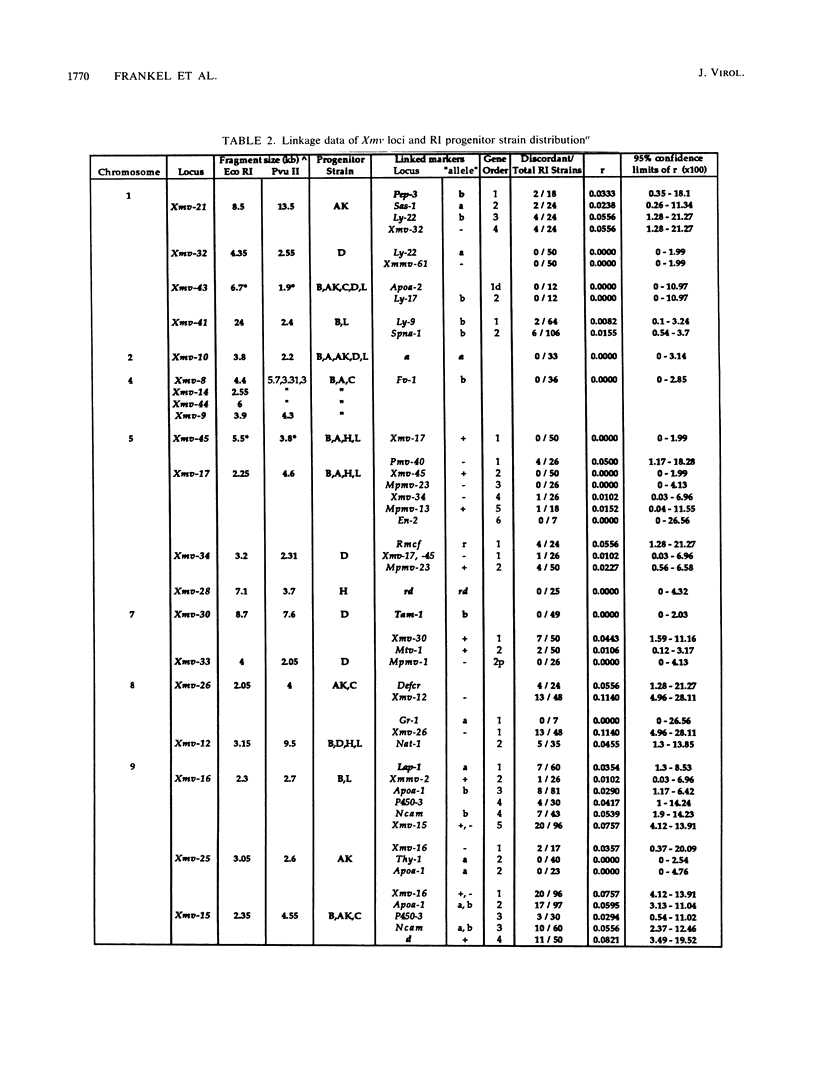
We have defined 40 endogenous xenotropic virus (Xmv) loci from several common inbred strains of mice by examining provirus-cell DNA junction fragments in recombinant inbred mice. Some inbred strains carried unique proviruses, but most Xmv loci were present in several strains, indicating that many Xmv integration events preexisted modern inbreeding. It was also clear that most Xmv junction fragment variation between inbred strains resulted from independent integration events and not modification or restriction site polymorphism following integration. Chromosomal assignments were determined for 32 Xmv loci by comparing their recombinant inbred strain distribution patterns to those of known genetic markers. The Xmv loci were generally dispersed throughout the genome, but several chromosomal regions contained more than one provirus. Furthermore, several close genetic associations with cellular genes were discovered. Four Xmv loci were closely linked to Fv-1b, a dominant viral resistance gene present in C57BL/6J, BALB/cJ, A/J, and several other strains. Xmv-28 was closely linked to rd (retinal degeneration), and Xmv-10 was closely linked to a (non-agouti), both of which are old mutations as inferred from their broad distribution in mice. We suggest that Xmv integration contributed to genetic diversity in the past and that much of this diversity exists today in common laboratory strains.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bishop C. E., Boursot P., Baron B., Bonhomme F., Hatat D. Most classical Mus musculus domesticus laboratory mouse strains carry a Mus musculus musculus Y chromosome. Nature. 1985 May 2;315(6014):70–72. doi: 10.1038/315070a0. [DOI] [PubMed] [Google Scholar]
- Blatt C., Mileham K., Haas M., Nesbitt M. N., Harper M. E., Simon M. I. Chromosomal mapping of the mink cell focus-inducing and xenotropic env gene family in the mouse. Proc Natl Acad Sci U S A. 1983 Oct;80(20):6298–6302. doi: 10.1073/pnas.80.20.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone L. R., Glover P. L., Innes C. L., Niver L. A., Bondurant M. C., Yang W. K. Fv-1 N- and B-tropism-specific sequences in murine leukemia virus and related endogenous proviral genomes. J Virol. 1988 Aug;62(8):2644–2650. doi: 10.1128/jvi.62.8.2644-2650.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S. V., Nadeau J. H., Tanzi R. E., Watkins P. C., Jagadesh J., Taylor B. A., Haines J. L., Sacchi N., Gusella J. F. Comparative mapping of DNA markers from the familial Alzheimer disease and Down syndrome regions of human chromosome 21 to mouse chromosomes 16 and 17. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6032–6036. doi: 10.1073/pnas.85.16.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland N. G., Hutchison K. W., Jenkins N. A. Excision of the DBA ecotropic provirus in dilute coat-color revertants of mice occurs by homologous recombination involving the viral LTRs. Cell. 1983 Jun;33(2):379–387. doi: 10.1016/0092-8674(83)90419-1. [DOI] [PubMed] [Google Scholar]
- Hieter P. A., Hollis G. F., Korsmeyer S. J., Waldmann T. A., Leder P. Clustered arrangement of immunoglobulin lambda constant region genes in man. Nature. 1981 Dec 10;294(5841):536–540. doi: 10.1038/294536a0. [DOI] [PubMed] [Google Scholar]
- Hoggan M. D., O'Neill R. R., Kozak C. A. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J Virol. 1986 Dec;60(3):980–986. doi: 10.1128/jvi.60.3.980-986.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin A., Keshet E. A novel retroviruslike family in mouse DNA. J Virol. 1986 Aug;59(2):301–307. doi: 10.1128/jvi.59.2.301-307.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itin A., Keshet E. Diverse long terminal repeats are associated with murine retroviruslike (VL30) elements. Mol Cell Biol. 1986 Apr;6(4):1276–1282. doi: 10.1128/mcb.6.4.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Dilute (d) coat colour mutation of DBA/2J mice is associated with the site of integration of an ecotropic MuLV genome. Nature. 1981 Oct 1;293(5831):370–374. doi: 10.1038/293370a0. [DOI] [PubMed] [Google Scholar]
- Jenkins N. A., Copeland N. G., Taylor B. A., Lee B. K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982 Jul;43(1):26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolicoeur P., Rassart E. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J Virol. 1980 Jan;33(1):183–195. doi: 10.1128/jvi.33.1.183-195.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A. S. Nucleotide sequence analysis establishes the role of endogenous murine leukemia virus DNA segments in formation of recombinant mink cell focus-forming murine leukemia viruses. J Virol. 1984 Jun;50(3):864–871. doi: 10.1128/jvi.50.3.864-871.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A., O'Neill R. R. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J Virol. 1987 Oct;61(10):3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak C. A. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single dominant locus on chromosome 1. J Virol. 1985 Sep;55(3):690–695. doi: 10.1128/jvi.55.3.690-695.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattano S. S., Erickson R. P., Nesbitt M. N., Weber W. W. Linkage of Nat and Es-1 in the mouse and development of strains congenic for N-acetyltransferase. J Hered. 1988 Nov-Dec;79(6):430–433. doi: 10.1093/oxfordjournals.jhered.a110546. [DOI] [PubMed] [Google Scholar]
- O'Neill R. R., Buckler C. E., Theodore T. S., Martin M. A., Repaske R. Envelope and long terminal repeat sequences of a cloned infectious NZB xenotropic murine leukemia virus. J Virol. 1985 Jan;53(1):100–106. doi: 10.1128/jvi.53.1.100-106.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill R. R., Khan A. S., Hoggan M. D., Hartley J. W., Martin M. A., Repaske R. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J Virol. 1986 May;58(2):359–366. doi: 10.1128/jvi.58.2.359-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou C. Y., Boone L. R., Koh C. K., Tennant R. W., Yang W. K. Nucleotide sequences of gag-pol regions that determine the Fv-1 host range property of BALB/c N-tropic and B-tropic murine leukemia viruses. J Virol. 1983 Dec;48(3):779–784. doi: 10.1128/jvi.48.3.779-784.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips S. J., Birkenmeier E. H., Callahan R., Eicher E. M. Male and female mouse DNAs can be discriminated using retroviral probes. Nature. 1982 May 20;297(5863):241–243. doi: 10.1038/297241a0. [DOI] [PubMed] [Google Scholar]
- Pincus T., Hartley J. W., Rowe W. P. A major genetic locus affecting resistance to infection with murine leukemia viruses. IV. Dose-response relationships in Fv-1-sensitive and resistant cell cultures. Virology. 1975 Jun;65(2):333–342. doi: 10.1016/0042-6822(75)90039-2. [DOI] [PubMed] [Google Scholar]
- Quint W., Boelens W., van Wezenbeek P., Cuypers T., Maandag E. R., Selten G., Berns A. Generation of AKR mink cell focus-forming viruses: a conserved single-copy xenotrope-like provirus provides recombinant long terminal repeat sequences. J Virol. 1984 May;50(2):432–438. doi: 10.1128/jvi.50.2.432-438.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rein A. Interference grouping of murine leukemia viruses: a distinct receptor for the MCF-recombinant viruses in mouse cells. Virology. 1982 Jul 15;120(1):251–257. doi: 10.1016/0042-6822(82)90024-1. [DOI] [PubMed] [Google Scholar]
- Rommelaere J., Donis-Keller H., Hopkins N. RNA sequencing provides evidence for allelism of determinants of the N-, B- or NB-tropism of murine leukemia viruses. Cell. 1979 Jan;16(1):43–50. doi: 10.1016/0092-8674(79)90186-7. [DOI] [PubMed] [Google Scholar]
- Rossomando A., Meruelo D. Viral sequences are associated with many histocompatibility genes. Immunogenetics. 1986;23(4):233–245. doi: 10.1007/BF00373018. [DOI] [PubMed] [Google Scholar]
- Silver J., Buckler C. E. Statistical considerations for linkage analysis using recombinant inbred strains and backcrosses. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1423–1427. doi: 10.1073/pnas.83.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver J. Confidence limits for estimates of gene linkage based on analysis of recombinant inbred strains. J Hered. 1985 Nov-Dec;76(6):436–440. doi: 10.1093/oxfordjournals.jhered.a110140. [DOI] [PubMed] [Google Scholar]
- Siracusa L. D., Russell L. B., Eicher E. M., Corrow D. J., Copeland N. G., Jenkins N. A. Genetic organization of the agouti region of the mouse. Genetics. 1987 Sep;117(1):93–100. doi: 10.1093/genetics/117.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen J. B., Robins D. M. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell. 1988 Oct 21;55(2):247–254. doi: 10.1016/0092-8674(88)90047-5. [DOI] [PubMed] [Google Scholar]
- Stoye J. P., Coffin J. M. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J Virol. 1988 Jan;62(1):168–175. doi: 10.1128/jvi.62.1.168-175.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. P., Coffin J. M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987 Sep;61(9):2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. P., Fenner S., Greenoak G. E., Moran C., Coffin J. M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988 Jul 29;54(3):383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Suzuki S. FV-4: a new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn J Exp Med. 1975 Dec;45(6):473–478. [PubMed] [Google Scholar]
- Wejman J. C., Taylor B. A., Jenkins N. A., Copeland N. G. Endogenous xenotropic murine leukemia virus-related sequences map to chromosomal regions encoding mouse lymphocyte antigens. J Virol. 1984 Apr;50(1):237–247. doi: 10.1128/jvi.50.1.237-247.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. K., Kiggans J. O., Yang D. M., Ou C. Y., Tennant R. W., Brown A., Bassin R. H. Synthesis and circularization of N- and B-tropic retroviral DNA Fv-1 permissive and restrictive mouse cells. Proc Natl Acad Sci U S A. 1980 May;77(5):2994–2998. doi: 10.1073/pnas.77.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]