Abstract
Salmonella enterica serovar Typhimurium is capable of swarming over semi-solid surfaces. Although its swarming behavior shares many readily observable similarities with other swarming bacteria, the phenomenon remains somewhat of an enigma in this bacterium since some attributes skew away from the better characterized systems. Swarming is quite distinct from the classic swimming motility, as there is a prerequisite for cells to first undergo a morphological transformation into swarmer cells. In some organisms, swarming is controlled by quorum sensing, and in others, swarming has been shown to be coupled to increased expression of important virulence factors. Swarming in serovar Typhimurium is coupled to elevated resistance to a wide variety of structurally and functionally distinct classes of antimicrobial compounds. As serovar Typhimurium differentiates into swarm cells, the pmrHFIJKLM operon is up-regulated, resulting in a more positively charged LPS core. Furthermore, as swarm cells begin to de-differentiate, the pmr operon expression is down-regulated, rapidly reaching the levels observed in swim cells. This is one potential mechanism which confers swarm cells increased resistance to antibiotics such as the cationic antimicrobial peptides. However, additional mechanisms are likely associated with the cells in the swarm state that confer elevated resistance to such a broad spectrum of antimicrobial agents.
Keywords: Drug Resistance, Salmonella enterica, Physiological Processes
Introduction
Salmonella enterica serovar Typhimurium (S. typhimurium) is a bacterium capable of infecting a wide range of mammalian hosts. Although it can cause typhoid-like fever in mice, it is usually associated with a self-limiting gastroenteritis in humans (1). In some cases, however, the disease can manifest as a chronic infection in humans where the biliary duct and gall bladder may serve as the potential reservoirs for re-infection (2, 3). In addition to being a clinically significant pathogen, S. typhimurium has also long served as an important bacterial model system for studying many aspects of general microbial physiology and molecular biology. One such aspect is chemotaxis and swimming motility in aqueous environments (4-9). More recently, it was demonstrated that S. typhimurium is also capable of swarming over semi-solid surfaces (10). These two distinct forms of motility are driven by the same physical (i.e. flagella) and chemotactic machineries (10, 11), however, swarming is strictly preceded by a morphological differentiation of short swimmer cells into elongated, multinucleated and hyperflagellated swarmer cells.
Although the mechanisms that drive the surface swarming behavior in S. typhimurium remain largely uncharacterized, several intriguing phenotypes have been reported to be associated with swarming motility in other organisms. Cell-density is a key factor that triggers swarmer differentiation and migration (10, 12), which suggests that swarming is a co-ordinated multicellular behavior. Co-ordinated behaviors can be mediated by intricate communication mechanisms between individual cells within a given population, a phenomenon generally referred to as quorum sensing (13, 14). Indeed, the disruption of the quorum sensing systems in a variety of organisms has been shown to abolish their ability to swarm (15-18). In the uropathogen Proteus mirabilis, the transition from the swimmer to swarmer cells plays an important role during the invasion of uroepithelial cells (19), which is largely due to the increased expression of important virulence factors coupled to swarm-cell differentiation (20). Taken together, swarming is a clinically significant population behavior, which promotes co-ordinated, yet rapid community migration and expansion. These observations warrant studies to unravel other physiological phenomena that are linked to the swarming behavior.
By utilizing a transposon-based mutagenesis approach, we isolated a swarm-defective mutant of S. typhimurium (21). The transposition site was determined to be in an operon associated with resistance to cationic peptides. E-test analyses revealed that swarm-cell differentiation in wild-type S. typhimurium results in elevated resistance to not only cationic peptides, but also against several other classes of antibiotics. In this report, we provide an in-depth methods supplement to the E-test analyses, in addition to other experimental approaches to further validate that swarm cell differentiation in S. typhimurium is coupled to multiple antibiotic resistance.
Materials and Methods
Bacterial strain and media
S. typhimurium 14028 is a wild-type virulent strain (ATCC 14028). All media components were from Difco, or as noted otherwise. For routine culturing, Luria broth (LB) and agar were used, and when necessary, kanamycin (Sigma) was added at 50 μg/ml. For swarm and swim motility assays, 0.5 μl of an overnight culture was spotted in the middle of a swarm plate (Nutrient broth [NB], 0.5% [w/v] glucose, 0.5% [w/v] agar) or a swim plate (NB, 0.5% glucose, 0.25% agar) and allowed to dry for 1 h at room temperature. All plates were incubated at 37°C for 10 h, or as noted otherwise.
Construction of the pmrHFIJKLM operon transcriptional reporter system
The intergenic region between the divergently oriented pmrG gene and the pmrHFIJKLM operon was PCR amplified from S. typhimurium 14028 by primers Pmr1 (5’-AGTCGGATCCACTACCGGATGCTGCTTC-3’) containing a BamH1 restriction site and Pmr2 (5’-AGTCCTCGAGATTGCCAGTTAGCCGACA-3’) with an XhoI restriction site. Plasmid pCS26 is a low-copy vector that contains a kanamycin resistance gene cassette, and a multiple cloning site (MCS) upstream of a promoterless luciferase operon (luxCDABE) (22). Plasmid pU220 is identical to pCS26, except that the orientation of the MCS is reversed (i.e. BamH1-XhoI-lux operon orientation). The PCR product containing the pmr operon promoter (Ppmr) was digested with BamH1 and XhoI, and subsequently cloned into pU220 to generate the Ppmr::lux transcriptional reporter plasmid pPMRH. Plasmid pPMRH was then transformed into strain 14028. Molecular techniques were performed as described by Sambrook et al. (23), and all enzymes were obtained from Invitrogen.
pmr operon expression analysis
The strain 14028 harbouring the plasmid pPMRH was used to monitor the expression of the pmr operon. 0.5 μL of an overnight culture was spotted onto NBG (NB with 0.5% glucose) plates containing 0, 0.25, 0.5, 0.7, 1.0, or 1.5% agar. Following 10 h of incubation at 37°C, cells were harvested with sterile toothpicks and inoculated into NBG, and the optical density at 620nm (OD620) of all samples were equilibrated to 0.08-0.09 in 200 μl of fresh NBG. The luciferase activity in the cell preparations and the OD were measured in clear-bottom 96 well black plates (Nunc) with the Victor2 (Wallac) luminometer, as described by Bjarnason et al. (22).
To monitor the changes in the pmr operon expression during de-differentiation of swarmer cells into swimmer cells, the same procedures were followed as described above, except that measurements were taken at 30 minute intervals over 24 h at 37°C. Sterile mineral oil (50 μl) was added to individual wells to prevent evaporation during the assay.
All assays were performed with or without the presence of kanamycin in NBG (+/- agar) and no significant differences were observed in luciferase activity. The low-copy plasmid pU220 is therefore stable even in the absence of kanamycin. All assays were repeated four times to ensure reproducibility.
Comparison of antibiotic Resistance using E-test strips
E-test strips (AB Biodisk, Solna, Sweden) were used to compare the relative resistances of swarming and non-swarming cells of strain 14028 to a variety of antibiotics. Non-swarming (vegetative) cells were analyzed on either solid (NBG, 1.5% agar) or swim plates. For solid plate analysis, 200 μl inocula containing 104-105 cells were evenly spread and allowed to dry at room temperature for 30 min, and the E-test strip was placed in the middle of each plate. On swim plates, 0.5 μl of the seed culture was inoculated at two spots and allowed to dry, and the E-test strip was placed between the two spots. Swarm plates were inoculated in the same manner as the swim plates. All plates were incubated overnight at 37°C, and the E-test results were interpreted as recommended by the manufacturer, or as noted otherwise. All assays were repeated at least two times to ensure reproducibility.
Monitoring changes in antibiotic resistance during de-differentiation of swarmer cells into swimmer cells
To determine the swimmer cells’ minimum inhibitory concentration (MIC) for polymyxin and kanamycin, an overnight LB culture of strain 14028 was diluted in fresh LB medium and 104-105 cells were inoculated into 96 well plates, with each well containing 100 μl LB medium supplemented with a series of two-fold dilutions of polymyxin and kanamycin at concentrations ranging from 500 to 0.000125 μg/ml. The plates were incubated with shaking at 37°C overnight, and the minimum concentration that inhibited visible growth was declared as the MIC (kanamycin 31.25 μg/ml, polymyxin 8 μg/ml).
To monitor the changes in resistance during de-differentiation, cells were harvested from swarm plates with 3 ml LB broth by gentle shaking on a rotating platform and inoculated into a flask containing 250 ml LB at a final OD600 of 0.01. The flask was incubated with shaking (200 rpm) at 37°C, and starting at the time of initial transfer, 1-5 ml aliquots were removed at 30 minute intervals over a period of 8 h. At each interval, the aliquot was diluted back to the initial OD600 with fresh LB. In three 1.5 ml microfuge tubes each containing 1 ml LB, cells were exposed to either 62.5 μg/ml kanamycin or 15 μg/ml polymyxin for 30 min at room temperature. For the reference culture, three additional aliquots were incubated at each time point in the same manner as above, but without exposure to antibiotics. Following exposure, all samples were immediately diluted by a series of 10-fold dilutions in 96 well plates with fresh LB to dilute out the antibiotic, and 5 μl aliquots from each well were spot-plated onto Luria agar plates, allowed to dry, and incubated at 37°C overnight. Colonies were counted and averaged from the three independent exposures to each antibiotic, and divided by the number of colonies obtained from unexposed samples for each time point to generate the survival curve.
Results and Discussion
Lipopolysaccharide (LPS) plays an important role in swarming, as mutations in numerous loci involved in LPS biosynthesis and modification result in a defective swarming phenotype (24). The pmrHFIJKLM operon in S. typhimurium encodes enzymes that add 4-amino-4-deoxy-L-arabinose (L-AraN) residues to the lipid A portion of LPS (25, 26). This modification results in a more positively charged outer membrane, reducing the binding potential of positively charged antimicrobial peptides such as polymyxin. A mutation in the pmr operon also abolishes swarming motility in S. typhimurium 14028 (21). To determine whether swarm-cell differentiation leads to altered regulation of the pmr operon, we constructed a pmr operon promoter (Ppmr)-luciferase transcriptional fusion to compare its expression under swim, swarm, and non-motile conditions. When grown in NBG medium containing 0.5-1.0% (w/v) agar, S. typhimurium undergoes swarmer differentiation and exhibits surface motility (Fig. 1). The pmr operon is significantly up-regulated under conditions that promote swarming, and the highest expression was observed in the presence of 0.5% agar, which coincides with the most proficient swarming motility. Although the pmr operon is also expressed in non-swarming (vegetative) populations, its expression was consistently lower than in those conditions which promote swarmer differentiation (Fig. 1). The expression profile is in agreement with β-galactosidase activities measured under similar conditions in a strain containing a lacZ insertion (MudJ) in the pmrK gene (21). The up-regulation of the pmr operon in swarm cells suggests that cells in the swarm state should be more resistant to polymyxin than those in the vegetative state.
Fig. 1. Expression profiling of the pmrHFIJKLM operon in swarm and vegetative cells of S. typhimurium 14028.
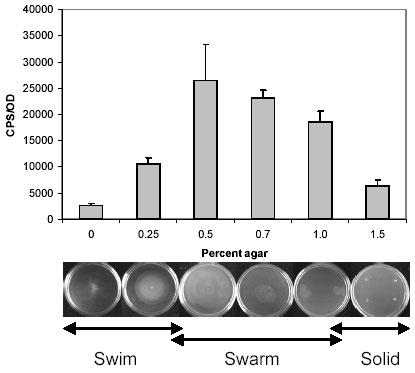
Luciferase activity of the Ppmr-luxCDABE reporter was measured in cells grown in various agar concentrations with NBG (Nutrient broth, 0.5% glucose) as the basal medium, and normalized with OD620nm. The lower images show that cells are swimming (0, 0.25%), swarming (0.5-1.0%), or non-motile (1.5%) depending on the agar concentration. The normalized luciferase activities (CPS/OD) were averaged from four independent experiments, and the error bars represent the standard deviation.
It was not feasible to compare polymyxin resistance between vegetative and swarm cells by the conventional overnight minimum inhibitory concentration (MIC) assays, since swarm cells de-differentiate into vegetative cells when grown in liquid media. To overcome this issue, we utilized polymyxin-coated E-test strips (AB Biodisk) directly on swarm plates and compared relative MIC differences against swim and solid plates. Unlike the standard antibiotic diffusion disks, E-test strips contain a defined immobilized antibiotic gradient, generating a continuous antibiotic gradient when placed on an agar surface (27). In agreement with the pmr operon expression data, swarm cells exhibited greater tolerance to polymyxin at least an eight-fold higher concentration in comparison to the cells in the vegetative state (Fig. 2). Remarkably, the E-test strips generated very similar results between the swim and solid plates, indicating that the relative differences in the agar concentrations (i.e. wetness of the plates) did not significantly distort the immobilized antibiotic gradient.
Fig. 2. E-test strip comparison of polymyxin and kanamycin resistance of strain 14028 in vegetative and swarm states.
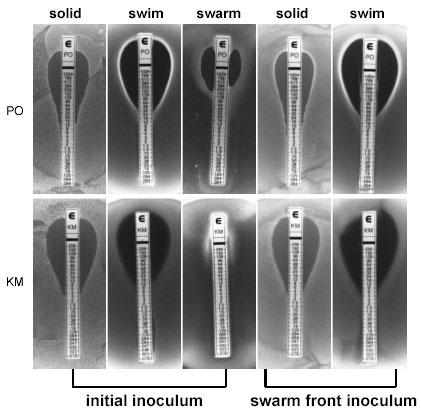
Resistance to polymyxin (PO) is shown in the upper level and kanamycin (KM) is shown below. Initial inoculum: cells were grown overnight in NBG then spread-plated on NBG containing 1.5% agar (solid) or spot-plated on 0.25% (swim), or 0.5% (swarm) agar. Swarm front inoculum: cells were harvested from the swarm front at the highest concentration of the antibiotic from the initial inoculum (i.e. middle images) and grown overnight in NBG, then inoculated on either solid or swim plates as done initially.
Given the relative simplicity of utilizing the E-test strips, we compared the MICs of 25 other antibiotics against the swarm and vegetative cells of strain 14028 (21). Detailed results are presented in Kim et al. (21) and are summarized below. Against penicillin G, oxacillin, bacitracin, clarithromycin, erythromycin, clindamycin, and trimethoprim, both populations of cells grew at concentrations beyond the maximum range of the strips. Ampicillin, amoxicillin, amoxicillin-clavulanic acid, imipenem, fosfomycin, tetracycline, and doxycycline, representing different generations of β-lactams and tetracyclines, yielded identical results between the swarm and vegetative cells. These results further support the notion that the MIC differences observed between swarm and vegetative cells are genuinely the result of the swarm-cell differentiation. Swarm cells exhibited elevated resistance to 11 antibiotics, with cellular targets extending beyond the cell envelope, including those involved in protein synthesis, DNA replication, and transcription. Greater than five-fold increase in resistance was observed against colistin (polymyxin E) and three of the newer generation β-lactams. Swarm cells were greater than 20-fold more resistant against the aminoglycosides, quinolones and rifampin, and with the exception of ciprofloxacin (93-fold increase), the swarm front migrated beyond the maximum range of the strips (Fig. 2). Distinct waves of swarm fronts were observed against increasing levels of several antibiotics (Fig. 3), suggesting that we may be selecting for resistant mutants. To address this issue, cells were harvested from the swarm fronts at the highest concentrations from seven different antibiotics, including those shown in Fig. 3, and incubated overnight in fresh nutrient broth supplemented with glucose (NBG). Swarm cells de-differentiate into vegetative cells when transferred from swarm plates to liquid media. The cell preparations were spread onto fresh solid NBG plates and spotted onto fresh swim plates, then re-exposed to a respective set of E-test strips. As represented by polymyxin and kanamycin strips in Fig. 2, identical MIC values were generated in comparison to the initial inocula, for all seven antibiotics tested. Therefore, the elevated antibiotic resistance phenotype observed in swarm cells is reflective of adaptive changes associated with this transient physiological state rather than selective mutations. The distinct waves of swarm fronts (Fig. 3) are somewhat similar in appearance to the concentric rings produced by Proteus spp., which represent phases of consolidation, where swarm cells repeatedly de-differentiate and re-differentiate in spatially defined cycles (12). However, Salmonella spp. do not typically exhibit phases of consolidation (10), thus, the distinct fronts may indeed represent phases of adaptation that are unique to the cells in the swarm state in response to increasing levels of antibiotics. Hence, we hypothesize that distinct fronts will form against the aminoglycosides at concentrations higher than the maximum range of the E-test strips.
Fig. 3. Distinct waves of swarm fronts formed against several antibiotics.
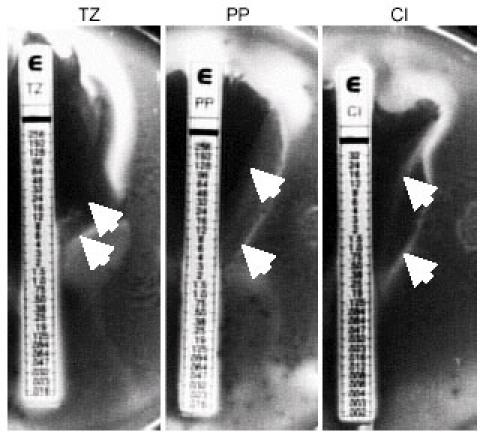
More than one zone of growth inhibition was seen on swarm plates (NBG, 0.5% agar) against ceftazidime (TZ), piperacillin (PP), and ciprofloxacin (CI). The arrows indicate the presence of initial and secondary swarm fronts.
In order to obtain further insight into the de-differentiation phase, we monitored the activity of the pmr operon during the transition from swarm to swim cells. Similar to the experiment shown in Fig. 1, the Ppmr-luciferase reporter strain was grown under swim, swarm and non-motile conditions. Cells from individual growth conditions were harvested and inoculated into fresh liquid NBG, and luciferase activities in each cell types were measured over time. The expression of the pmr operon in cells grown on 0.5% agar (i.e. swarm plates used in E-test analyses) dropped approximately 50% just thirty minutes following transfer, and continued to decrease rapidly until 60 minutes post-transfer (Fig. 4). These results suggest that de-differentiation begins rapidly, and the protective L-Ara4N LPS-modification is most likely diluted away in the population at a similar rate. In agreement with the data shown in Fig. 1, the pmr operon expression profile was highest at the moment of transfer (t=0), under growth conditions that favor swarm-cell differentiation (Fig. 4).
Fig. 4. Expression of the pmr operon during the de-differentiation phase.
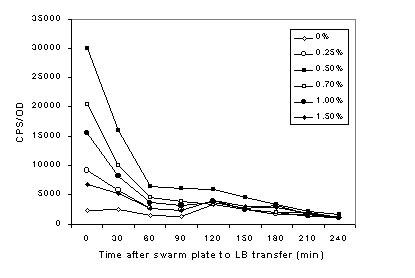
Cells grown for 10 h in various agar concentrations (shown in the legend) were inoculated into fresh NBG, and the luciferase activity of the Ppmr-luxCDABE reporter was measured over time. The data points shown at time 0 were obtained immediately following the transfer of cells from their respective plates to NBG. The graph is representative of four independent experiments.
Albeit at a much reduced amplitude, the same expression profile can be seen up to 90 minutes post-transfer. Regardless of the agar concentration, pmr activity continues to drop until 210 minutes post-transfer, when the activity from all cell types reach a similar level to that detected in NBG without agar. Similar expression profiles were seen as those at 210 and 240 minutes post-transfer throughout the period of 24 hours.
Only the cationic peptide strips generated sharp and pronounced zones of growth inhibition on the swarm plates (Fig. 2). With the exception of E-test strips that permitted the swarm front to migrate beyond their maximum range, and those that produced no difference between the swarm and vegetative cells, multiple swarm fronts were observed (Fig. 3). These observations, in addition to the wide range of structural differences and the cellular targets represented by the wide variety of antimicrobial compounds, suggest that swarm-cell differentiation is likely coupled to multiple resistance mechanisms. In addition to the LPS modification resulting from the up-regulation of the pmr operon, the dramatic morphological changes associated with swarm-cell differentiation may be indicative of an altered cell envelope structure. This could decrease the effectiveness of the cell wall-targeting compounds and the influx-rate of those that target the components within the cytoplasm (28, 29).
The rapid decrease in the expression of the pmr operon (Fig. 4) suggests that the swarm cells’ susceptibility to polymyxin should be rapidly restored to that of vegetative cells when forced into the de-differentiation phase. To test this hypothesis, swarm cells were inoculated into fresh LB to trigger de-differentiation and the transferred cells were exposed to polymyxin at a concentration that was previously determined to be lethal to cells grown in LB. Immediately following transfer, less than 10% of the population survived the polymyxin treatment and 100% death was observed when cells were exposed after 30 minutes post-transfer (Fig. 5). The rapid killing rate is in agreement with the rapid decrease in the pmr operon expression. The post-transfer time indicated on the x-axes in Figs. 4 and 5 are not in frame with one another, since cells sampled at each time point was exposed to polymyxin for an additional 30 minutes prior to plating out the cells. The sharp zone of growth inhibition generated by the polymyxin E-test strips against the swarm front (Fig. 2) may be reflective of its rapid killing mechanism, as evidenced by the sharp killing curve (Fig. 5). There are at least five other genetic loci in Salmonella that have been implicated in conferring resistance to polymyxin and other antimicrobial peptides, by unknown mechanisms that are independent of the L-Ara4N LPS modification (30, 31). Whether or not these genes play physical and/or regulatory roles in polymyxin resistance, some may also behave differently during the transition into the de-differentiation phase. In contrast, virtually the entire population survived lethal dose exposures to kanamycin shortly after transfer (Fig. 5).
Fig. 5. Changes in polymyxin and kanamycin resistance during the de-differentiation phase.
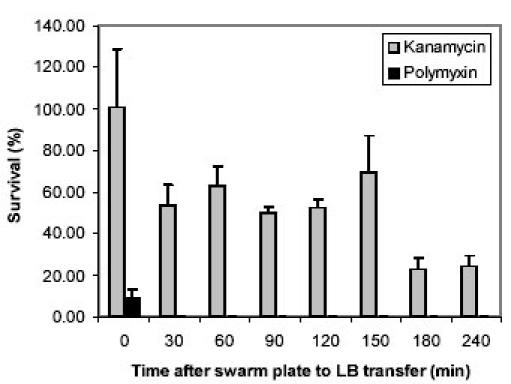
Swarm cells were inoculated into fresh Luria broth (LB) and incubated with shaking at 37°C. Time 0 represents the time of transfer from the swarm plates to LB. At each time point, aliquots were removed from the LB culture and exposed to lethal concentrations of polymyxin (15 μg/ml) or kanamycin (62.5 μg/ml) for 30 min. At each time point, an additional aliquot was left unexposed for 30 min. After the 30 min incubation, cells were immediately diluted in fresh LB (also diluting the lethal dose of the antibiotic) and plated on Luria agar plates, and colonies were counted the next day. To calculate the percentage of surviving cells, the colony counts from the exposed samples were divided by the unexposed samples for individual time points. The results were averaged from three independent exposures and the error bars indicate the standard deviation.
There was more significant killing at 30 minutes post-transfer, where only about 50% of the population survived the exposure. Between 30 to 150 minute post-transfer, there were no dramatic changes in the survival of the population to kanamycin exposure, but viability dropped down to less than 25% after 180 and 240 minutes post-transfer. The kanamycin survival profile may reflect the presence of a biphasic, or at least two independent resistance mechanisms in the swarm population of S. typhimurium. The first significant killing at 30 minutes post-transfer may be related to the dramatic reduction in the pmr operon expression, which would lead to a less positively charged cell surface. Since aminoglycosides are also positively charged, the pmr-induced modification may provide the initial protection. Numerous other changes may occur in the outer membrane, since the influx rate of aminoglycosides have been shown to increase in the presence of other cell-wall targeting antibiotics (32) and other alterations in the outer membrane stability (28). Alternatively, extracellular components have been shown to bind and block the entry of the another aminoglycoside antibiotic, tobramycin (29). The second significant killing observed at 180 minutes post-transfer may reflect the loss of these and/or other yet-to-be-characterized resistance mechanisms. Variability was quite high at certain time points, but this is due to the fact that the results were averaged from three independent exposures for each time point.
Conclusions
The use of E-test strips on non-traditional media revealed that swarm-cell differentiation in S. typhimurium is intimately associated with elevated antibiotic resistance, whereby at least one mechanism was traced down to the level of gene regulation. In the instruction manuals provided with the E-test strips, the manufacturer (AB Biodisk) instructs the user to disregard the swarm fronts of Proteus spp. when declaring the MIC. Unlike most bacteria that are capable of swarming, Proteus spp. represent a rather unique class in that they undergo differentiation and exhibit surface motility even on standard solid (i.e. 1.5% agar) plates (12) that are the standard medium for E-test strip applications. Interestingly, the absence of L-Ara4N modification of the LPS was shown to affect swarming motility and to increase sensitivity to cationic antimicrobial peptides in P. mirabilis (33). Thus, elevated antibiotic resistance may be a general phenotype associated with other swarming bacteria and may share similar resistance mechanisms.
The secondary swarm fronts that were observed in response to some of the E-test strips add another level of interest. In addition to comparing swarm-cells and vegetative cells, such subpopulations can be separately harvested and compared to the remaining population for subtractive analyses of differential gene regulation and biochemical modifications. This approach may provide insight into novel resistance mechanisms and antimicrobial targets. Biofilm cells are believed to possess a variety of antibiotic resistance mechanisms, however, there are some convincing evidence that the major resistance mechanism stems from the fact that biofilm cells grow very slowly, and most antibiotics are most effective against rapidly growing cells (34, 35). Unlike in a biofilm, S. typhimurium grows exponentially in the swarm state, which suggests that swarming S. typhimurium may possess a distinct set of resistance mechanisms. Conversely, these two populations of cells may also share a similar set of resistance mechanisms, most likely those associated with alterations in the cell envelope and extracellular components.
This study supports the view that bacteria are highly adaptive, and their robustness may be achieved by numerous transient physiological states. A swarming population of bacteria represents a unique physiological state that promotes rapid population expansion and migration. Swarming in Salmonella may potentially be a clinically significant state, since the cells are well equipped to withstand high levels antimicrobial compounds that are either artificially introduced or naturally generated by the hosts. In addition to the utility of E-test strips, the luciferase-based reporters provide a simple and sensitive means for monitoring gene regulation in real-time. The methods described here can be easily implemented into studying other swarming bacteria in addition to other interesting phenomena, where a bacterium undergoes a transition from one physiological state to another.
Acknowledgments
We thank D. Woods and J. Davies for providing E-test strips and members of the Surette laboratory for comments on the manuscript and helpful suggestions. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR). M.G.S. is supported as an Alberta Heritage Foundation for Medical Research Senior Scholar and Canada Research Chair in Microbial Gene Expression.
Appendix
Protocols
Swarm and swim motility assays
Solutions and media:
Difco Nutrient Broth (NB).
NBG: NB with 0.5% glucose
Glucose stock: 50% (w/v) in dH2O, filter sterilized
Swim plate: NBG, 0.25% agar
Swarm plate: NBG, 0.5% agar
Solid plate: NBG, 1.5% agar
Protocol:
From a frozen stock, streak out strain on a solid plate and incubate overnight at 37°C
Inoculate an isolated colony in a test tube with 5 ml NB and incubate overnight
Autoclave swarm and swim media and cool down in a 50°C water bath. On a stir plate, add glucose and cool down as much as possible while stirring. Pipet exactly 30 ml volumes into petri dishes. Solidify plates at room temperature overnight (do not invert).
Inoculate 0.2-1 μL of the overnight NB culture in the center of the plates. When inoculating swim plates, pierce the surface with the pipet tip. When inoculating swarm plates, release the inoculum on the surface.
Leave the plate lid slightly open until the inoculum is completely absorbed by the medium. Rotate the lid to maintain uniform drying.
Once the inoculum is completely dry, incubate the plates at 37°C for 8-12 h. Do not invert or stack plates. Make sure to cover the plates with a tray to prevent drying during incubation. It may be necessary to use an incubator with humidity control.
Note: The protocol may need modifications depending on the humidity of the laboratory. Our experience has been that conditions need to be optimized for each laboratory. For Salmonella, swarming patterns and rates are very sensitive to plate conditions. If the working area is very dry, let the plates solidify for 2 hours then cover with a tray to maintain humidity. For more humid environments, longer drying times may be necessary. Any liquid remaining on the surface of the plates will result in rapid spreading of the bacteria and not represent swarming. This is usually easily distinguished by the observed patterns. Never stack plates during the entire procedure, as this will vary the drying from plate to plate and reproducible swarms will not be observed on the different plates. It is our experience that with careful control of the parameters it is possible to obtain consistent and reproducible swarm patterns. It should also be stressed that the different patterns can be obtained with only small changes in plate conditions.
E-test strip analysis on Swim and Swarm plates
Protocol:
Prepare swarm, swim, and solid plates as described above.
For solid plate inoculations, follow the instructions provided by the manufacturer.
Inoculate swim and solid plates as described above, but rather than inoculating the center of the plate, inoculate two spots away from one another closer to the edges of the plate.
Let the media absorb the inocula as described above.
Place the E-test strip between the two inocula. Make sure that no air bubbles get trapped between the strip and the medium.
Incubate overnight at 37°C as described above.
Follow the instructions provided by the manufacturer to interpret the MIC results on solid and swim plates. For swarm plates, declare the MIC where the swarm front is inhibited from migrating.
References
- Finlay BB, Brumell JH. Salmonella interactions with host cells: in vitro to in vivo. PhilTrans R Soc Lond B. 2000;355:623–631. doi: 10.1098/rstb.2000.0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BB. Molecular and cellular mechanisms of Salmonella pathogenesis. Curr Top Microbiol. 1994;192:163–185. doi: 10.1007/978-3-642-78624-2_8. [DOI] [PubMed] [Google Scholar]
- Prouty AM, Schwesinger WH, Gunn JS. Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect Immun. 2002;70:2640–2649. doi: 10.1128/IAI.70.5.2640-2649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry RM, Armitage JP. The bacterial flagella motor. Adv Microb Physiol. 1999;41:291–337. doi: 10.1016/s0065-2911(08)60169-1. [DOI] [PubMed] [Google Scholar]
- Bouret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–9628. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:594–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist FW. Amplification of signaling events in bacteria. Sci STKE. 2002;132:PE24. doi: 10.1126/stke.2002.132.pe24. [DOI] [PubMed] [Google Scholar]
- Stock JB, Surette MG. Chemotaxis. In: Neidhardt FC, Curtis III R, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schacchter M, Umbarger HE, editors. Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. Washington, DC: ASM Press; 1996; p. 1103-1129.
- Stock JB, Levit MN, Wolanin PM. Information processing in bacterial chemotaxis. Sci STKE. 2002;132:PE25. doi: 10.1126/stke.2002.132.pe25. [DOI] [PubMed] [Google Scholar]
- Harshey RM, Matsuyama T. Dimorphic transition in Escherichia coli and Salmonella typhimurium: surface-induced differentiation into hyperflagellate swarmer cells. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkart M, Toguchi A, Harshey RM. The chemotaxis system, but not chemotaxis, is essential for swarming motility in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GM, Hughes C. Swarming motility. Curr Opin Microbiol. 1999;2:630–635. doi: 10.1016/S1369-5274(99)00033-8. [DOI] [PubMed] [Google Scholar]
- Bassler BL. Small talk. Cell-to-cell communication in bacteria. Cell. 2002;109:421–424. doi: 10.1016/s0092-8674(02)00749-3. [DOI] [PubMed] [Google Scholar]
- Fuqua C, Parsek MR, Greenberg EP. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Ann Rev Genet. 2001;35:439–468. doi: 10.1146/annurev.genet.35.102401.090913. [DOI] [PubMed] [Google Scholar]
- Lindum PW, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-L-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang S, Horng YT, Shu JC, Soo PC, Liu JH, Yi WC, Lai HC, Luh KT, Ho SW, Swift S. The role of RsmA in the regulation of swarming motility in Serratia marcescens. J Biomed Sci. 2001;8:160–169. doi: 10.1007/BF02256408. [DOI] [PubMed] [Google Scholar]
- Reimman C, Ginet N, Michel L, Keel C, Michaux P, Krishnapillai V, Zala M, Heurlier K, Triandafillu K, Harms H, Defago G, Haas O. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiol. 2002;148:923–932. doi: 10.1099/00221287-148-4-923. [DOI] [PubMed] [Google Scholar]
- Huber B, Riedel K, Hentzer M, Heydorn A, Gotschlich A, Givskov M, Molin S, Eberl L. The cepcep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiol. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- Allison C, Coleman N, Jones PL, Hughes C. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect Immun. 1992;60:4740–4746. doi: 10.1128/iai.60.11.4740-4746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GM, Claret L, Furness R, Gupta S, Hughes C. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiol. 2002;148:2191–2201. doi: 10.1099/00221287-148-7-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Killam T, Sood V, Surette MG. Swarm-cell differentiation in Salmonella enterica serovar Typhimurium results in elevated resistance to multiple antibiotics. J Bacteriol. 2003;185:3111–3117. doi: 10.1128/JB.185.10.3111-3117.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnason J, Southward CM, Surette MG. Genomic profiling of iron-responsive genes in Salmonella enterica serovar Typhimurium by high-throughput screening of a random promoter library. J Bacteriol. 2003;185:4973–4982. doi: 10.1128/JB.185.16.4973-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 1989.
- Toguchi A, Siano M, Burkart M, Harshey RM. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J Bacteriol. 2000;182:6308–6321. doi: 10.1128/JB.182.22.6308-6321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS, Ryan SS, Van Velkinburgh JC, Ernst RK, Miller SI. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2000;68:6139–6146. doi: 10.1128/IAI.68.11.6139-6146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trent MS, Ribeiro AA, Lin S, Cotter RJ, Raetz CR. An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J Biol Chem. 2001;276:43122–43131. doi: 10.1074/jbc.M106961200. [DOI] [PubMed] [Google Scholar]
- Brown DF, Brown L. Evaluation of the E test, a novel method of quantifying antimicrobial activity. J Antimicrob Chemother. 1991;27:185–190. doi: 10.1093/jac/27.2.185. [DOI] [PubMed] [Google Scholar]
- Macfarlane ELA, Kwasnicka A, Hancock REW. Role of Pseudomonas aeruginosa PhoP-PhoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiol. 2000;146:2543–2554. doi: 10.1099/00221287-146-10-2543. [DOI] [PubMed] [Google Scholar]
- Nichols WW, Dorrington SM, Slack MP, Walmsley HL. Inhibition of tobramycin diffusion by binding to alginate. Antimicrob Agents Chemother. 1988;32:518–523. doi: 10.1128/aac.32.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IE, Ernst RK, Miller SI, Falkow S. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J Bacteriol. 2002;184:3203–3213. doi: 10.1128/JB.184.12.3203-3213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Ryan SS, McCoy AJ, Gunn JS. Identification and genetic characterization of PmrA-regulated genes involved in polymyxin B resistance in Salmonella enterica serovar Typhimurium. Infect Immun. 2002;70:6770–6778. doi: 10.1128/IAI.70.12.6770-6778.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JW. Aminoglycoside resistance in enterococci. Clin Infect Dis. 2000;31:586–589. doi: 10.1086/313949. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Liu H, Falla TJ, Gunn JS. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob Agents Chemother. 2001;45:2030–2037. doi: 10.1128/AAC.45.7.2030-2037.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- Spoering AL, Lewis K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J Bacteriol. 2001;183:6746–6751. doi: 10.1128/JB.183.23.6746-6751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


