Abstract
Enhancers are defined by their ability to stimulate gene activity from remote sites and their requirement for promoter-proximal upstream activators to activate transcription. Here we demonstrate that recruitment of the p300/CBP-associated factor PCAF to a reporter gene is sufficient to stimulate promoter activity. The PCAF-mediated stimulation of transcription from either a distant or promoter-proximal position depends on the presence of an upstream activator (Sp1). These data suggest that acetyltransferase activity may be a primary component of enhancer function, and that recruitment of polymerase and enhancement of transcription are separable. Transcriptional activation by PCAF requires both its acetyltransferase activity and an additional activity within its N terminus. We also show that the simian virus 40 enhancer and PCAF itself are sufficient to counteract Mad-mediated repression. These results are compatible with recent models in which gene activity is regulated by the competition between deacetylase-mediated repression and enhancer-mediated recruitment of acetyltransferases.
In higher eukaryotes, gene expression often is influenced by enhancers that stimulate, independent of their orientation, gene activity from remote positions. This enhancement of transcription requires upstream activators immediately adjacent to the core promoter region (1). Enhancers contain multiple transcription factor binding sites, and many transcription factors can stimulate gene expression as a component of either promoter or enhancer. These observations have led to a model in which enhancer-bound elements synergize with upstream activators, bound at the immediate vicinity of the TATA box, to recruit RNA polymerase II complexes to the transcription start site. This recruitment model of gene activation is compatible with the enhancer-mediated increase in the rate of transcriptional initiation (2, 3) and/or an increase in the number of templates actively engaged in transcription (ref. 4 and references therein).
Enhancers may function to relieve chromatin-mediated repression of promoters (5). The binding of transcription factors to their respective promoter recognition sequence is impeded by chromosomal barriers imposed by nucleosomal arrays (reviewed in ref. 6). The stimulation of promoter activity requires the alteration of repressive chromatin structures by remodeling activities or by posttranslational modification of the core histones (reviewed in refs. 7 and 8).
The activity of enhancers, including the simian virus 40 (SV40), Ig heavy chain, polyoma, and insulin enhancers, is suppressed by the adenoviral protein E1a (9, 10). The analysis of E1a mutants indicates that E1a repression of enhancer function requires its N-terminal domain (reviewed in ref. 11). This domain interacts with the transcriptional coactivators p300 and cAMP response element binding protein [CREB binding protein, (CBP); ref. 12], which are very similar in sequence and function. Overexpression of p300 mutants that do not associate with E1a abrogates the E1a-mediated repression of SV40 enhancer activity (12). This result indicates that p300/CBP is an important component of SV40 enhancer function. P300/CBP interacts with a variety of transcriptional activators (reviewed in ref. 13) and has been found stably associated with a fraction of RNA polymerase II holoenzyme (14).
P300/CBP also binds to PCAF (p300/CBP-associated factor); this interaction is disrupted when E1a is overexpressed (15). These observations suggest that disruption of the p300/CBP-PCAF complex could be the molecular basis of SV40 enhancer suppression by E1a. Both p300/CBP and PCAF contain histone acetyltransferase (HAT) activity in vitro (15–17). The HAT domains of p300/CBP and/or PCAF are required for the activity of several transcription factors, including MyoD (18), the retinoic acid receptor, and CREB (19). These results have been interpreted to suggest that both p300/CBP and PCAF mediate their effect on transcriptional regulation partially through histone acetylation. However, the recent discovery of acetylated forms of transcriptional activators such as p53 (20) and of general transcription factors TFIIE and TFIIF (21) extends the range of potential pathways by which an acetyltransferase might regulate transcription.
In contrast to acetyltransferases, deacetylases are known to repress transcription. Repressors with sequence specific DNA binding activity, including Mad, unliganded nuclear receptor, YY1, and Ume6, associate with a protein complex including the mSin3 protein, the nuclear coreceptor protein N-CoR/SMRT, the mSin3-associated polypeptide SAP18, and histone deacetylase 1 (HDAC1) or its homologues (reviewed in refs. 22–24).
Below, we present evidence that transcriptional enhancement from a promoter-distant position uses a mechanism fundamentally different from transcriptional activation at a promoter-proximal site. We show that the recruitment of the HAT PCAF is sufficient to enhance transcription from a distance, but is not sufficient to activate transcription when recruited to the vicinity of the TATA box. The PCAF-mediated stimulation of transcription from either a distant or promoter–proximal position requires the presence of an upstream activator, suggesting that PCAF does not recruit RNA polymerase. Thus, in contrast to gene activation by promoter-proximally bound upstream activators (25–27), transcriptional enhancement from a distance does not comprise the ability of transcription factors to recruit RNA polymerase. We also show that the SV40 enhancer and PCAF itself are sufficient to counteract Mad-mediated repression. In combination, these results support a model in which gene activity is regulated by the competition between deacetylase-mediated repression and enhancer-mediated recruitment of acetyltransferases (28).
MATERIALS AND METHODS
Plasmids and Constructs.
The constructs SP1-TATA-MG and SP1-TATA-SVE are described in ref. 29. The reporter construct SP1-TATA-MG-Gal was generated by inserting an oligonucleotide containing three Gal4-DNA recognition sequences into the Aat II recognition site of SP1-TATA-MG. The construct TATA-MG-Gal lacking recognition sequences for cellular transcription factors was derived from SP1-TATA-MG-Gal through excision of the Sp1 binding sites by HpaI and BglII and insertion of an oligonucleotide containing two recognition sites for the prokaryotic LexA protein. Expression vectors for Gal4-PCAF fusion proteins were generated by inserting a PCR fragment coding for the Gal4-DNA binding domain into the unique EcoRI site of pCX-PCAF (15). PCAF deletion mutants were generated by site-directed mutagenesis. Expression vectors coding for Gal4-VP16, Gal4-VP16mad, and Gal4-VP16madpro are described in ref. 30. LexVP16mad and LexVP16madpro were constructed by inserting a PCR fragment containing the coding region for the mSin3-interacting domain of Mad into the 5′ region of the LexVP16 hybrid gene driven by the cytomegalovirus (CMV) promoter.
Transient Transfection Experiments.
The procedure for transient transfection of mouse melanoma B78 cells using the calcium phosphate precipitation method is described in detail in ref. 29. The total amount of transfected DNA (27.5 μg DNA per 15-cm dish) was kept constant through addition of empty vector DNA. Experiments were performed several times, and representative results are shown. For treatment with Trichostatin A (TSA), transfected cells first were pooled 24 hr postaddition of DNA, then split onto two 15-cm dishes. TSA then was added to a final concentration of 100 ng/ml for 16 hr.
Nuclease S1 Assays.
Analysis of steady-state RNA was done with S1-nuclease protection experiments as described in ref. 29. Briefly, an end-labeled probe extending from position +131 to −2073 relative to the transcription initiation site was hybridized to 5 or 10 μg RNA. After digestion with S1-nuclease, the samples were run on a 8% denaturing acrylamide/urea gel.
Western Analyses.
Aliqouts of transfected cells from a 150-mm tissue culture dish were lysed in RIPA buffer containing protease and phosphatase inhibitors (10 mM Tris, pH 7.4/0.15 M NaCl/1% Nonidet P-40/1% deoxycholate/0.1% SDS/10 μg/ml each of aprotonin, leupeptin, and pepstatin/50 mM NaF/1 mM vanadate). Extracts were quantified, electrophoresed on 8% polyacrylamide gels, and transferred to poly(vinylidene difluoride) membranes. Membranes were incubated with primary antibodies. Proteins were visualized with horseradish peroxidase-conjugated secondary antibodies followed by ECL according to the manufacturer (Amersham).
HAT Assay.
DNA fragments coding for wild-type and mutant PCAF proteins were subcloned into a baculovirus expression vector. FLAG-tagged PCAF derivatives were expressed and affinity-purified. The analysis of HAT was performed as described in ref. 15.
RESULTS
Inhibition of Deacetylases Does Not Increase SV40 Enhancer-Stimulated Transcription.
Highly acetylated histones have been found associated with transcriptionally active chromatin, whereas hypoacetylated histones generally are correlated with transcriptionally inert chromatin (reviewed in refs. 31 and 32). However, the increase in transcriptional activity in response to inhibitors of deacetylation such as butyrate or TSA is restricted to only a subset of the genome (22). The molecular basis for this specificity remains unknown. We have determined the influence of TSA on the transcription of two previously characterized DNA templates (29). Both reporter constructs consist of a myc-globin (MG) fusion gene and are driven by the same Sp1 promoter composed of multiple SP1 binding sites upstream of a consensus TATA box. The SP1-TATA MG-SVE template additionally contains the SV40 enhancer 3.1 kb downstream of the transcription start site. Transcripts from the enhancer-less reporter construct SP1*-TATA-MG are distinguished from the enhancer-containing template SP1-TATA-MG-SVE through insertion of a 30-nt oligomer at +38. These constructs were transfected into mouse B78 cells, as described (29).
After transfection, transcription of the templates was measured with S1-nuclease assays within 48 hr. Consistent with our previous analysis (29), the Sp1 promoter of the enhancer-less construct SP1*-TATA-MG mediates only low-level transcription of the linked MG reporter gene (Fig. 1, lanes 1, 3, and 5). The addition of the HDAC inhibitor TSA results in a 10-fold increase in transcriptional activity (Fig. 1, lanes 2, 4, and 6). In contrast to its strong stimulation of the enhancer-less construct, TSA does not significantly increase the activity of the enhancer-containing reporter construct SP1-TATA-MG-SVE. Transfection experiments with varying amounts of SP1-TATA-MG-SVE demonstrate that the failure of TSA to activate the SV40 enhancer-containing plasmid is not caused by a limitation of cellular factors/activities (Fig. 1). The differential effect of TSA on the enhancerless- and enhancer-containing templates suggests that the activation of transcription by the deacetylase inhibitor TSA and by enhancer elements share the same molecular pathway.
Figure 1.
SV40 enhancer-activated transcription is not stimulated in the presence of the deacetylation inhibitor TSA. Nuclease S1 analysis of RNA from mouse B78 cells transfected with the reporter constructs SP1*-TATA-MG and SP1-TATA-MG-SVE in the presence and absence of the deacetylase inhibitor TSA. Cells were transfected with 12.5 μg of SP1*-TATA-MG and different amounts of SP1-TATA-MG-SVE (lanes 1 and 2, 12.5 μg; lanes 3 and 4, 2.5 μg; lanes 5 and 6, 0.5 μg). Half of the transfected cells was treated with TSA (100 ng/μl) for 16 hr, and RNA from treated and nontreated cells was subjected to nuclease S1 analysis using an end-labeled probe derived from the SP1*-TATA-MG template (see Materials and Methods). The transcribed regions of SP1-TATA-MG-SVE (SVE) and SP1*-TATA-MG (MG*) differ through a 30-nt sequence; therefore, transcripts can be distinguished by using the same probe.
Direct Recruitment of the Acetyltransferase PCAF Activates Transcription From a Distance.
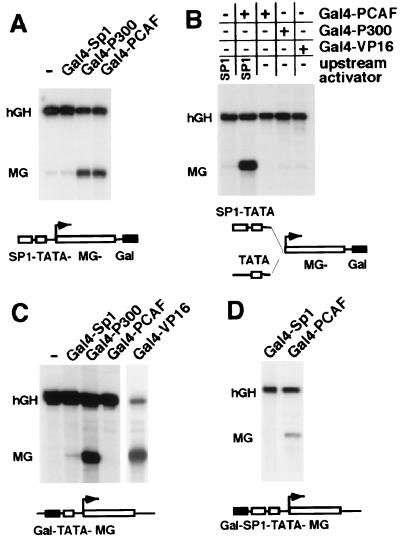
Stimulation of gene activity by the SV40 enhancer requires the transcriptional coactivator p300/CBP (12). P300/CBP and its associated factor PCAF possess acetyltransferase activity that catalyzes the acetylation of nucleosomes and nonhistone proteins (15–17, 20, 21). Clearly, these activities could account for the lack of SV40 enhancer-containing templates to respond to treatment with TSA (Fig. 1). Thus, we determined whether PCAF and/or p300/CBP, like the SV40 enhancer, stimulate promoter activity from a distant position and whether they require upstream activators in the vicinity of the TATA box for their activities. The SV40 enhancer, located 3.1 kb downstream of the transcription start site within the reporter construct SP1-TATA-MG-SVE (29), was replaced with Gal4-DNA binding sites. The ability of Gal4-p300 and Gal4-PCAF fusion proteins to stimulate transcription of this reporter was tested after transfection into mouse B78 cells. As shown in Fig. 2A, Gal4-p300 stimulated Sp1-promoter activity from a downstream position. Similarly, full-length PCAF fused to the Gal4 DNA binding domain was sufficient to enhance promoter activity. Quantitative PhosphorImager analysis of several independent experiments revealed that Gal 4-PCAF stimulated the Sp1-promoter on average 13-fold, whereas Gal4-p300 increases MG-RNA levels 19-fold.
Figure 2.
Direct recruitment of PCAF enhances Sp1-promoter activity similar to the SV40 enhancer. (A) Stimulation of the SP1-TATA-MG-Gal reporter construct by the acetyltransferase PCAF fused to the Gal4-DNA binding domain was compared with the transcriptional enhancement by the known transactivators Gal4-Sp1 and Gal4-p300. The reporter construct SP1-TATA-MG-Gal was transiently transfected with empty vector DNA, Gal4-Sp1, Gal4-p300, or Gal4-PCAF and with the CMV-hGH gene as an internal control for efficiency of transfection. Steady-state RNA levels of the MG fusion genes and the human growth hormone (hGH) gene were determined with the S1-nuclease assay as described in Materials and Methods. (B) The activity of the reporter gene TATA-MG-Gal, lacking upstream-Sp1 binding sites, in the presence of Gal4-VP16, Gal4-p300, and Gal4-PCAF, is shown. For comparison, the stimulation of transcription by Gal4-PCAF in the presence of Sp1 upstream activating sequences is shown in the left two lanes. (C) Gal4-PCAF does not activate transcription by itself. The steady-state levels of the transfected Gal-TATA-MG construct in the presence of Gal4-PCAF were compared with the activity of the reporter gene after cotransfection of empty vector DNA, Gal4-Sp1, Gal4-p300, and Gal4-VP16. The samples derived from cotransfections with different Gal4-fusion proteins were mapped with the same end-labeled probe in the nuclease-S1 assay; however, the Gal4-VP16 derived sample was run on a separate gel. (D) Stimulation by Gal4-PCAF bound at a proximal position, adjacent to the Sp1 upstream activator sites. The reporter gene Gal-SP1-TATA-MG, which contains three copies of Gal4 recognition sites just upstream of multiple Sp1-sites, was cotransfected with Gal4-Sp1 or the Gal4-PCAF expression plasmid.
PCAF Activates Transcription From a Distance But Does Not Recruit RNA Polymerase.
We determined whether the activation by Gal4-PCAF and Gal4-p300 requires RNA polymerase loading onto the template by promoter-proximal activators, similar to conventional enhancers. The dependence on upstream activators was investigated initially by using a reporter gene containing Gal4-DNA binding sites distant from the TATA box and lacking DNA binding sites for endogenous, cellular transcriptional activators (TATA-MG-Gal, Fig. 2B). In contrast to its ability to transcriptionally activate the SP1-TATA-MG-Gal template, Gal4-PCAF does not activate transcription from a distance on reporter genes that lack binding sites for promoter-proximal activators (TATA-MG-Gal template, Fig. 2B). In addition, neither Gal4-VP16 nor Gal4-p300 significantly transactivate transcription of the TATA-MG-Gal template.
P300 and VP16 can activate transcription when recruited to the vicinity of a TATA box. To determine whether PCAF also can stimulate transcription from an upstream position, we compared the ability of Gal4-PCAF, Gal4-p300, and Gal4-VP16 to activate transcription when recruited to a position immediately adjacent to the TATA box in a template devoid of binding sites for an upstream activator (the Gal-TATA-MG template). As shown in Fig. 2C, Gal4-p300 and Gal4-VP16 activate transcription to a high level when recruited to the vicinity of the TATA box on the Gal-TATA-MG template, whereas Gal4-PCAF does not. However, Gal4-PCAF can activate transcription more than 15-fold when it is recruited to a position immediately adjacent to the Sp1 upstream activator in the Gal-SP1-TATA-MG template (Fig. 2D). Thus, in contrast to Gal4-p300 and Gal4-VP16, Gal4-PCAF activates transcription in the presence, but not absence, of Sp1, even when recruited to the core promoter region. The PCAF stimulation of Sp1-activated transcription requires tethering of PCAF to the promoter, because overexpression of PCAF without the Gal4-DNA tether does not stimulate Sp1-promoter activity (data not shown). Recruitment of the Gal4-Sp1 fusion protein to the Gal-SP1-TATA-MG and SP1-TATA-MG-Gal templates, respectively, had no effect on the transcriptional activity of these reporter genes (Fig. 2 D and A, respectively).
In summary, these results demonstrate that recruitment of Gal4-PCAF to the template is sufficient to stimulate promoter activity, and that this stimulation depends on the presence of an upstream activator (Sp1). In contrast, and similar to Gal4-VP16, Gal4-p300 can stimulate transcription from the proximal upstream site in the absence of Sp1. These differences suggest that, unlike Gal4-PCAF, Gal4-p300 may associate with transcription activators and basal transcription factors (reviewed in ref. 13) or directly contact/recruit RNA polymerase II holoenzyme (14).
HAT Activity of PCAF Is Not Sufficient for Promoter Stimulation.
To map protein domains required for PCAF function, we determined the ability of PCAF deletion mutants to stimulate promoter activity (Fig. 3). The mutant Gal4-PCAFΔHAT(574–608) contains a deletion within the HAT domain, and Gal4-PCAFΔN(65–112) is deleted in the N-terminal region. The HAT activity of these mutants was tested with in vitro histone acetylation assays (Fig. 3B). Whereas the deletion in the N terminus does not affect the ability of PCAF to acetylate histones in vitro, the HAT activity of PCAF is completely abolished by deletion of amino acids 574–608. As shown in Fig. 3A, this deletion within the HAT domain also abrogates the ability of Gal4-PCAF to stimulate transcription from a distance. Surprisingly, the deletion of amino acids 65–112 in the N terminus also results in loss of enhancer activity, although this mutation does not affect the ability of the mutant protein to acetylate histones in vitro (Fig. 3B). Western analyses of extracts from transiently transfected cells demonstrate that the Gal4-fusion proteins containing the wild-type or the mutant PCAF proteins are expressed at equivalent levels (data not shown). These results demonstrate that the HAT domain of PCAF is necessary, but not sufficient, to enhance the activity of the Sp1 promoter (see Discussion).
Figure 3.
The HAT domain of PCAF is required, but not sufficient, to stimulate promoter activity. (A) Deletions within the N or C terminus of PCAF abrogate the ability of PCAF to enhance the Sp1-driven reporter gene. Two deletions within Gal4-PCAF were tested: ΔHAT(574–608), a Gal4-PCAF mutant containing a deletion within the HAT domain of the C-terminal portion of PCAF; and ΔN(65–112), a Gal4-PCAF mutant containing a deletion of amino acids 65–112 within the N-terminal portion of PCAF. RNA from transfected B78 cells was tested with S1-nuclease protection assays. (B) HAT activity in vitro. Wild-type and mutant PCAF proteins were tested for acetyltransferase activity in vitro.
P300/CBP-, But Not PCAF-, Mediated Enhancement of Transcription Is Repressed by E1a.
As mentioned in the Introduction, the disruption of the p300/CBP-PCAF complex could be the molecular basis of enhancer suppression by E1a. In this model, enhancer-bound transcription factors interact with p300/CBP, which in turn recruits PCAF. This model further predicts that Gal4-p300 induced stimulation of Sp1 promoter activity would be suppressed in the presence of E1a, whereas Gal4-PCAF mediated enhancement should remain unaffected.
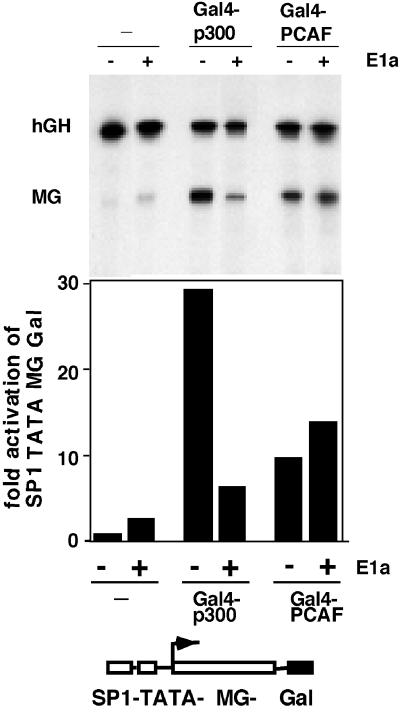
To test these predictions, we analyzed the activation of transcription by Gal4-p300 and Gal4-PCAF in the presence and absence of E1a. As shown in Fig. 4, coexpression of E1a reduces Gal4-p300 induced stimulation of the SP1-TATA-MG-Gal reporter construct 4-fold. In contrast, Gal4-PCAF-mediated enhancement of transcription remains unaffected or is slightly increased in the presence of E1a. Western blot analyses demonstrate that E1a is expressed at the same level in both transient transfection experiments (data not shown). In summary, these results are consistent with a model in which PCAF is an important component of the SV40-enhancer-mediated long-distance activation of promoter activity.
Figure 4.
Gal4-PCAF-mediated stimulation of transcription is resistant to suppression by the adenoviral protein E1a. B78 cells were transfected with the reporter construct SP1-TATA-MG-Gal and expression vectors for Gal4-p300 and Gal4-PCAF. The transfection mixtures contained the CMV-promoter driven human growth hormone (hGH) gene as an internal control. The E1a gene was expressed from the Rous sarcoma virus (RSV) promoter in pRc/RSV (Invitrogen). (Upper) S1-nuclease assay using end-labeled probes homologous to the hGH gene and the MG reporter gene. (Lower) Quantitative PhosphorImager analyses of the S1-nuclease assay reveals a 4-fold decrease in Gal4-p300 mediated activation in the presence of E1a. In contrast, Gal4-PCAF stimulated transcription remains unaffected or increases slightly.
The SV40 Enhancer and PCAF Acetyltransferase Counteract Mad/mSin3-Mediated Repression.
To further investigate whether gene activity actually can be influenced by changes in the acetylation/deacetylation balance, we determined whether the SV40 enhancer can counteract repression mediated by deacetylases recruited by Mad. A 35-residue region of Mad known to interact with mSin3 (Sin3 interacting domain or SID) is sufficient to repress transactivation domains of a linked VP16 activator (30). Two mutations in the SID domain (L12P/A16P) of Gal4-VP16mad (Gal4-VP16madpro) abolish binding to mSin3 and restore full transactivation potency of the fused VP16 protein.
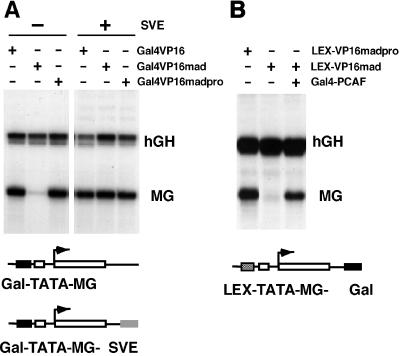
The ability of Gal4-VP16, Gal4-VP16mad, or Gal4-VP16- madpro to activate reporter constructs with and without SV40 enhancer (Gal-TATA-MG-SVE and Gal-TATA-MG, respectively) was determined in transiently transfected B78 cells (Fig. 5A). Consistent with previous observations (30), Gal4-VP16 and Gal4-VP16madpro highly stimulated transcription of the Gal-TATA promoter, whereas Gal4-VP16mad only marginally activates the Gal-TATA-MG construct (Fig. 5A, lanes 1–3). Remarkably, the insertion of SV40 enhancer sequences 2.5 kb downstream of the transcription start site of the reporter construct Gal-TATA-MG-SVE also permits a high level of promoter activity in the presence of Gal4-VP16mad (Fig. 5B, lanes 4–6). Thus, the SV40 enhancer counteracts Mad/mSin3-mediated transcriptional repression.
Figure 5.
The SV40 enhancer or the recruitment of PCAF counteracts HDAC-mediated repression. (A) The reporter constructs Gal-TATA-MG (Left, −SVE) and Gal-TATA-MG-SVE (Right, +SVE), which differ through SV40 enhancer sequences (SVE) downstream of the transcribed MG gene, were transfected into mouse B78 cells along with the CMV-hGH gene as an internal control and with Gal4-VP16, Gal4-VP16 mad, or Gal4-VP16 madpro as transactivators. Gal4-VP16 mad contains the Sin3 interaction domain (SID) of the Mad protein, whereas Gal4-VP16 madpro is mutated within SID, and therefore does not interact with Sin3 (30). RNA from transfected cells was analyzed with S1-nuclease assays. Transactivation by Gal4-VP16 mad is significantly less (12-fold) than transactivation by Gal4-VP16 and Gal4-VP16 madpro. (B) Gal4-PCAF counteracts Mad-mediated repression of VP16 transactivation. The LEX-TATA-MG-Gal reporter construct was cotransfected into B78 cells with expression vectors encoding Lex-VP16 madpro (left lane), LexVP16 mad (middle and right lanes), and Gal4-PCAF (right lane). The CMV-hGH plasmid was included as an internal control for transfection efficiency. RNA was analyzed in S1-nuclease assays.
To determine whether the direct recruitment of an acetyltransferase is sufficient to overcome the Mad-mediated repression of VP16 activity, Gal4-PCAF was tethered downstream of the transcription unit via Gal4-DNA binding sites. For this experiment, the activating region of VP16 with the normal or mutated Mad domain was linked to the LexA DNA binding domain. As expected, Lex-VP16madpro is a potent transactivator of the LEX-TATA-MG-Gal reporter gene containing Lex-DNA-binding sequences upstream of the TATA box and Gal4 DNA binding sites downstream of the MG reporter gene (Fig. 5B, lane 1). Lex-VP16mad, however, is inactive (Fig. 5B, lane 2). Although coexpression of the Gal4 DNA binding domain or Gal4-Sp1 does not reverse the Mad/mSin3-mediated repression of transactivation (data not shown), the expression of Gal4-PCAF renders Lex-VP16mad active (Fig. 5B, lane 3). Thus, the direct recruitment of PCAF is sufficient to overcome the Mad/mSin3-mediated repression of gene activity.
DISCUSSION
We have presented several lines of evidence that suggest a primary role for acetyltransferases in transcriptional enhancement from a distance: (i) Inhibition of deacetylation fails to further stimulate enhancer-activated transcription. (ii) The direct recruitment of PCAF via a Gal4-DNA binding domain can enhance transcription from a distance. PCAF-mediated transcriptional enhancement requires the presence of upstream activators to recruit RNA polymerases to the promoter. (iii) Both the SV40 enhancer and PCAF itself can counteract the repressive effect of deacetylases.
The mechanisms by which transcription activators enhance promoter activity from a distance appear to differ from transcriptional activation close to the TATA box. Activator proteins bound in the vicinity of the TATA box interact with components of the holoenzyme complex to recruit RNA polymerase to the transcription start site (25–27). In contrast, transcriptional stimulation from a distance may involve the activity of acetyltransferases recruited by enhancer-bound activator proteins, but does not comprise the ability to recruit RNA polymerase complexes. Consistent with this model, transcription activators bound at the proximal promoter can function autonomously, but transcriptional stimulation from a distance additionally requires activators bound in the vicinity of the TATA box. For example, the strong acidic activator Gal4-VP16 and Gal4-p300/CBP transactivate expression of a linked reporter gene when bound in the vicinity of the TATA box (Fig. 2; refs. 33 and 34). The mechanism by which VP16 and p300/CBP increase transcriptional activity from a promoter-proximal position may rely on their ability to contact the general transcription factor TBP (TATA box-binding protein) (35) and recruit RNA polymerase holoenzyme (36). However, despite their ability to contact RNA polymerase II, Gal4-p300 and Gal4-VP16 have very little transactivation potential when recruited to a downstream position in the absence of promoter-proximal transactivators. Thus, the ability of these factors to stimulate transcription from a distance and their ability to recruit RNA polymerase II complexes appear to be separable phenomena. This functional distinction is exemplified by Gal4-PCAF which, in contrast to Gal4-p300 and Gal4-VP16, fails to transactivate from either the promoter or downstream position in the absence of an upstream activator, but strongly stimulates transcription from a distal position in the presence of upstream activators. The notion PCAF does not activate transcription via recruitment of initiation-competent RNA polymerase is supported further by the recent demonstration that PCAF does not interact with the nonphosphorylated form of RNA polymerase (37).
Previous analyses have indicated that the HAT activity of the yeast counterpart of PCAF, Gcn5p, is critical for transcriptional activation of Gcn5p-dependent genes (38, 39). The analysis of mutant PCAF proteins (Fig. 3) indicates that the acetyltransferase activity, located in the C-terminal portion of PCAF, is required but not sufficient to stimulate Sp1-driven transcription from a distance. Although a mutant with a deletion in the N-terminal region efficiently acetylates histones in vitro, it does not transactivate Sp1 promoter activity. Thus, this region contains an as-yet-unknown stimulatory activity or associates with other factors required for transcriptional enhancement from a distance.
The observation that acetyltransferase activity is the primary component of enhancer function is consistent with our recent finding that the Ig heavy chain 3′Cα enhancer mediates a significant increase in histone acetylation along a linked gene (40). Significantly, the increase in acetylation was not restricted to nucleosomes within the promoter region, but also was apparent both far upstream and downstream of the transcription start site. In addition, synergistic activation by the interferon β enhanceosome requires the transcription coactivator p300/CBP (41), and SV40 enhancer function depends on p300/CBP (12). Furthermore, the adenoviral protein E1a, which disrupts p300/CBP-PCAF interaction (15), inhibits transcriptional activation from a distance by the SV40 enhancer (9, 10). These results suggest that the partial repression of enhancer activity by E1a is caused by disruption of the interaction between PCAF or similar factors with p300/CBP. Consistent with this model, the direct recruitment of PCAF via the Gal4-DNA binding domain bypasses the repressive effect of E1a (Fig. 4).
Enhancers have been suggested to relieve chromatin-mediated repression of weak promoters (5). Consistent with this, the SV40 enhancer counteracts repression mediated by HDACs recruited by Mad. The HDAC-mediated inhibition is partly reversed by treatment with TSA (42) or by direct recruitment of the Gal4-PCAF fusion protein, suggesting that an acetyltransferase activity associated with the SV40 enhancer is responsible for promoter derepression. In contrast to transcriptional activation by Gal4-PCAF from a distance, the counteraction of Mad-mediated-repression does not require the N-terminal region of PCAF. Curiously, deletion of the HAT domain within the Gal4-PCAF fusion protein reduced the ability to counteract Mad-mediated repression to only 50 percent (data not shown). Although the reason for this result is unclear, it is reminiscent of the partial inhibition of transcription mediated by a mutated mSin3 protein deficient in interaction with HDAC (42).
The precise target of acetyltransferase activities of PCAF or its associated proteins is not known. Nucleosomes are acetylated by PCAF in vitro and, consistent with the observations of others (reviewed in ref. 43), the transiently transfected templates in our experiments are associated with nucleosomes (data not shown). Hyperacetylation of histones facilitates the access of transcriptional activators to the promoter (reviewed in refs. 22–24, 31, and 32), which could explain the enhancer-mediated increase in transcription initiation. However, enhancers, such as the SV40 and the Ig heavy chain 3′Cα, increase gene expression to a large extent by modulation of the elongation competence of RNA polymerase II (29, 44). Importantly, the TSA-mediated activation of an episomal c-myc gene also is caused primarily by an increase in the transcriptional elongation efficiency of RNA polymerases, rather than to an increase in transcription initiation (40). The interaction of PCAF with the elongation-competent, highly phosphorylated form of RNA polymerase II is also consistent with a model in which acetyltransferases alter the elongation competence of already recruited transcription complexes (37). In combination, these results suggest that the recruitment of acetyltransferases by enhancers may activate transcription by modulating the elongation competence of the transcription complex. This increase in elongation efficiency could be accomplished by the acetylation of histones and/or components of the transcriptional machinery itself. Clearly, insights into the mechanisms by which PCAF and other acetyltransferases affect gene expression await identification of their targets in the various assays used to examine acetyltransferase function, as well as characterization of the function and the factors that associate with the N-terminal and/or acetyltransferase domains of PCAF.
Acknowledgments
Special thanks go to Jennifer Stout and Mary Peretz for their time and excellent technical help; Carol Laherty, Don Ayer, Pier P. Claudio, and Elizabeth Moran for providing plasmids; and R. Eisenman and S. Hahn for their critical reading of this manuscript. This work was supported by National Cancer Institute Grant CA54337.
ABBREVIATIONS
- CBP
cAMP response element binding protein (CREB) binding protein
- PCAF
p300/CBP-associated factor
- SV40
simian virus 40
- HAT
histone acetyltransferase
- CMV
cytomegalovirus
- TSA
Trichostatin A
- MG
myc-globin
- HDAC
histone deacetylase
References
- 1.Green M R, Treisman R H, Maniatis T. Cell. 1983;35:137–148. doi: 10.1016/0092-8674(83)90216-7. [DOI] [PubMed] [Google Scholar]
- 2.Treisman R, Maniatis T. Nature (London) 1985;315:73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- 3.Weber F, Schaffner W. Nature (London) 1985;315:75–78. doi: 10.1038/315075a0. [DOI] [PubMed] [Google Scholar]
- 4.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 5.Majumder S, Miranda M, DePamphilis M L. EMBO J. 1993;12:1131–1140. doi: 10.1002/j.1460-2075.1993.tb05754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenfeld G. Nature (London) 1992;355:219–224. doi: 10.1038/355219a0. [DOI] [PubMed] [Google Scholar]
- 7.Tsukiyama T, Wu C. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 8.Kingston R E, Bunker C A, Imbalzano A N. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 9.Borelli E R, Hen R, Chambon P. Nature (London) 1984;312:608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- 10.Velcich A, Ziff E. Cell. 1985;40:705–716. doi: 10.1016/0092-8674(85)90219-3. [DOI] [PubMed] [Google Scholar]
- 11.Moran E. Semin Virol. 1994;5:327–340. [Google Scholar]
- 12.Eckner R, Ewen M E, Newsome D, Gerded M, DeCaprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 13.Goldman P S, Goodman R H. Recent Prog Horm Res. 1997;52:103–119. [PubMed] [Google Scholar]
- 14.Kee B L, Arias J, Montminy M R. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 15.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 16.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 17.Ogryzko R, Schiltz L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 18.Puri P L, Sartorelli V, Yang X J, Hamamori Y, Ogryzko V V, Howard B H, Kedes L, Wang J Y J, Graessmann A, Nakatani Y, Levrero M. Mol Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 19.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 20.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 21.Imhof A, Yang X J, Ogryzko V V, Nakatani Y, Wolffe A P, Ge H. Curr Biol. 1997;7:689–692. doi: 10.1016/s0960-9822(06)00296-x. [DOI] [PubMed] [Google Scholar]
- 22.Pazin M J, Kadonaga J T. Cell. 1997;89:325–328. doi: 10.1016/s0092-8674(00)80211-1. [DOI] [PubMed] [Google Scholar]
- 23.Wolffe A P. Nature (London) 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- 24.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 25.Gaudreau L, Adam M, Ptashne M. Mol Cell. 1998;1:913–916. doi: 10.1016/s1097-2765(00)80090-8. [DOI] [PubMed] [Google Scholar]
- 26.Keaveney M, Ptashne M. Mol Cell. 1998;1:917–924. doi: 10.1016/s1097-2765(00)80091-x. [DOI] [PubMed] [Google Scholar]
- 27.Koh S S, Ansari A Z, Ptashne M, Young R A. Mol Cell. 1998;1:895–904. doi: 10.1016/s1097-2765(00)80088-x. [DOI] [PubMed] [Google Scholar]
- 28.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalski M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 29.Krumm A, Hickey L B, Groudine M. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 30.Ayer D E, Laherty C D, Lawrence Q A, Armstrong A P, Eisenman R N. Mol Cell Biol. 1996;16:5772–5781. doi: 10.1128/mcb.16.10.5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizzen C A, Allis C D. Cell Mol Life Sci. 1998;54:6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 33.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 34.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 35.Abraham S E, Lobo S, Yaciuk S, Wang H H, Moran E. Oncogene. 1993;8:1639–1647. [PubMed] [Google Scholar]
- 36.Nakajima T, Uchida C, Anderson S F, Lee C, Hurwitz J, Parvin J D, Montminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 37.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Candau R, Zhou J X, Allis C D, Berger S L. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo M H, Zhou J, Jambeck P, Churchill M E, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madisen L, Krumm A, Hebbes T R, Groudine M. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merika M, Williams A J, Chen G, Collins T, Thanos D. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 42.Laherty C D, Yang W M, Sun J M, Davie J R, Seto E, Eisenman R N. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- 43.Smith C, Hager G. J Biol Chem. 1997;272:27493–27496. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 44.Madisen L, Groudine M. Genes Dev. 1994;8:2212–2226. doi: 10.1101/gad.8.18.2212. [DOI] [PubMed] [Google Scholar]